Abstract
Mating pheromones promote cellular differentiation and fusion of yeast cells with those of the opposite mating type. In the absence of a suitable partner, high concentrations of mating pheromones induced rapid cell death in ∼25% of the population of clonal cultures independent of cell age. Rapid cell death required Fig1, a transmembrane protein homologous to PMP-22/EMP/MP20/Claudin proteins, but did not require its Ca2+ influx activity. Rapid cell death also required cell wall degradation, which was inhibited in some surviving cells by the activation of a negative feedback loop involving the MAP kinase Slt2/Mpk1. Mutants lacking Slt2/Mpk1 or its upstream regulators also underwent a second slower wave of cell death that was independent of Fig1 and dependent on much lower concentrations of pheromones. A third wave of cell death that was independent of Fig1 and Slt2/Mpk1 was observed in mutants and conditions that eliminate calcineurin signaling. All three waves of cell death appeared independent of the caspase-like protein Mca1 and lacked certain “hallmarks” of apoptosis. Though all three waves of cell death were preceded by accumulation of reactive oxygen species, mitochondrial respiration was only required for the slowest wave in calcineurin-deficient cells. These findings suggest that yeast cells can die by necrosis-like mechanisms during the response to mating pheromones if essential response pathways are lacking or if mating is attempted in the absence of a partner.
INTRODUCTION
Programmed cell death (PCD) occurs in metazoans as a means of eliminating unwanted cells during development and removing damaged, weak, infected, or malignant cells from the organism to avoid potentially harmful consequences (Danial and Korsmeyer, 2004). PCD is highly coordinated and regulated at multiple levels. Inputs from a variety of sources can impact on a core set of enzymes that coordinate destruction of key cellular components necessary for cell survival. Apoptosis, one form of PCD, typically requires activation of cysteine-aspartyl proteases (caspases) by signaling factors derived from mitochondria or the plasma membrane. Conservation of PCD factors among all animals (Koonin and Aravind, 2002) is consistent with a very early origin of the PCD mechanism, potentially even before the divergence of animals and fungi.
The occurrence of PCD in fungi has received support from numerous studies using the budding yeast Saccharomyces cerevisiae (reviewed in Madeo et al., 2002, 2004; Longo et al., 2005). Several so-called “hallmarks” of apoptosis can be observed in populations of yeast cells that have been mortally wounded by environmental stresses such as high heat, high salt, hypertonic shock, DNA damaging agents, food preservatives, and hydrogen peroxide. Starvation and aging also seem to induce apoptosis-like cell death in a minority of cells in the dying population (reviewed in Longo et al., 2005). Expression of mammalian Bax in yeast also leads to mitochondrial dysfunction, accumulation of reactive oxygen species (ROS), and cell death (reviewed in Priault et al., 2003). To date, yeast homologues of mammalian caspase (Mca1), cytochrome c (CYC; Cyc1 and Cyc7), and several other factors have been implicated in one or more of these forms of apoptosis-like cell death. The molecular interactions between all these factors and the sequence of their actions have not been thoroughly investigated.
Recently, apoptosis-like cell death was reported in yeast cells engaged in sexual conjugation or “mating” (Severin and Hyman, 2002). Diploid yeast cells (a/α-cells) can propagate by mitosis and in certain conditions will undergo meiosis to produce haploid cells (a-cells and α-cells), which are capable of mitosis as well as mating. To initiate this process, a- and α-cells secrete mating pheromones (a-factor and α-factor) that bind serpentine receptors expressed on the opposite cell type (reviewed in Sprague and Thorner, 1992; Dohlman and Thorner, 2001). Engaged receptors activate a heterotrimeric G-protein and mitogen-activated protein (MAP)-kinase cascade, which result in induction of mating-specific genes, arrest in G1-phase of the cell division cycle, cell wall remodeling, and extension of a mating projection from the cell body toward the pheromone source. After cell–cell contact and agglutination, the intervening cell wall continues to be degraded and remodeled to permit membrane fusion, followed by subsequent mixing of cytoplasmic contents, fusion of haploid nuclei, and resumption of cell division. Even under optimal mating conditions, many a- and α-cells fail to find a partner or successfully mate. Haploid cells that fail to mate eventually become desensitized to the mating pheromones and resume vegetative growth. In mixed cultures of a- and α-cells, ∼6% of the nonmating cells were found dead (Severin and Hyman, 2002). In the presence of high α-factor and in the absence of mating partners, ∼30% of a-cells died, and these cells exhibited several morphological hallmarks of apoptosis (Severin and Hyman, 2002). Because cell death in these conditions also required CYC and an undefined target of cyclosporin A, the authors proposed that an apoptosis-like form of PCD occurred in populations of mating yeast cells and speculated that PCD could benefit the species by eliminating the weakest individuals from the mating population and reducing their ability to compete with healthier cells for mates or other resources.
However, the high levels of cell death reported in the recent study conflicted with earlier studies that failed to detect more than ∼1% cell death in populations of wild-type yeast cells responding to mating pheromones (Iida et al., 1990; Cyert et al., 1991; Cyert and Thorner, 1992; Foor et al., 1992; Iida et al., 1994; Ono et al., 1994; Moser et al., 1996; Fischer et al., 1997; Paidhungat and Garrett, 1997; Withee et al., 1997; Muller et al., 2001). These earlier studies also reported nearly complete cell death in yeast mutants that lack components of a calcium signaling pathway, such as the high-affinity Ca2+ influx channel (Cch1, Mid1), calmodulin (Cmd1), or calcineurin (CN; Cnb1, Cna1-Cna2). This broadly conserved calcium signaling pathway was also found to prevent cell death in response to a variety of natural and synthetic fungistatic drugs (Del Poeta et al., 2000; Marchetti et al., 2000; Bonilla et al., 2002; Cruz et al., 2002; Edlind et al., 2002; Bonilla and Cunningham, 2003; Onyewu et al., 2003; Sanglard et al., 2003; Kaur et al., 2004; Steinbach et al., 2004). In contrast to the recent report (Severin and Hyman, 2002), compounds that directly bind and inhibit CN strongly increased the observed incidence of cell death.
Here we reexamine the roles of CN, CYC, and other factors associated with cell death in yeast during the response to mating pheromones in order to resolve the discrepancies and to define the underlying regulatory mechanisms. Instead of just one wave of cell death occurring in response to mating pheromones, three distinct waves were distinguishable by pharmacological, genetic, and kinetic criteria. None of the three waves of cell death was related to apoptosis-like cell death or was dependent on yeast metacaspase, but all were preceded by accumulation of ROS. The fastest wave of cell death involved ROS accumulation from nonmitochondrial sources and was dependent on the plasma membrane protein Fig1, which we identify here as the first fungal member of the PMP-22/EMP/MP20/Claudin superfamily of four-spanner transmembrane proteins. Though Fig1 promotes Ca2+ influx and elevation of cytosolic free Ca2+ concentrations in these conditions (Muller et al., 2003), similar to the effects of Stargazin on ionotropic glutamate receptors (Letts et al., 1998), Fig1 promoted cell death independent of Ca2+. The other two waves of cell death were slower and observed only in cells lacking CN or the MAP kinase Slt2/Mpk1 (or their upstream regulators). Rather than undergoing altruistic suicide, we suggest instead that yeast cells die by necrosis-like processes relating to either the inappropriate execution of mating steps in the absence of a mating partner (Fig1-dependent fast death) or the failure to perform essential functions that are necessary for cell survival in mating conditions (slow deaths in CN- and mitogen-activate protein kinase [MPK]-less mutants).
MATERIALS AND METHODS
Media, Reagents, Yeast Strains, Plasmids, and Growth Conditions
All yeast strains were cultured in rich YPD medium or synthetic SC medium containing 2% glucose as described (Sherman et al., 1986). Synthetic α-factor (U.S. Biomedical, New York, NY), cyclosporin A (Sigma-Aldrich, St. Louis, MO) and FK506 (Fujisawa Healthcare, Deerfield, IL) were dissolved in dimethyl sulfoxide. Antimycin and oligomycin (both from Sigma-Aldrich) were dissolved in ethanol. d-Glucosamine (Sigma-Aldrich) and potassium-BAPTA (Invitrogen, Carlsbad, CA) were dissolved in water, and all the reagents described above were added to the culture medium at indicated concentrations.
All yeast strains used in this study (Table 1) were derived from wild-type W303-1A (MATa ade2-1 can1-100 his3-1 leu2-3112 trp1-1 ura3-1; Wallis et al., 1989) or BY4741 (MATa his3-1 Leu2-2 met15-0 ura3-0) (Brachmann et al., 1998) parent strains using standard molecular procedures and/or genetic crosses. Knockout mutations for CYC, Cyc3, Cpr1, Cpr3, Mca1, Cox6, Fus1, Fus2, Rvs161, Cnb1, Lrg1, Fig1, Nuc1, Bck1, and Atp2 were generated by homologous recombination of appropriate PCR products, selection for proper drug resistance or amino acid markers, and PCR screening of genomic loci as described (Sambrook et al., 1989). Replacement of SST1 with sst1::URA3 was also made by homologous recombination using the 5.6-kb fragments of the pJGsst1 plasmid after digestion with EcoRI and SalI (Elion et al., 1993).
Table 1.
List of yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| K601 | W303-1A | Cunningham and Fink (1996) |
| K603 | cnb1::LEU2 | Cunningham and Fink (1996) |
| NZY010 | cyc1::G418r cyc7::HIS3 | This study |
| NZY011 | cyc1::G418r cyc7::HIS3 cnb1::LEU2 | This study |
| NZY107 | cpr3::G418r | This study |
| NZY106 | cpr1::G418r | This study |
| NZY001 | mca1::G418r | This study |
| DDY42 | nuc1::HIS3 | This study |
| JSY5140 | fis1::HIS3 | Mozdy et al. (2000) |
| JSY5138 | dnm1::HIS3 | Mozdy et al. (2000) |
| JSY5145 | mdv1::HIS3 | Mozdy et al. (2000) |
| W303–1A Δcoq1 | coq1::LEU2 | Gin and Clarke (2005) |
| NZY012 | cyc3::NATr | This study |
| NZY105 | cox6::G418r | This study |
| NZY080 | atp2::LEU2 | This study |
| NZY082 | fus1::G418r | This study |
| EMY232 | fus2::G418r | This study |
| EMY237 | rvs161::G418r | This study |
| NZY079 | fig1::G418r | This study |
| #409 | prm1::HIS3 | This study |
| NZY084 | lrg1::G418r | This study |
| EMY157 | bni1::LEU2 | Evangelista et al. (1997) |
| #411 | prm1::HIS3 fig1::G418r | This study |
| K410 | sst1::URA3 | Elion et al. (1993) |
| K1584 | sst1::URA3 cnb1::NATr | This study |
| EMY206 | sst1::URA3 fig1::G418r | This study |
| NZY027 | sst1::URA3 fig1::G418r cnb1::NATr | This study |
| K665 | pmc1::TRP1 vcx1Δ | Cunningham and Fink (1996) |
| NZY036 | pmc1::TRP1 vcx1Δ fig1::G418r | This study |
| NZY091 | bck1::TRP1 | This study |
| NZY093 | bck1::TRP1 fus2::G418 | This study |
| K1251 | BY4741 | Research Genetics |
| RG01328 | bck1::G418r | Research Genetics |
| NZY100 | bck1::G418 fig1::NATr | This study |
| LMY006 | fig1::NATr | This study |
| NZY101 | sst1::URA3 | This study |
| NZY102 | sst1::URA3 bck1::G418r | This study |
| NZY103 | sst1::URA3 bck1::G418r fig1::NATr | This study |
| NZY104 | sst1::URA3 fig1::NATr | This study |
Cell Death Assays
Yeast strains were inoculated into YPD medium, serially diluted, and cultured overnight at 30°C. Log-phase cultures were selected, diluted to a concentration of 3 × 106 cells/ml (OD600 = 0.1) using fresh YPD medium and treated with α-factor and other compounds as described in Results. At various times of incubation at 30°C, 100 μl of cells were harvested by centrifugation (1 min, 13,000 rpm, room temperature [RT]), washed with 200 μl of SC medium, and resuspended in 10 μl of fresh SC medium containing 200 μg/ml methylene blue (Sigma), spotted onto microscope slides, imaged using bright-field microscopy, and scored as live (unstained) or dead (blue-stained) as described previously (Muller et al., 2001). At least 200 cells were scored for every strain at each time point. Similar results were obtained after 5 min staining with 0.4 mg/ml phloxine B (Sigma Chemical) in YPD medium (unpublished data; Severin and Hyman, 2002), or after 10 min staining with 1 μg/ml propidium iodide (PI; Sigma Chemical) in YPD medium and imaging with epifluorescence illumination (Nikon Diaphot inverted microscope, Melville, NY; 540-nm excitation and 620-nm emission).
Time-Lapse Microscopy
High-throughput time-lapse imaging of individual cells was done in a microfluidic chip that allowed us to maintain chemostatic conditions for long time periods (Groisman et al., 2005). The microfluidic device was made of a silicon elastomer polydimethylsiloxane (PDMS; RTV 615 by General Electric, Cleveland, OH) chip sealed to a no. 1.5 microscope coverglass, with inlets for introduction of media and cells into the channels and test chambers of the chip. Cells were grown in YPD medium to midlog phase, loaded into test chambers of the chip, and grown at RT with continuous perfusion of fresh culture medium. After a 4-h incubation, the cells were perfused with YPD medium containing 2 μM α-factor and 1 μg/ml PI and incubated an additional 8 h. Multiple fields of cells were imaged using phase-contrast microscopy (10-min intervals) and epifluorescence microscopy (30-min intervals) using a Nikon Eclipse TE2000-U inverted microscope equipped with an electronically controlled shutter and stage (X, Y, and Z directions), a 40×/0.75 objective, and a filter cube appropriate for detection of PI (540/25-nm excitation and 605/55-nm emission filters with a 505-nm dichroic mirror). Time-lapse movies of each field were generated from the stacked images and analyzed manually.
ROS Accumulation Assays
Cells producing ROS were visualized and counted as described previously (Madeo et al., 1999), except with slight modifications. Briefly, log-phase cultures were diluted to a concentration of 3 × 106 cells/ml and exposed to α-factor in YPD medium at 30°C. At each time point, 100 μl of cells was harvested by centrifugation, resuspended in 100 μl of fresh SC medium, and incubated with 10 μg/ml 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA, Molecular Probes, Eugene, OR) at 30°C for 10 min. Cells were concentrated by centrifugation and resuspended in 10 μl of fresh SC medium. 5μL of cells were loaded onto slides and observed immediately under epifluorescence microscopy (495-nm excitation and 525-nm emission). At least 200 cells per sample were scored manually as fluorescent or nonfluorescent.
TUNEL Assays
Log-phase cultures were diluted to 3 × 106 cells/ml in YPD medium and treated with α-factor. About 0.25 OD cells were harvested after incubation for 3, 6, and 9 h at 30°C. Cells were resuspended in 0.5 ml phosphate-buffered saline (PBS) and fixed for 1 h at RT with 3.7% formaldehyde (J. T. Baker). After the fixation, cells were washed with 0.5 ml PBS and resuspended in 1 ml SPM (1.2 M sorbitol, 50 mM K-phosphate, pH 7.3, 1 mM MgCl2). After digestion with a combination of 300 μg/ml Zymolyase 100T (Seikagaku, Tokyo, Japan) and 12 μg/ml lyticase (Sigma) in SPM for 40 min at 30°C, cells were gently harvested (5000 rpm, 1 min), washed with SPM, and resuspended in 100 μl SPM. Suspended cells (40 μL) were transferred into polylysine-coated wells of a microscope slide and allowed to settle for 20 min at RT. The slide was washed three times by SPM to remove the enzymes. Each well was incubated with 40 μl fresh permeabilization solution (0.1% Triton X-100 in a 0.1% sodium citrate solution) for 2 min at 4°C, rinsed with PBS, and incubated with 10 μl of 5 μg/ml DNase-free RNase (Boehringer Mannheim) for 30 min at 37°C. This last step was found to remove background staining in the cytoplasm that obscured nuclear staining. Each well was then rinsed three times with PBS and incubated with 10 μl TUNEL reaction mixture (In Situ Cell Death Detection Kit, POD, Roche, Indianapolis, IN) for 60 min at 37°C in a humidified container (Madeo et al., 1999). Slides were then rinsed three times with PBS and mounted under coverslips with Prolong Gold antifade solution (Molecular Probes). At least 200 cells per sample were scored as containing or lacking fluorescent nuclei using a Zeiss Axioplan/Coolsnap epi-fluorescence microscope (Thornwood, NY) with FITC channel. Though no fluorescent nuclei were observed in cells treated with α-factor at any time point, more than 90% of nuclei in H2O2-treated cells and nearly 100% of nuclei in cells exposed to DNAse I (Sigma, 0.1 μg/μl for 10 min, RT) stained TUNEL positive.
Aequorin Luminescence Measurements
Yeast strains were transformed with plasmid pKC147 (2 μ URA3 PMA-aequorin; Muller et al., 2001) or pEVP11/AEQ89 (2 μ LEU2 ADH-aequorin; Batiza et al., 1996), and independent transformants were grown to log phase in SC medium lacking uracil or leucine. Cells were harvested by centrifugation, resuspended in fresh medium, and loaded with 25 μg/ml coelenterazine (Molecular Probes) for 20 min at RT. Loaded cells were washed with fresh YPD medium and raised in YPD to OD600 = 0.25. After recovering at 30°C roller for 80 min, 600 μl from each sample was transferred to an luminometer tube, loaded with α-factor, and monitored for luminescence in a LB9507 luminometer (EG&G Wallac, Turku, Finland). Aequorin expression and loading in each strain was similar as judged by measuring total luminescence units after cell lysis using digitonin (Sigma).
Data Analysis
Data from several experiments were fit to a standard sigmoid equation (Y = min + (max − min)/(1 + (mid/X) slope), where min, max, mid, and slope are variables corresponding to the minimum value of Y, maximum value of Y, value of X corresponding to the midpoint of Y, and slope of line as it crosses the midpoint) using nonlinear regression. Values for min and max were constrained to >0 and <100%, respectively. Additionally, some data were fit to the sum of or difference between two sigmoid equations.
RESULTS
Two Waves of Cell Death Are Differentially Regulated by CN and Concentration of α-factor
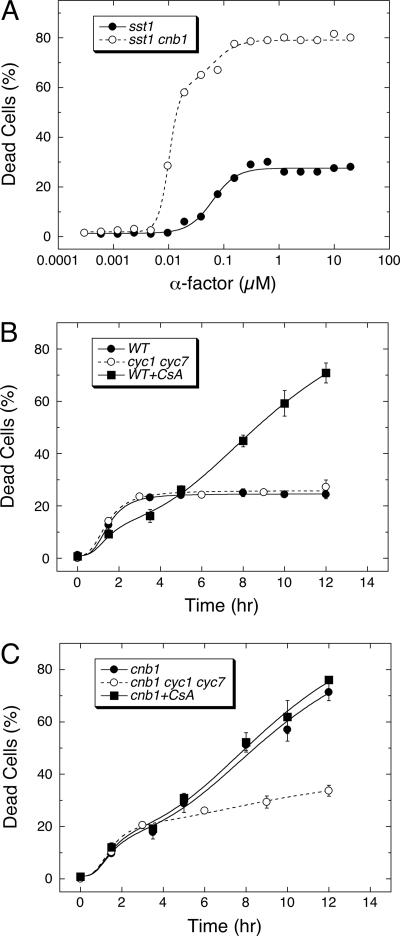
The many discrepancies reported in studies of yeast cell death during the response to mating pheromones might be attributed to the 10-fold higher concentrations of α-factor used in the recent study (Severin and Hyman, 2002). To test this idea, the death of yeast cells containing or lacking CN was measured after a 10-h exposure to a wide range of α-factor concentrations (Figure 1A). Strains lacking the secreted protease Sst1 were used in this experiment to avoid extracellular degradation of the added α-factor (Sprague and Herskowitz, 1981; Chan and Otte, 1982). At saturating concentrations of α-factor, up to ∼27% of the sst1 mutant cells (●) and over 80% of the sst1 cnb1 double mutant (○) cells died within the allotted window of time. The concentrations of α-factor resulting in 50% maximal death of sst1 mutants and sst1 cnb1 double mutants were estimated at 62 and 10 nM, respectively, by nonlinear regression of the dose–response data to standard sigmoid equations (see Materials and Methods). This sixfold difference in sensitivity to α-factor was also observed in wild-type and cnb1 mutants expressing the Sst1 protease, though the 50% lethal doses were ∼200-fold higher than the Sst1-deficient strains (unpublished data). However, CN-deficient mutants were not more sensitive to α-factor in a variety of other measurable responses (Moser et al., 1996; Withee et al., 1997). Therefore, CN completely prevented death at moderate concentrations of α-factor but not at high concentrations.
Figure 1.
Two waves of cell death during the response to α-factor are independently regulated. (A) Cell death was measured in populations of sst1 mutants and cnb1 sst1 double mutants lacking CN activity using methylene blue staining after 10 h of treatment with the indicated concentrations of α-factor. Dose–response curves for wild-type and cnb1 mutant populations were shifted to the right by ∼200-fold (unpublished data). (B and C) Populations of wild-type cells (WT) or CN-deficient cells (cnb1) lacking cytochrome c (−CYC) or experiencing 50 μM cyclosporin A (+CsA) were treated with 60 μM α-factor and monitored for cell death at the indicated times. Averages of three replicate experiments (±SD) are shown. Monophasic and biphasic patterns were evident in the data, of which the maxima, minima, and midpoints were estimated by nonlinear regression using the sum of one or two standard sigmoid equations. A fast wave of cell death was observed in ∼18–27% of all populations, and a slow wave of cell death was observed only in CN-deficient populations. The fast wave of cell death required sixfold higher concentrations of α-factor than CN-less death. Cytochrome c was important for the slow wave but not the fast wave.
The deaths of wild-type and CN-deficient cells were also analyzed at various times during the response to excess (60 μM) α-factor. Approximately 25% of wild-type cells died very rapidly (median lifespan of ∼1.5 h), as determined by nonlinear regression of the data to a standard sigmoid equation (Figure 1B). The cnb1 mutants exhibited two waves of cell death that were described very well by the sum of two sigmoid equations (Figure 1C). In this experiment, ∼17% of the population died in the first wave (median lifespan of 1.5 h) and the remainder died in the second wave (median lifespan of 9.7 h). The faster wave of cell death in cnb1 mutants was decreased to undetectable levels when lower concentrations of α-factor were used (6 μM; Moser et al., 1996; Withee et al., 1997) or when certain mating genes were eliminated (see below). Because cnb1 mutants do not die in the absence of mating pheromones, these findings suggest that low concentrations of α-factor stimulate a slow CN-sensitive cell death and that high α-factor concentrations stimulate a fast cell death in a fraction of the population that is largely insensitive to the presence or absence of CN.
The recent study of fast cell death at high α-factor concentrations demonstrated a dependence on CYC and a sensitivity to cyclosporin A (Severin and Hyman, 2002) even though cyclosporin A is known to inhibit CN. However, we find that fast cell death did not depend on CYC and was only slightly inhibited by cyclosporin A or the loss of CN (Figure 1B). Cyclosporin A also stimulated a slow wave of cell death in wild-type populations (Figure 1B) while having no detectable effect on either the rate or extent of cell death in cnb1 mutant populations (Figure 1C). All these effects of cyclosporin A were mimicked by the loss of CN and by the addition of FK506, a chemically distinct inhibitor of CN (unpublished data), indicating that these compounds modulate cell death solely by their activity against CN. Though CYC had no detectable effect on the fast wave of cell death in either wild-type or CN-deficient cells, the loss of CYC dramatically decreased the slow wave of cell death in cnb1 mutants (Figure 1C). The best-fit sigmoid equations indicate that the loss of CYC decreases the extent of slow death by about fourfold (from 81 to 18% of the population). Thus, CYC specifically increased slow CN-sensitive cell death without affecting the faster CN-insensitive type of cell death. Collectively, these findings suggest that at least two waves of cell death can occur during response to mating pheromones. The two waves differ markedly in their sensitivity to α-factor, kinetics, penetrance in the population, sensitivity to CN, and dependence on CYC.
Fast and Slow Waves of Cell Death are Morphologically and Mechanistically Distinct from Apoptosis-like Cell Death
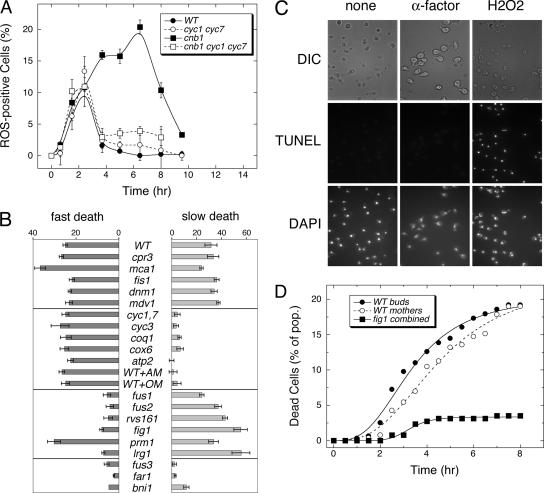
ROS accumulation and chromatin fragmentation, considered hallmarks of apoptosis in yeast (Madeo et al., 1999; Frohlich and Madeo, 2001), have been associated with death of yeast cells responding to α-factor (Severin and Hyman, 2002). To determine whether ROS accumulation precedes fast, slow, or both waves of cell death, CN- and/or CYC-deficient cells were treated with high α-factor and periodically stained for ROS accumulation using the fluorogenic probe H2DCFDA. The frequency of ROS-positive cells in wild-type populations peaked at ∼11% during the response to high α-factor and then declined to very low levels (Figure 2A, ●). The CYC-deficient cyc1 cyc7 double mutants behaved similarly (○). Time-lapse video microscopy of wild-type cells responding to high α-factor demonstrated that all of the fast cell deaths were preceded by ROS accumulation and that the surviving cells fail to accumulate detectable ROS (unpublished data). Therefore, fast cell death was closely associated with accumulation of ROS from a CYC-independent source.
Figure 2.
Fast and slow waves of cell death are associated with ROS accumulation but not chromatin fragmentation. (A) ROS accumulation was measured in populations of wild-type (WT) and CN-deficient (cnb1 mutant) cells that contain or lack cytochrome c (−CYC) at various times after treatment with 60 μM α-factor. (B) Mutants lacking either cyclophilin D (cpr3), metacaspase (mca1), mitochondrial fission factors (fis1, dnm1, mdv1), cytochrome c (cyc1 cyc7), cytochrome-c heme lyase (cyc3), coenzyme-Q (coq1), complex-IV (cox6), complex-V (atp2), cell wall degradation factors (fus1, fus2, rvs161), low-affinity Ca2+ influx (fig1), membrane fusion factor (prm1), Rho-GAP of the CWI signaling pathway (lrg1), or factors involved in pheromone signaling (fus3, far1, bni1) were treated with 60 μM α-factor for 10 h in the presence or absence of CN inhibitor (2.5 μM FK506) and assayed for cell death. Wild-type cells treated with inhibitors of complex-III (1 μg/ml antimycin A; AM) or complex-V (1 μg/ml oligomycin; OM) were also analyzed. Fast cell death was estimated as the frequency of cell death occurring in the absence of FK506. Slow cell death was estimated as the net increase of cell death occurring in the presence of FK506. (C) Chromatin fragmentation was monitored in wild-type cells after 3-h treatment with 60 μM α-factor or 1 mM H2O2 as indicated. Similar results were obtained using CN-deficient cnb1 mutants and longer periods of treatment. (D) Time-lapse video microscopy was used to monitor the fate of mother cells and buds of sst1 mutant (WT) and sst1 fig1 double mutant (fig1) populations after treatment with 2 μM α-factor at RT. Dead cells were identified using PI instead of methylene blue in other experiments and the results for fig1 mothers and buds were combined. Data were fit to standard Sigmoid equations.
Slow cell death of CN-deficient mutants was also associated with ROS accumulation. The frequency of ROS-positive cells in the population of cnb1 mutants peaked at ∼20% of the population ∼3 h before the midpoint of the slow wave of cell death (Figure 2A, ■). The loss of CYC in CN-deficient cells abolished the second wave of ROS accumulation but had no effect on the first wave (□). These findings indicate that CYC stimulates and CN inhibits a second wave of ROS accumulation that precedes and contributes to CN-less death.
CYC may promote ROS accumulation and CN-less death by several distinct mechanisms, such as stimulation of apoptosis-like processes or stimulation of mitochondrial respiration. To discriminate between these possibilities, we measured cell death in a panel of respiration- and apoptosis-deficient mutants after 10-h treatment with high α-factor or high α-factor plus FK506 to mimic the CN-deficient state (Figure 2B). Apoptosis-deficient mutants lacking metacaspase (mca1), mitochondrial cyclophilin D (cpr3), or mitochondrial fission factors (dnm1, fis1, mdv1) all exhibited wild-type levels of fast and slow cell death. Respiration-deficient mutants lacking coenzyme Q (coq1), CYC-heme lyase (cyc3), CYC-oxidase (cox6), and ATP synthase (atp2) behaved exactly like CYC-deficient mutants, showing wild-type levels of fast cell death with little or no slow cell death. Because respiration-deficient mutants have adapted to the loss of respiratory function, we also examined the effects of complex III inhibitors (antimycin) and complex V inhibitors (oligomycin) on wild-type cells. When added at the same time as α-factor, all of these respiration inhibitors strongly diminished slow death of CN-deficient cells but had no effect on fast death of wild-type cells (Figure 2B). Antimycin also abolished the second wave of ROS accumulation in cnb1 mutants responding to α-factor (unpublished data). Thus, functional mitochondrial respiratory complexes III, IV, and V were as important as CYC and coenzyme-Q in promoting slow death of CN-deficient cells. The data suggest that respiring mitochondria directly or indirectly generate ROS in CN-deficient cells and that ROS play a major role in their death.
To determine if chromatin fragmentation is associated with fast, slow, or both waves of cell death, standard TUNEL assays were performed on wild-type and cnb1 mutant cells at several times during the response to high α-factor. After 3 h of response, nearly 100% of cells in both populations stained TUNEL-positive (unpublished data). However, TUNEL fluorescence was observed in the cytoplasm of these cells and not restricted to the nuclear DNA. The cytoplasmic fluorescence was completely abolished by a brief treatment of the fixed and permeabilized cells with RNase before the TUNEL reaction, indicating the standard TUNEL assay was subject to high background of RNA. Using a modified TUNEL assay that included RNase, no TUNEL-positive nuclei were detected in wild-type or cnb1 mutant populations at any time after treatment with α-factor (Figure 2C). As a control for sensitivity of the modified TUNEL method, we confirmed that the vast majority of wild-type and cnb1 mutant cells stain positive for chromatin fragmentation after treatment with H2O2. Thus, neither fast nor slow waves of cell death were associated with chromatin fragmentation or stimulated by factors involved in apoptosis-like cell death.
Random Occurrence of Fast Cell Death in Mother Cells and Newly Budded Daughter Cells
Fast cell death was restricted to only ∼27% of the population of wild-type cells in our experimental conditions. Log-phase cultures of yeast are expected to arrest in response to α-factor with a population structure of ∼50% daughter cells, ∼25% mother cells, and ∼25% grandmother cells that have produced two or more daughters. Previous studies have shown that replicatively aged grandmother cells die more quickly than younger cells (Herker et al., 2004). To test if one of these subpopulations is more susceptible to fast cell death, the fates of 576 sst1 mutant cells responding to α-factor were followed by time-lapse videomicroscopy. Asynchronous log-phase cells were placed in a microfluidic chamber with flowing YPD medium, incubated at RT, and photographed every 10 min using phase-contrast microscopy in order to track cell lineages. After 3 h of incubation at RT, the medium was replaced with YPD medium containing excess α-factor and 1 μM PI. Fields of cells were photographed as before and also photographed every 30 min using epifluorescence microscopy to detect dead (PI-positive) cells. The resulting time-lapse movies allowed tracking of 96% of cells in the population (44% mother and grandmother cells, 44% immature daughters, and 8% mature daughters, which remained attached to budded mother cells). As shown in Figure 2D, immature daughters and their mother cells were observed to die at slightly different rates (median lifespan at 3.5 and 4.5 h, respectively), but after 8 h the levels of cell death in these two subpopulations was indistinguishable (20.6 and 19.1%, respectively). These frequencies of cell death were not significantly different from each other or the overall population (19%) but were significantly higher than those of fig1 mutants (5% cell death in the overall population). The lifespan of cells undergoing fast cell death was longer in this experiment, probably because of decreased incubation temperature. Importantly, the observed frequency of dead mother–daughter pairs (3.6%) was not significantly different from the frequency expected if cell death in these two populations occurred at random (0.191 × 0.206 = 3.9%). Thus, the rate and extent of fast cell death was not correlated with any particular cell age or lineage and such deaths appeared to occur randomly in the population. As an alternative explanation, the low and random incidence of fast cell death in wild-type populations may result from stochastic variations in pro– and anti–death regulatory pathways.
Factors That Promote Fast Cell Death
To identify factors that specifically regulate fast but not slow cell death, we first tested many factors known to participate in the primary response to mating pheromones (receptors, heterotrimeric G-proteins, and MAP-kinase cascades) and in secondary responses such as cell cycle arrest and polarized morphogenesis. All “sterile” mutants that fail to mount the full primary response to α-factor were completely deficient in both fast and slow cell death (unpublished data). Mutants lacking the ability to arrest in G1-phase of the cell cycle (fus3 and far1 mutants) or to undergo polarized morphogenesis (bni1, pea2, and spa2 mutants) were also profoundly deficient in both fast and slow cell death (Figure 2B and unpublished data). Several factors involved in cell–cell fusion and low-affinity Ca2+ influx appear to function downstream of these secondary factors (Brizzio et al., 1998; Muller et al., 2003; Fitch et al., 2004). Remarkably, mutants lacking the downstream factors Fus1, Fus2, or Rvs161 were deficient in fast cell death but were fully capable of slow cell death (Figure 2B) even at saturating α-factor when Sst1 protease was also eliminated (unpublished data). Thus, fast and slow waves of cell death were genetically distinguishable on the basis of their requirements for cell fusion factors and mitochondrial respiratory factors.
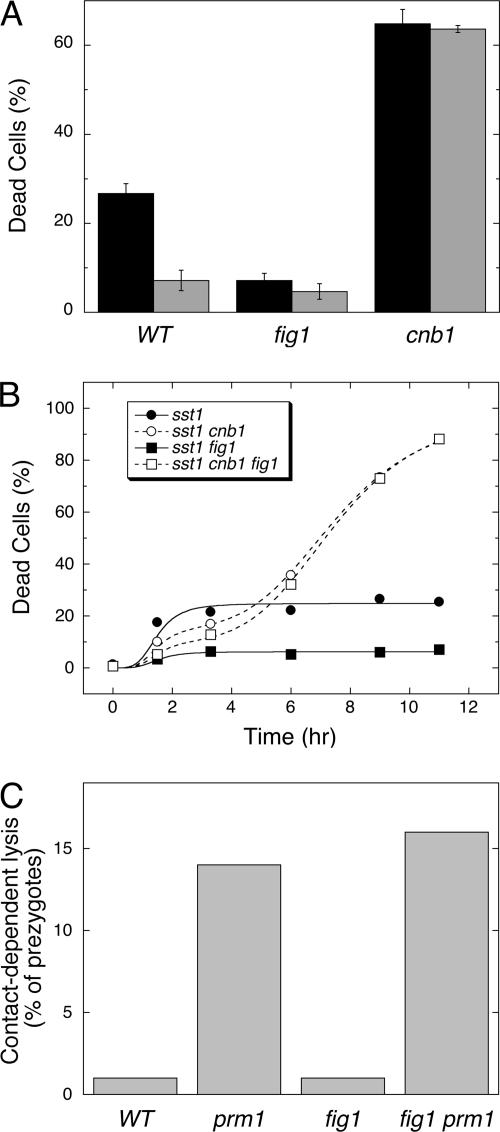
Fus1, Fus2, and Rvs161 are localized to the tip of mating projections and are thought to promote mating by facilitating the focal secretion of cell wall hydrolases, which remove barriers to the membrane by locally degrading the chitin and glucan components of the cell wall (Brizzio et al., 1998). To test if cell wall degradation is necessary for fast cell death, chitin biosynthesis was induced about fivefold during the response to α-factor by the addition of its biosynthetic precursor glucosamine (15 mM) to the culture medium (Bulik et al., 2003). Glucosamine strongly suppressed fast death of wild-type cells but did not suppress slow death of cnb1 mutants (Figure 3A). Therefore, cell wall degradation appeared to be necessary for fast cell death.
Figure 3.
Fast but not slow cell death requires dissolution of cell wall polymers. (A) Cell death was measured in wild-type, fig1 mutant, and cnb1 fig1 double mutant populations after 8.5-h treatment with 60 μM α-factor in the presence (gray bars) or absence (black bars) of 15 mM glucosamine. (B) Cell death was measured in single, double, and triple mutants at various times after exposure to 60 μM α-factor. The averages of three replicate experiments were plotted and fit to standard Sigmoid equations as described in Figure 1. (C) Contact-dependent lysis of wild-type, prm1 mutant, fig1 mutant, and fig1 prm1 double mutant prezygotes was measured as described previously (Jin et al., 2004).
The four-spanner transmembrane protein Fig1 functions downstream of Fus1, Fus2, and Rvs161 as part of a low-affinity Ca2+ influx system (Erdman et al., 1998; Muller et al., 2003). Interestingly, fig1 mutants also exhibited little or no fast cell death whereas cnb1 fig1 double mutants exhibited high levels of slow cell death (Figures 2B and 3B). In contrast, a five-spanner transmembrane protein Prm1 that promotes fusion of cell membranes during mating after cell wall removal (Heiman and Walter, 2000) had no effect on fast or slow cell death (Figure 2B). Recently, ∼10% of prm1 mutant cells engaged in conjugation but arrested as late prezygotes were observed to die by a process termed contact-dependent lysis (Jin et al., 2004). Contact-dependent lysis was not dependent on Fig1 (Figure 3C) and was insensitive to antimycin (unpublished data), suggesting regulation by an unknown mechanism. Fast cell death specifically required factors involved in cell wall degradation (Fus1, Fus2, and Rvs161) and additionally required a function of Fig1.
Fig1 Is a Member of the PMP-22/EMP/MP20/Claudin Superfamily and Promotes Cell Death Independent of Ca2+
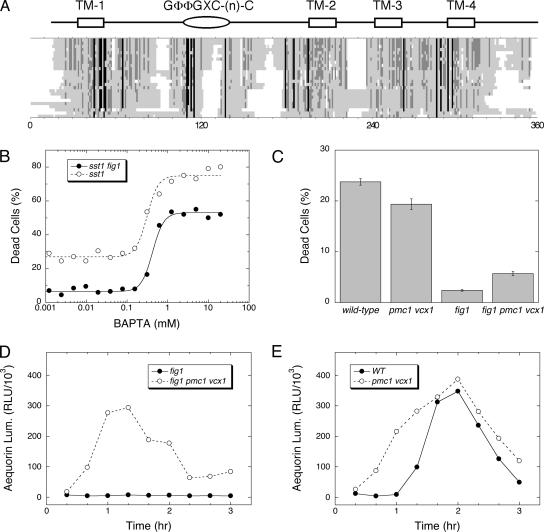
BLAST searches of protein and DNA databanks revealed orthologues of Fig1 in other species of fungi. A multiple sequence alignment of the best homolog from 30 diverse fungal species revealed conservation of size and transmembrane topology but only one strongly conserved motif (denoted GΦΦGXC(n)C, where Φ = YFLM and 8 < n < 20) located in the extracellular loop between the first and second predicted transmembrane spans (Figure 4A). To identify more distantly related proteins, the yeast Fig1 sequence was used to seed a PSI-BLAST search of all eukaryotic proteins (Altschul et al., 1997). Starting with the 11th iteration, metazoan proteins belonging to PMP-22/EMP/MP20/Claudin superfamily of proteins (pfam00822) were recovered with significant E-values. Nearly all members of this diverse superfamily possess four predicted transmembrane segments of similar spacing and a highly conserved GLWXXC(n)C motif in the same location as that of the Fig1 family. More than 40 members of this superfamily have been described in humans, most of which are thought to interact with themselves and/or other transmembrane proteins in the same or adjacent membranes. Eight members of the superfamily in humans are thought to bind and regulate functions of either voltage-gated Ca2+ channels (e.g., VGCC-γ subunit) or glutamate-sensitive ion channels (e.g., stargazin; Burgess et al., 2001). In most neuronal cell types, excessive Ca2+ influx via either pathway can trigger excitotoxic cell death or necrosis (Arundine and Tymianski, 2004).
Figure 4.
Fig1 is a member of the stargazin/VGCC-γ/claudin superfamily but promotes fast cell death independent of [Ca2+]c elevation. (A) Fig1 orthologues from yeast and 24 other fungi were identified by BLAST searches of genome databases and aligned using CLUSTALW algorithm. Sites of high, moderate, and poor sequence conservation are shaded black, dark gray, and light gray, respectively. White spaces indicate gaps. The positions of four predicted transmembrane segments and a conserved GΦΦGXC(n)C motif are indicated. (B) Cell death was measured in sst1 mutant and sst1 fig1 double mutant populations after 3-h treatment with 60 μM α-factor in the presence of varying amounts of BAPTA, a chelator of divalent metals such as Ca2+. Data were fit to standard sigmoid equations. (C) Cell death was measured in wild-type, pmc1 vcx1 double mutant, fig1 mutant, and fig1 pmc1 vcx1 triple mutant populations after 3.5-h treatment with 60 μM α-factor. (D and E) Luminescence of cytoplasmic aequorin was measured in the same strains at various times after treatment with 60 μM α-factor.
Fig1 promotes Ca2+ influx and elevation of cytosolic free Ca2+ concentrations ([Ca2+]c) during the response to α-factor by activating a low-affinity system that is independent of the high-affinity Ca2+ influx system homologous to VGCC’s (Muller et al., 2003). To test if Ca2+ influx and/or [Ca2+]c elevation are required for fast cell death, a cell-impermeant chelator with high affinity for Ca2+ (BAPTA) was added to the culture medium bathing cells responding to high α-factor, and cell death was measured. No concentration of BAPTA was able to diminish death of wild-type cells relative to that of fig1 mutants (Figure 4B), indicating that Fig1-dependent fast cell death was not dependent on Ca2+ influx. Instead, BAPTA concentrations greater than 0.3 mM strongly stimulated the death of wild-type cells and fig1 mutants. This effect can be explained by the ability of BAPTA to prevent activation of calmodulin, which is also necessary for activation of Ca2+/calmodulin-dependent protein kinases that promote cell survival in these conditions independent of CN (Moser et al., 1996; Withee et al., 1997). Therefore, Fig1 appeared to promote fast cell death independent of its ability to promote Ca2+ influx and [Ca2+]c elevation.
To test if forced [Ca2+]c elevation can restore the death of fig1mutants, we measured cell death in mutants lacking the vacuolar Ca2+ pump Pmc1 and the vacuolar Ca2+/H+ exchanger Vcx1, enzymes that redundantly serve to lower [Ca2+]c in yeast (Cunningham and Fink, 1994, 1996). The loss of Pmc1 and Vcx1 in fig1 mutants restored [Ca2+]c to near wild-type levels and augmented [Ca2+]c in cells expressing Fig1 to even higher levels (Figure 4, D and E). The loss of Pmc1 and Vcx1 slightly increased the death of fig1 mutants to much lower levels than those of wild-type cells and slightly decreased the death of Fig1–proficient cells (Figure 4C). These findings reveal no important role for Ca2+ in fast cell death but do not rule out the possibility that Fig1 promotes influx of other toxic ions.
Factors That Inhibit Fast Cell Death
During the normal response to α-factor, cell wall degradation activates the cell wall integrity (CWI) signaling pathway involving the rho-type GTPase Rho1 and Pkc1, Bck1, Mkk1, Mkk2 cascade of protein kinases that culminate with activation of the MAP kinase Mpk1/Slt2 (Buehrer and Errede, 1997). The activated CWI signaling pathway stimulates chitin and glucan synthases, which synthesize new cell wall polymers and compensate for the cell wall degradation (Levin, 2005). To test the prediction that CWI signaling can inhibit fast cell death, we first measured cell death in lrg1 mutants where CWI signaling may be hyperactivated because of the loss of Lrg1, a GTPase-activating protein for Rho1 (Fitch et al., 2004). Indeed, similar to fus1, fus2, and rvs161 mutants, the lrg1 mutants exhibited greatly diminished levels of fast cell death (Figure 2B).
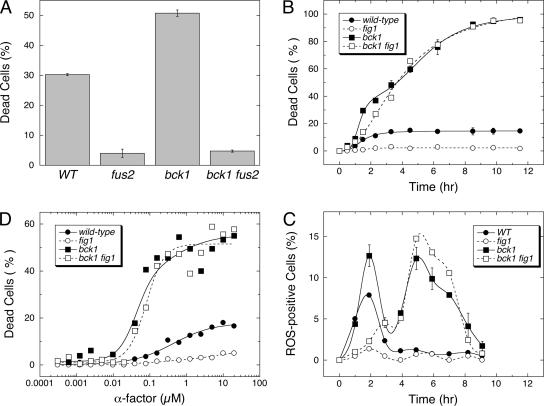
Cells deficient in CWI signaling are known to exhibit very high levels of cell death in response to mating pheromones (Errede et al., 1995), but the kinetics, sensitivity to α-factor, and dependence on cellular factors have not been examined. If this enhanced cell death represents enhanced fast cell death, the death of CWI-deficient mutants should be dependent on Fig1, Fus1, Fus2, and Rvs161. Remarkably, the high level of cell death observed for bck1 mutants (lacking the MAPKKK in the CWI pathway) was partially blocked by glucosamine (unpublished data) and completely abolished by the loss of Fus2 (Figure 5A). However, the death of bck1 mutants was not blocked by the loss of Fig1 (Figure 5B). Careful inspection of the data indicated two waves of cell death in bck1 mutants and only one wave of cell death in bck1 fig1 double mutants. The monophasic death curve of bck1 fig1 double mutants was described very well by a sigmoid equation where all cells died at an intermediate rate (median lifespan of ∼4.5 h). The death of bck1 mutants was best described by the sum of two sigmoid equations where 40% of the population died rapidly (median lifespan of 1.4 h) and the remainder of the population died at or near the intermediate rate (median lifespan of ∼3.6 h). Two peaks of ROS accumulation corresponding to the two waves of cell death were observed in bck1 mutants (Figure 5C). The loss of Fig1 abolished the first peak of ROS and the gain of Bck1 abolished the second peak (Figure 5C). These findings are consistent with two waves of cell death occurring in CWI-deficient cells, a faster wave that is dependent on Fig1 and a slower one that is independent of Fig1 and dependent on cell wall degradation. The slower wave in bck1 mutants and bck1 fig1 double mutants was triggered by about sevenfold lower concentrations of α-factor than the faster wave in wild-type cells (Figure 5D) and was mechanistically distinct from that of cnb1 and cnb1 fig1 mutants in that it could not inhibited by antimycin A (unpublished data). The findings that Fig1-dependent fast cell death increased about threefold in bck1 mutants and decreased more than fivefold in lrg1 mutants relative to wild-type cells (∼13% in this strain background) indicates that this process may be negatively regulated by CWI signaling.
Figure 5.
CWI signaling inhibits fast cell death and an intermediate cell death. (A) Cell death was measured in wild-type, fus2 mutant, bck1 mutant, and bck1 fus2 double mutant populations after 3-h treatment with 60 μM α-factor. (B) Dead cells were measured in wild-type, bck1 mutant, fig1 mutant, and bck1 fig1 double mutant populations at various times after treatment with 60 μM α-factor. The average of three independent measurements were plotted (±SD) and fit to the sum of one or two standard sigmoid equations. (C) Percentages of ROS positive cells were measured over time in three replicate cultures of wild-type, bck1 mutant, fig1 mutant, and bck1 fig1 double mutant strains after treatment with 60 μM α-factor and plotted (mean ± SD). (D) Cell death was measured in sst1 mutant, sst1 bck1 double mutant, sst1 fig1 double mutant, and sst1 bck1 fig1 triple mutant populations after 3-h treatment with various concentrations of α-factor. Data were fit to the sum of one or two standard sigmoid equations.
DISCUSSION
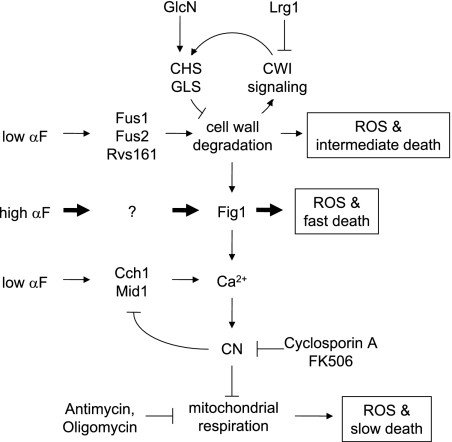
The findings described here support a working model where at least three genetically independent regulatory pathways contribute to the death of yeast cells responding to mating pheromones (see Figure 6). The CN and CWI signaling pathways become activated during the response to low and high mating pheromones and independently perform functions essential for long-term cell survival. CN- and CWI-deficient cells appear to die by two distinct processes because only the former can be suppressed by blockade of mitochondrial respiration and only the latter can be suppressed by glucosamine-enhanced cell wall biosynthesis. Even in wild-type cells that are protected by the CN and CWI pathways, Fig1 promotes a third, fast wave of death in response to high concentrations of mating pheromones. It is possible that all three forms of death involve common pathways. However, the different kinetics and genetic requirements we documented are most easily accounted for by distinct prodeath or antisurvival mechanisms playing a role. Because it is not yet known if any of these pathways regulate cell death in normal development or physiological matings in the wild, we cannot conclude that any of them represent physiological mechanisms of PCD. Nevertheless, endogenous mating pheromones acting through their G-protein–coupled receptors are frequently considered physiological stimuli in yeasts. For the sake of clarity, we will first discuss independently each of the three regulatory pathways and their possible evolutionary relationships to cell death mechanisms operating in mammalian cells and later consider the possibility that all three pathways might be interrelated.
Figure 6.
Working model of regulatory mechanisms controlling cell death in response to mating pheromones. Low concentrations of α-factor (αF) activate regulatory pathways leading to cell wall degradation (CW deg.) and stimulation of the high-affinity Ca2+ influx system (Cch1, Mid1) but not the Fig1-dependent low-affinity Ca2+ influx system. The resulting rise in [Ca2+]c activates CN, which prevents ROS accumulation by either inhibiting ROS production by the mitochondrial respiratory chain or stimulating ROS dissipation. The diminished cell wall strength may lead to increased ROS accumulation and intermediate lifespan of cells were it not for the activation of the CWI signaling pathway, which increases biosynthesis of new cell wall material by increasing the activities of chitin and glucan synthases (CHS and GLS). Because CN and CWI signaling pathways operate in wild-type cells, low concentrations of α-factor fail to induce significant amounts of cell death. High concentrations of α-factor activate the above processes but also induce Fig1-dependent Ca2+ influx, early ROS accumulation, and rapid cell death in a fraction of the population. Glucosamine (GlcN) supplement and deficiencies of Fus1, Fus2, or Rvs161 block all these responses and additionally prevent activation of CWI signaling. Fig1 deficiency does not affect cell wall degradation or CWI signaling.
Death of CN-Deficient Mutants (“CN-less death”)
CN-less death was first discovered after chelating extracellular Ca2+ during the response to α-factor and later was found to be mimicked by mutations that eliminate the high-affinity Ca2+ influx channel, Ca2+/calmodulin, and CN (Iida et al., 1990; Cyert et al., 1991; Cyert and Thorner, 1992; Foor et al., 1992; Iida et al., 1994; Ono et al., 1994; Moser et al., 1996; Fischer et al., 1997; Paidhungat and Garrett, 1997; Withee et al., 1997; Muller et al., 2001). This pathway does not desensitize cells to mating pheromones or promote recovery after withdrawal of mating pheromones (Moser et al., 1996) and therefore implicates CN and its upstream regulators as key components of an antideath or prosurvival pathway in cells responding to mating pheromones. Here we demonstrate that many cells undergoing CN-less death transiently accumulate ROS before death. ROS accumulation may occur in all cells undergoing CN-less death, and the brief appearance of ROS-positive cells may reflect some combination of a transient ROS production, active ROS dissipation, and the loss of ROS and fluorescent probes upon cell death. Therefore, CN can be firmly positioned as an upstream inhibitor of ROS accumulation in these conditions. CYC, coenzyme Q, and complexes III, IV, and V of the mitochondrial respiratory chain were all required for efficient CN-less death and for accumulation of ROS. Whether CN regulates these factors or other factors involved in ROS production or dissipation remains to be elucidated.
CN-less death was not prevented by cyclosporin A or dependent on mitochondrial cyclophilin D (Figures 1 and 2), suggesting that the mitochondrial permeability transition pore may not have as strong a role as previously suggested (Severin and Hyman, 2002). CN-less death also did not require Fus1, Fus2, Rvs161, Lrg1, or Fig1 but did require Bni1, Spa2, and Pea2 for maximum effectiveness (Figure 2B). The latter factors are all components of the polarisome, a regulator of actin dynamics and cell polarity during yeast mating (Chenevert et al., 1994; Valtz and Herskowitz, 1996; Evangelista et al., 1997; Bidlingmaier and Snyder, 2004). Recently, polarisome function was shown to be important for maximum activation of the pheromone signaling pathway and for mating (Qi and Elion, 2005), so its roles in CN-less death and Fig1-dependent death are not unexpected. A dominant-negative fragment of Spa2 recently isolated as a high-dosage suppressor of CN-less death (Noma et al., 2005) may also depress pheromone signaling. Because of the pleiotropic effects of the polarisome on mating and multiple death pathways, it seems an unlikely target of CN regulation. Obviously, more work will be needed to determine precisely how CN and the other pheromone-responsive pathways regulate CN-less death.
Interestingly, the dependence of CN-less death on mitochondrial respiration closely resembles “Bax-death,” which arises in yeast as a consequence of heterologous overexpression of human Bax (Harris et al., 2000). Both CN-less death and Bax-death are sensitive to respiration inhibitors but insensitive to other factors involved in apoptosis-like death such as Mca1 (Guscetti et al., 2005). It is therefore reasonable to speculate that a Bax-like factor endogenous to yeast might be activated in response to mating pheromones and counteracted by CN. In mammals, CN dephosphorylates at least one regulator of mammalian apoptosis (Bad), which then stimulates Bax-induced apoptosis by interfering with Bax inhibitors (Wang et al., 1999). However, no obvious homologues of Bax or Bad are evident in the genomes of modern fungi. Fis1, a strongly conserved protein involved in fission of yeast and mammalian mitochondria (Okamoto and Shaw, 2005), has been proposed as a surrogate of Bcl-2 or Bax in yeast cells undergoing apoptosis-like cell death (Fannjiang et al., 2004). Though Fis1 can regulate apoptosis-like cell death in other conditions (Ivanovska and Hardwick, 2005), Fis1 did not detectably modulate CN-less death in response to mating pheromones (Figure 2B). Therefore, CN-less death in yeast seems mechanistically distinct from apoptosis in animals and apoptosis-like cell death in fungi.
Death of CWI-Deficient Mutants
CWI-deficient mutants responding to low concentrations of α-factor also die, but their manner of death appears quite distinct from that of CN-deficient mutants. Unlike CN-less death, the death of CWI-deficient cells did not depend on respiration and did depend on the functions of Fus1, Fus2, and Rvs161, which promote localized degradation of cell wall polymers at the tips of mating projections (Fitch et al., 2004). In wild-type cells, activation of CWI signaling during the response to α-factor leads to increased biosynthesis of new cell wall polymers (Bulik et al., 2003) and prevention of cell death. As expected for an inducer of cell wall chitin biosynthesis, glucosamine suppressed death of CWI-deficient cells (but not CN-deficient cells). FK506 increased the rate of death of bck1 mutants and bck1 fig1 double mutants (unpublished observations), and independent effects of MPK and CN have been noted previously (Zhao et al., 1998). Although the death of CWI-deficient mutants responding to α-factor was originally considered to be a consequence of lysis due to cell wall failure (Errede et al., 1995), we observed a peak of ROS-positive cells in the dying population, and their death was not avoided by environmental osmotica such as sorbitol and salt (unpublished observations). Therefore, CWI signaling may prevent death during the response to α-factor, primarily by inhibiting a nonmitochondrial source of ROS. It may be impossible to prove that “MPK-less death” and CN-less death are completely independent until their direct and indirect targets are fully elucidated.
Fig1-Dependent Death
High concentrations of α-factor triggered rapid death of wild-type cells, in striking contrast to low concentrations that induced much slower cell deaths in CN-deficient and CWI-deficient mutants but not wild-type cells. The fast high-α-factor cell death was further distinguished from the slow low-α-factor cell deaths (CN- and MPK-less deaths) by its complete dependence on Fig1, a plasma membrane protein that is strongly induced by the response to mating pheromones (Erdman et al., 1998; Muller et al., 2003). A Fig1-dependent fast wave of cell death was detected even in CN-deficient and CWI-deficient cells responding to high concentrations of α-factor, suggesting that Fig1 can promote cell death independent of CN and CWI signaling. However, mathematical deconvolution of the biphasic death curves indicated a mild stimulation of Fig1-dependent death by CN signaling and a strong inhibition of Fig1-dependent death by CWI signaling. The effect of CN may be insignificant because it prevents a large majority of cells from succumbing to CN-less death and therefore may increase the number of cells susceptible to Fig1-dependent death. On the other hand, the effect of CWI signaling appeared much more robust: the loss of CWI signaling in bck1 mutants increased Fig1-dependent death to ∼40% of the population, whereas the gain of CWI signaling in lrg1 mutants decreased Fig1-dependent death to 2% or less. Factors that promote cell wall degradation (Fus1, Fus2, and Rvs161) were required for Fig1-dependent death and factors that prevent cell wall degradation (glucosamine) also prevented Fig1-dependent death. Because all these factors are interconnected in a negative feedback loop (see Figure 6), the incidence of fast cell death in a population may reflect the outcome of a race between the Fig1-dependent prodeath pathway and a CWI-dependent antideath pathway (that probably involves cell wall repair). The apparently random occurrence of fast cell death can be explained by cell-to-cell variations in either the pheromone signaling pathways (Colman-Lerner et al., 2005) or the CWI signaling pathway as observed for other MAP kinase pathways (Ferrell and Machleder, 1998). Small strain-to-strain variations in either pathway also may be responsible for the varying degrees of fast cell death in different strain backgrounds. Therefore, the negative feedback loop provides possible explanations for why only a fraction of yeast cells die by the Fig1-dependent process, why this fraction differs in various strain backgrounds, and why the cell death appears random in the population with respect to cell age. Because Fig1 also performs functions that promote cell fusion during mating (Erdman et al., 1998), fast cell death may reflect a lethal outcome of attempting late steps of the mating program in the absence of a mating partner. These findings provide an alternative to the view of altruistic PCD triggered by mating pheromones.
The most conserved region of Fig1 and its closest homologues in fungi mapped to the GΦΦGXC(n)C motif in the first extracellular loop. A similar motif in the PMP-22/EMP/MP20/Claudin superfamily frequently mediates homophilic and heterophilic interactions with transmembrane proteins in the same or adjacent cell membrane (Van Itallie and Anderson, 2006). Fig1 does not function like a VGCC-γ subunit in yeast because Fig1 was not required for normal activation of the VGCC-related high-affinity Ca2+ influx channel (Muller et al., 2003). Although Fig1 resembles stargazin in its ability to promote low-affinity Ca2+ influx and Ca2+ influx may contribute to yeast cell death in other circumstances (Courchesne, 2002; Gupta et al., 2003; Pozniakovsky et al., 2005), Ca2+ influx was not required for fast cell death in yeast. However, we cannot rule out other possible ion fluxes as the cause of Fig1-dependent death. Other members of the PMP-22/EMP/MP20/Claudin superfamily termed PERP have been shown to induce apoptosis of mammalian cells by unknown mechanisms (Ihrie et al., 2003). Overexpression of PMP-22 or EMP1,2,3 proteins stimulates apoptosis in HEK293 cells, possibly through interactions with P2X7 ion channels (Wilson et al., 2002). Identification of factors that function together with or downstream of Fig1 may provide additional insights into its roles and possible conservation in mammals.
Apoptosis and PCD in Yeast?
Apoptosis is a well-characterized form of programmed cell death occurring in all animals. Orthologues of the key proapoptosis factors caspase/Ced-3, Apaf-1/Ced-4, and Bax/Ced-9 are all absent from all fungal genomes sequenced to date. Some “apoptosis-like” cell deaths in yeast may be regulated by a distantly related caspase-like protease termed Mca1 (Madeo et al., 2004) and certain mitochondrial factors (Fannjiang et al., 2004). We found no detectable role of these factors and no evidence for chromatin fragmentation (a hallmark of apoptosis-like cell death in yeast) during any of the three waves of cell death reported here. Reports to the contrary (Severin and Hyman, 2002) utilized a TUNEL assay method that we find susceptible to background from an RNase-sensitive factor. Previous findings that cyclosporin A can prevent fast cell death during the response to mating pheromones (Severin and Hyman, 2002) were not reproducible in our hands. All three manners of cell death studied here result in uptake of methylene blue and PI, suggesting the cells become metabolically inactive and lose their integrity, which seems very different from apoptosis and apoptosis-like cell death in which only cell viability/proliferation is lost. Finally, death of CN-deficient and CWI-deficient mutants appears morphologically dissimilar from apoptosis (Errede et al., 1995; Withee et al., 1997). Collectively, these findings argue against the idea that mating pheromones trigger apoptosis-like cell death in yeast.
PCD in animals, whether apoptotic or not, implies an altruistic behavior that benefits the surviving cells. It has been speculated that death of yeast cells responding to mating pheromones may represent an altruistic behavior where, for example, older or weaker cells sacrifice themselves to preserve resources (nutrients and/or mating partners) for younger or healthier cells (Severin and Hyman, 2002). We have directly tested one aspect of that idea and determined that mother cells and newborn daughter cells are equally likely to undergo Fig1-dependent death and that death of mother-daughter pairs is not more common or less common than that expected by chance. Thus, Fig1-dependent death was apparently independent of cell aging. One might argue that only weak cells undergo Fig1-dependent death to eliminate their inferior genomes from the mating pool. However, genetically weakened mutants lacking CN or CYC did not show higher levels of Fig1-dependent death. Fig1-dependent death in a-cells also required much higher concentrations of α-factor than other types of cell death, conditions that might occur naturally only when potential mates are in abundance or when mating cells are closely apposed. However, Fig1-dependent death was not detectable in mixed populations of a- and α-cells undergoing mating or in prezygotes arrested at the membrane fusion step of mating. Thus, Fig1-dependent death may be too infrequent in normal conditions to provide a significant advantage to the surviving kin.
As an alternative to altruistic cell suicide, Fig1-dependent death may represent the execution of an ordinary event in yeast cell mating at an inappropriate time or place, resulting in lethal consequences. Low concentrations of mating pheromones trigger early events in mating such as agglutination of a- and α-cells and formation of pre-zygotes while high concentrations of mating pheromones are necessary for dissolution of the cell wall material that prevents close apposition of cell membranes before membrane fusion (Brizzio et al., 1996; Dorer et al., 1997). Mutants lacking Fig1 exhibit mild but detectable defects in dissolution of cell walls of prezygotes and zygotes (Erdman et al., 1998). Therefore, high mating pheromones may trigger Fig1-dependent dissolution of cell wall material, which promotes fusion of prezygotes and possibly lysis of cells that lack a mating partner. Thus, there is no need to speculate the existence of altruistic behavior of yeast cells in these circumstances.
CN-less death and MPK-less death did not occur at appreciable levels in populations of wild-type cells but occur in nearly 100% of cells responding to low concentrations of α-factor (unless they have already undergone Fig1-dependent death). Although these putative cell death pathways have the potential to eliminate certain types of weakened cells from the mating pool, the time required for death is longer than the time required for successful conjugation, and mutants lacking CN can mate as efficiently as wild-type cells (Cyert et al., 1991; Cyert and Thorner, 1992). One interpretation of these findings is that CN and MPK each perform functions that are essential for adaptation of cells exposed to mating pheromones for long periods of time. The essential function of CWI signaling may be the repair of damaged cell walls or suppression of nonmitochondrial ROS production, though other pathways have not been excluded. In the case of CN, its essential function appears to be suppression of mitochondrial ROS production or enhancement of ROS detoxification. Therefore, CN-less death and MPK-less deaths may be viewed more accurately as prosurvival pathways than antideath pathways, though such a view may change if bona fide prodeath pathways were identified in the future.
Independent of their potential relationships to mammalian processes, CN-less death and MPK-less death merit further study because similar processes may occur broadly in the fungal kingdom during the response to commonly prescribed antifungal antibiotics. Azole-class and echinocandin-class antibiotics do not kill the fungal species in diverse yeasts and fungi (Del Poeta et al., 2000; Marchetti et al., 2000; Bonilla et al., 2002; Cruz et al., 2002; Edlind et al., 2002; Bonilla and Cunningham, 2003; Kraus et al., 2003; Onyewu et al., 2003; Reinoso-Martin et al., 2003; Sanglard et al., 2003; Kaur et al., 2004; Steinbach et al., 2004). Like mating pheromones, the natural antifungal compound tunicamycin stimulates both CN-less death and MPK-less in yeast and probably other species (Bonilla et al., 2002; Chen et al., 2005). Therefore, chemical inhibitors of CN, MPK, or their targets in fungi are expected to vastly improve the fungicidal activity of most clinically relevant antibiotics and potentially diminish the acquisition of antibiotic resistance. Identification of the factors and processes downstream of CN, MPK, and Fig1 involved in each type of fungal cell death pathway should provide insights into the origins and diversity of cell death mechanisms in multicellular organisms.
ACKNOWLEDGMENTS
We are grateful to Janet Shaw, Charlie Boone, Cathy Clarke, and Lymarie Maldonado-Baez for yeast strains used in this study. We also thank Christian Martin and Hui Jin for excellent technical assistance. This work was supported by grants from the National Science Foundation (MCB-0331306 to A.L.), the American Cancer Society (RSG-05-205-01-MBC to E.G.), and the National Institutes of Health (GM072024 and RR020839 to A.L.; GM053082 and GM073038 to K.W.C.).
Abbreviations used:
- CN
calcineurin
- CWI
cell wall integrity
- CYC
cytochrome c
- MPK
mitogen-activate protein kinase
- PCD
programmed cell death
- ROS
reactive oxygen species.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0177) on May 31, 2006.
REFERENCES
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arundine M., Tymianski M. Molecular mechanisms of glutamate-dependent neurodegeneration in ischemia and traumatic brain injury. Cell. Mol. Life Sci. 2004;61:657–668. doi: 10.1007/s00018-003-3319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiza A. F., Schulz T., Masson P. H. Yeast respond to hypotonic shock with a calcium pulse. J. Biol. Chem. 1996;271:23357–23362. doi: 10.1074/jbc.271.38.23357. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier S., Snyder M. Regulation of polarized growth initiation and termination cycles by the polarisome and Cdc42 regulators. J. Cell Biol. 2004;164:207–218. doi: 10.1083/jcb.200307065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla M., Cunningham K. W. MAP kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell. 2003;14:4296–4305. doi: 10.1091/mbc.E03-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla M., Nastase K. K., Cunningham K. W. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002;21:2343–2353. doi: 10.1093/emboj/21.10.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brizzio V., Gammie A. E., Nijbroek G., Michaelis S., Rose M. D. Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J. Cell Biol. 1996;135:1727–1739. doi: 10.1083/jcb.135.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V., Gammie A. E., Rose M. D. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 1998;141:567–584. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer B. M., Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik D. A., Olczak M., Lucero H. A., Osmond B. C., Robbins P. W., Specht C. A. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell. 2003;2:886–900. doi: 10.1128/EC.2.5.886-900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D. L., Gefrides L. A., Foreman P. J., Noebels J. L. A cluster of three novel Ca2+ channel gamma subunit genes on chromosome 19q13.4, evolution and expression profile of the gamma subunit gene family. Genomics. 2001;71:339–350. doi: 10.1006/geno.2000.6440. [DOI] [PubMed] [Google Scholar]
- Chan R. K., Otte C. A. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol. Cell. Biol. 1982;2:11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Feldman D. E., Deng C., Brown J. A., De Giacomo A. F., Gaw A. F., Shi G., Le Q. T., Brown J. M., Koong A. C. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 2005;3:669–677. doi: 10.1158/1541-7786.MCR-05-0181. [DOI] [PubMed] [Google Scholar]
- Chenevert J., Valtz N., Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1296. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A., Gordon A., Serra E., Chin T., Resnekov O., Endy D., Pesce C. G., Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- Courchesne W. E. Characterization of a novel, broad-based fungicidal activity for the antiarrhythmic drug amiodarone. J. Pharmacol. Exp. Ther. 2002;300:195–199. doi: 10.1124/jpet.300.1.195. [DOI] [PubMed] [Google Scholar]
- Cruz M. C., Goldstein A. L., Blankenship J. R., Del Poeta M., Davis D., Cardenas M. E., Perfect J. R., McCusker J. H., Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002;21:546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. W., Fink G. R. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking. PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. W., Fink G. R. Calcineurin inhibits. VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in yeast. Mol. Cell. Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M. S., Kunisawa R., Kaim D., Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M. S., Thorner J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 1992;12:3460–3469. doi: 10.1128/mcb.12.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial N. N., Korsmeyer S. J. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Del Poeta M., Cruz M. C., Cardenas M. E., Perfect J. R., Heitman J. Synergistic antifungal activities of bafilomycin A1, fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob. Agents Chemother. 2000;44:739–746. doi: 10.1128/aac.44.3.739-746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J. W. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Dorer R., Boone C., Kimbrough T., Kim J., Hartwell L. H. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics. 1997;146:39–55. doi: 10.1093/genetics/146.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlind T., Smith L., Henry K., Katiyar S., Nickels J. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol. Microbiol. 2002;46:257–268. doi: 10.1046/j.1365-2958.2002.03165.x. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Satterberg B., Kranz J. E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Cade R. M., Yashar B. M., Kamada Y., Levin D. E., Irie K., Matsumoto K. Dynamics and organization of MAP kinase signal pathways. Reprod. Dev. 1995;42:477–485. doi: 10.1002/mrd.1080420416. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M. S., Chow C. J., Adames N., Pringle J. R., Peter M., Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fannjiang Y., Cheng W. C., Lee S. J., Qi B., Pevsner J., McCaffery J. M., Hill R. B., Basanez G., Hardwick J. M. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E., Jr., Machleder E. M. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- Fischer M., Schnell N., Chattaway J., Davies P., Dixon G., Sanders D. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 1997;419:259–262. doi: 10.1016/s0014-5793(97)01466-x. [DOI] [PubMed] [Google Scholar]
- Fitch P. G., Gammie A. E., Lee D. J., de Candal V. B., Rose M. D. Lrg1p is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics. 2004;168:733–746. doi: 10.1534/genetics.104.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foor F., Parent S. A., Morin N., Dahl A. M., Ramadan N., Chrebet G., Bostian K. A., Nielsen J. B. Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from alpha-factor arrest in yeast. Nature. 1992;360:682–684. doi: 10.1038/360682a0. [DOI] [PubMed] [Google Scholar]
- Frohlich K. U., Madeo F. Apoptosis in yeast: a new model for aging research. Exp. Gerontol. 2001;37:27–31. doi: 10.1016/s0531-5565(01)00177-2. [DOI] [PubMed] [Google Scholar]
- Gin P., Clarke C. F. Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J. Biol. Chem. 2005;280:2676–2681. doi: 10.1074/jbc.M411527200. [DOI] [PubMed] [Google Scholar]
- Groisman A., Lobo C., Cho H., Campbell J. K., Dufour Y. S., Stevens A. M., Levchenko A. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods. 2005;2:685–689. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- Gupta S. S., Ton V. K., Beaudry V., Rulli S., Cunningham K., Rao R. Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J. Biol. Chem. 2003;278:28831–28839. doi: 10.1074/jbc.M303300200. [DOI] [PubMed] [Google Scholar]
- Guscetti F., Nath N., Denko N. Functional characterization of human proapoptotic molecules in yeast S. cerevisiae. FASEB J. 2005;19:464–466. doi: 10.1096/fj.04-2316fje. [DOI] [PubMed] [Google Scholar]
- Harris M. H., Vander Heiden M. G., Kron S. J., Thompson C. B. Role of oxidative phosphorylation in Bax toxicity. Mol. Cell. Biol. 2000;20:3590–3596. doi: 10.1128/mcb.20.10.3590-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M. G., Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K. A., Maldener C., Frohlich K. U., Wissing S., Buttner S., Fehr M., Sigrist S., Madeo F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie R. A., Reczek E., Horner J. S., Khachatrian L., Sage J., Jacks T., Attardi L. D. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr. Biol. 2003;13:1985–1990. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- Iida H., Nakamura H., Ono T., Okumura M. S., Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol. Cell. Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H., Yagawa Y., Anraku Y. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. A study of [Ca2+]i in single Saccharomyces cerevisiae cells with imaging of fura-2. J. Biol. Chem. 1990;265:13391–13399. [PubMed] [Google Scholar]
- Ivanovska I., Hardwick J. M. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 2005;170:391–399. doi: 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Carlile C., Nolan S., Grote E. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot. Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Castano I., Cormack B. P. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 2004;48:1600–1613. doi: 10.1128/AAC.48.5.1600-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Kraus P. R., Fox D. S., Cox G. M., Heitman J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 2003;48:1377–1387. doi: 10.1046/j.1365-2958.2003.03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts V. A., Felix R., Biddlecome G. H., Arikkath J., Mahaffey C. L., Valenzuela A., Bartlett F. S., 2nd, Mori Y., Campbell K. P., Frankel W. N. The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat. Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- Levin D. E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V. D., Mitteldorf J., Skulachev V. P. Programmed and altruistic ageing. Nat. Rev. Genet. 2005;6:866–872. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- Madeo F., Engelhardt S., Herker E., Lehmann N., Maldener C., Proksch A., Wissing S., Frohlich K. U. Apoptosis in yeast: a new model system with applications in cell biology and medicine. Curr. Genet. 2002;41:208–216. doi: 10.1007/s00294-002-0310-2. [DOI] [PubMed] [Google Scholar]
- Madeo F., Frohlich E., Ligr M., Grey M., Sigrist S. J., Wolf D. H., Frohlich K. U. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Herker E., Wissing S., Jungwirth H., Eisenberg T., Frohlich K.-U. Apoptosis in yeast. Curr. Opin. Microbiol. 2004;7:655–660. doi: 10.1016/j.mib.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Marchetti O., Moreillon P., Glauser M. P., Bille J., Sanglard D. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 2000;44:2373–2381. doi: 10.1128/aac.44.9.2373-2381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M. J., Geiser J. R., Davis T. N. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 1996;16:4824–4831. doi: 10.1128/mcb.16.9.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M., Shaw J. M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. M., Locke E. G., Cunningham K. W. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics. 2001;159:1527–1538. doi: 10.1093/genetics/159.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. M., Mackin N. A., Erdman S. E., Cunningham K. W. Fig1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:38461–38469. doi: 10.1074/jbc.M304089200. [DOI] [PubMed] [Google Scholar]
- Noma S., Iida K., Iida H. Polarized morphogenesis regulator Spa2 is required for the function of putative stretch-activated Ca2+-permeable channel component Mid1 in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:1353–1363. doi: 10.1128/EC.4.8.1353-1363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Ono T., Suzuki T., Anraku Y., Iida H. The MID2 gene encodes a putative integral membrane protein with a Ca2+-binding domain and shows mating pheromone-stimulated expression in Saccharomyces cerevisiae. Gene. 1994;151:203–208. doi: 10.1016/0378-1119(94)90657-2. [DOI] [PubMed] [Google Scholar]
- Onyewu C., Blankenship J. R., Del Poeta M., Heitman J. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob. Agents Chemother. 2003;47:956–964. doi: 10.1128/AAC.47.3.956-964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M., Garrett S. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol. Cell. Biol. 1997;17:6339–6347. doi: 10.1128/mcb.17.11.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniakovsky A. I., Knorre D. A., Markova O. V., Hyman A. A., Skulachev V. P., Severin F. F. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J. Cell Biol. 2005;168:257–269. doi: 10.1083/jcb.200408145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault M., Camougrand N., Kinnally K. W., Vallette F. M., Manon S. Yeast as a tool to study Bax/mitochondrial interactions in cell death. FEMS Yeast Res. 2003;4:15–27. doi: 10.1016/S1567-1356(03)00143-0. [DOI] [PubMed] [Google Scholar]
- Qi M., Elion E. A. Formin-induced actin cables are required for polarized recruitment of the Ste5 scaffold and high level activation of MAPK Fus3. J. Cell Sci. 2005;118:2837–2848. doi: 10.1242/jcs.02418. [DOI] [PubMed] [Google Scholar]
- Reinoso-Martin C., Schuller C., Schuetzer-Muehlbauer M., Kuchler K. The yeast protein kinase C (PKC) cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot Cell. 2003;2:1200–1210. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanglard D., Ischer F., Marchetti O., Entenza J., Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Severin F. F., Hyman A. A. Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 2002;12:R233–R235. doi: 10.1016/s0960-9822(02)00776-5. [DOI] [PubMed] [Google Scholar]
- Sherman F., Hicks J B., Fink G. R. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sprague G. F., Jr., Herskowitz I. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J. Mol. Biol. 1981;153:305–321. doi: 10.1016/0022-2836(81)90280-1. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr., Thorner J. W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E. W., Pringle J. R., Broach J. R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- Steinbach W. J., Schell W. A., Blankenship J. R., Onyewu C., Heitman J., Perfect J. R. In vitro interactions between antifungals and immunosuppressants against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2004;48:1664–1669. doi: 10.1128/AAC.48.5.1664-1669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N., Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C. M., Anderson J. M. Claudins and epithelial paracellular transport. Annu. Rev. Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang H. G., Pathan N., Ethell I. M., Krajewski S., Yamaguchi Y., Shibasaki F., McKeon F., Bobo T., Franke T. F., Reed J. C. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- Wilson H. L., Wilson S. A., Surprenant A., North R. A. Epithelial membrane proteins induce membrane blebbing and interact with the P2X7 receptor C terminus. J. Biol. Chem. 2002;277:34017–34023. doi: 10.1074/jbc.M205120200. [DOI] [PubMed] [Google Scholar]
- Withee J. L., Mulholland J., Jeng R., Cyert M. S. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell. 1997;8:263–277. doi: 10.1091/mbc.8.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Jung U. S., Garrett-Engele P., Roe T., Cyert M. S., Levin D. E. Temperature-induced expression of yeast FKS2 is under the dual control of PKC and calcineurin. Mol. Cell. Biol. 1998;18:1013–1022. doi: 10.1128/mcb.18.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]