Abstract
Mesenchymal stem cells from adipose tissue can differentiate into mesodermal lineages. Differentiation potential, however, varies between clones of adipose stem cells (ASCs), raising the hypothesis that epigenetic differences account for this variability. We report here a bisulfite sequencing analysis of CpG methylation of adipogenic (leptin [LEP], peroxisome proliferator-activated receptor gamma 2 [PPARG2], fatty acid-binding protein 4 [FABP4], and lipoprotein lipase [LPL]) promoters and of nonadipogenic (myogenin [MYOG], CD31, and GAPDH) loci in freshly isolated human ASCs and in cultured ASCs, in relation to gene expression and differentiation potential. Uncultured ASCs display hypomethylated adipogenic promoters, in contrast to myogenic and endothelial loci, which are methylated. Adipogenic promoters exhibit mosaic CpG methylation, on the basis of heterogeneous methylation between cells and of variation in the extent of methylation of a given CpG between donors, and both between and within clonal cell lines. DNA methylation reflects neither transcriptional status nor potential for gene expression upon differentiation. ASC culture preserves hypomethylation of adipogenic promoters; however, between- and within-clone mosaic methylation is detected. Adipogenic differentiation also maintains the overall CpG hypomethylation of LEP, PPARG2, FABP4, and LPL despite demethylation of specific CpGs and transcriptional induction. Furthermore, enhanced methylation at adipogenic loci in primary differentiated cells unrelated to adipogenesis argues for ASC specificity of the hypomethylated state of these loci. Therefore, mosaic hypomethylation of adipogenic promoters may constitute a molecular signature of ASCs, and DNA methylation does not seem to be a determinant of differentiation potential of these cells.
INTRODUCTION
Stem cells have been identified in several adult mesenchymal tissues and are thought to be responsible for maintaining tissue homeostasis. Mesenchymal stem cells (MSCs) undergo self-renewing divisions but also give rise to more committed progenitor cells, which can differentiate into specific cell types. Bone marrow-derived MSCs can differentiate in vitro into primarily mesodermal lineages (Pittenger et al., 1999; Gronthos et al., 2003); however, a minor population seems to display greater multilineage differentiation potential (Jiang et al., 2002). Stem cells of stromal origin also can be obtained in large numbers from liposuction material (Zuk et al., 2001). These cells also display multilineage differentiation capacities (Zuk et al., 2001; Rodriguez et al., 2004; Boquest et al., 2005; Katz et al., 2005; Timper et al., 2006) and can promote neuronal (Kang et al., 2003) or osteogenic (Cowan et al., 2004) repair, restoration of hepatic function (Kim et al., 2003), and reconstitution of the immune system (Cousin et al., 2003; Fraser et al., 2006). We recently reported the identification, purification, and characterization of precursor cells with a CD34+CD105+CD45−CD31− phenotype from the stromal vascular fraction of human adipose tissue, which exhibit MSC properties upon culture (Boquest et al., 2005). CD31− adipose stem cells (ASCs) are relatively quiescent but reenter the cell cycle upon culture. They can be expanded clonally and differentiate into mesodermal lineages, including chondrogenic, adipogenic, and osteogenic cell types. Notably, clones derived from single ASCs, even when harboring the same genetic makeup, display variations in their differentiation potential (Boquest et al., 2005). This observation raises the hypothesis of an epigenetic basis for this variation. Because they can be collected in large numbers (>5 × 106 98% pure CD31− cells/100 ml of liposuction material), ASCs constitute an attractive source of multipotent cells suitable for epigenetic analyses, before and after culture.
Despite many reports on the differentiation potential of MSCs, little is known on the molecular premises of pluripotency of these cells and of ASCs in particular. Gene expression array-based attempts at defining stemness have been reported for embryonic stem cells (ESCs) (Ramalho-Santos et al., 2002), and gene expression profiles of ASCs have started to emerge (Urs et al., 2004; Boquest et al., 2005). The transcription profile of ASCs reveals expression of genes extending across all three germ layers, a feature coined as multilineage priming. Nevertheless, although such analyses may identify genes that potentially serve as pluripotency markers, there is to date no understanding of chromatin organization in ASCs, which may account for potential for gene activation or up-regulation upon differentiation.
Epigenetic modifications of DNA and histones contribute to the regulation of gene expression (Lachner and Jenuwein, 2002). DNA methylation consists in the addition of a methyl group to the 5′ position of cytosine in a CpG dinucleotide. DNA methylation is a heritable modification that favors genomic integrity and ensures proper regulation of gene expression. It largely contributes to gene silencing (Antequera, 2003) and is essential for development (Li et al., 1992), X chromosome inactivation (Panning and Jaenisch, 1998), and genomic imprinting (Li et al., 1993). Differentiation can also be associated with alterations in DNA methylation; however, only sporadic indications of DNA methylation changes have been reported upon stem or precursor cell differentiation (Brero et al., 2005; Deb-Rinker et al., 2005; Rodic et al., 2005).
Heterogeneity in the efficiency of differentiation of ASCs into mesodermal lineages in vitro raises the hypothesis of epigenetic variations at promoters required for lineage-specific differentiation. To begin addressing this issue, we examined the DNA methylation status of adipogenic and nonadipogenic genes in ASCs. This study reports a bisulfite sequencing analysis of DNA methylation in freshly isolated human ASCs and in undifferentiated and differentiated clonal populations of ASCs. Bisulfite sequencing enables identification of individual methylated cytosines in single DNA molecules (Grunau et al., 2001; Warnecke et al., 2002). Because ASCs are natural adipocyte precursors (Otto and Lane, 2005), we focused on four adipogenic gene promoters. Our results indicate that mosaic DNA hypomethylation established in adipogenic promoters in ASCs in vivo remains stable upon culture and in vitro differentiation. Nonadipogenic loci, however, are highly methylated. Furthermore, DNA methylation does not seem to be the sole determinant of differentiation potential of ASCs.
MATERIALS AND METHODS
ASC Isolation and Clonal Culture
Stromal vascular cells with a CD34+CD105+CD45−CD31− phenotype (ASCs) were isolated from human adipose tissue (Boquest et al., 2005). In short, tissue was obtained by liposuction from the hip and thigh regions of healthy women. After washing in Hank’s balanced salt solution (HBSS), the tissue was digested for 2 h at 37°C in HBSS with collagenase and DNase I. Adipocytes were separated from stromal vascular cells after sedimentation at 400 × g for 10 min and removed by aspiration. Erythrocytes were removed by resuspending stromal vascular cell pellets in lysis buffer (2.06 mg/ml Tris base, pH 7.2, and 7.49 mg/ml NH4Cl) for 10 min. After centrifugation, pellets were resuspended in HBSS containing 2% fetal bovine serum (FBS) (Sigma-Aldrich. St. Louis, MO) and passed through a 100-μm sieve and a 40-μm sieve. CD45+ cells were removed with paramagnetic beads conjugated to mouse anti-human CD45 monoclonal antibodies (Miltenyi Biotech, Bergish Gladbach, Germany) using a superMACS magnet (Miltenyi Biotech). Remaining CD45− cells were incubated with fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD31 antibodies (Serotec, Oxford, United Kingdom) at a concentration of 10 μl of antibody per 106 cells for 15 min at 4°C. Cells were washed and incubated with anti-FITC microbeads (Miltenyi Biotech) for 15 min at 4°C. CD31− and CD31+ cells were separated using an LS column (Miltenyi Biotech). CD31− cells were reexposed to a new LS column to eliminate any leftover contaminating CD31+ cells. Flow cytometry analysis of each cell subset from each donor indicated that purity was >98% (our unpublished data) (Boquest et al., 2005). Aliquots of each cell subset were immediately snap-frozen in liquid nitrogen for DNA and RNA isolations, or they were cultured.
CD31− clonal cell lines were generated by culturing single CD31− cells in each well of 48-well plates in DMEM/F-12 medium containing 50% FBS and antibiotics. After ∼16 h, the medium was replaced by DMEM/F-12 with 20% FBS. After ∼1 wk, colonies containing >10 cells were passaged by trypsinization and expanded. Only clonal lines that could be easily expanded were used in this study. Clones A1 and A2, and clones B1, B2, and B3 examined in this study were from two different female donors (age 27 and 39, respectively).
Adipogenic Differentiation
Clonal ASC lines generated from individual CD31− cells at passage 4 were cultured to confluence before differentiation. For adipogenic differentiation (Zuk et al., 2001), cells cultured in DMEM/F-12 with 10% FBS were stimulated for 3 wk with 0.5 mM 1-methyl-3 isobutylxanthine, 1 μM dexamethasone, 10 μg/ml insulin (Novo Nordisk, Copenhagen, Denmark), and 200 μM indomethacin (Dumex-Alpharma, Copenhagen, Denmark). To visualize lipid droplets, formalin-fixed cells were washed in 50% isopropanol and stained with Oil Red-O.
Gene Loci and Regions Analyzed by Bisulfite Sequencing
Supplemental Figure S1 illustrates the promoter regions of the genes analyzed by bisulfite sequencing in this study. We examined four adipogenic genes, including leptin (LEP) (Mason et al., 1998; Reseland et al., 2001), peroxisome proliferator-activated receptor gamma 2 (PPARG2) (Fajas et al., 1997), fatty acid-binding protein 4 (FABP4) (Ross et al., 1990; Graves et al., 1992), and lipoprotein lipase (LPL) (Bey et al., 1998; Merkel et al., 2002). We also examined genes unrelated to adipogenesis, such as myogenin (MYOG), a basic helix-loop-helix transcription factor required for myocyte differentiation (Massari and Murre, 2000); the endothelial marker gene CD31/PCAM-1 (Cao et al., 2002; Chi et al., 2003); and the constitutively expressed housekeeping gene GAPDH. The LEP promoter region analyzed was from nucleotides 2719–2937 (GenBank accession no. U43589) and spanned 27 potentially methylated cytosines in CpG dinucleotides starting 42 base pairs upstream of the ATG translational start site. The LEP proximal promoter activity is known to be regulated by DNA methylation (Melzner et al., 2002). The PPARG2 promoter region (Fajas et al., 1997) spanned nucleotides 108–587 (GenBank accession no. AB005520) and included 6 CpGs starting 264 base pairs upstream of the ATG. The FABP4 (GenBank accession no. NM_001442) promoter region examined was identified using ENSEMBL and encompassed four CpGs starting 130 base pairs upstream of the ATG. The LPL promoter region spanned bases 1321–1777 (GenBank accession no. X68111) and included 11 CpGs starting 134 base pairs upstream of the ATG. The MYOG region analyzed spanned nucleotides 1268–1484 (GenBank accession no. X62155) and included 16 CpGs starting 87 base pairs downstream of the ATG. The CD31 promoter region examined included nucleotides 1095–1480 (GenBank accession no. X96848) and included 18 CpGs ranging from nucleotide −352 to +34 relative to the ATG. The GAPDH promoter region spanned bases 1121–1337 (GenBank accession no. J04038) and encompassed 28 CpGs 116 base pairs upstream of the ATG.
Bisulfite Sequencing
DNA was purified either using the GenElute Mammalian Genomic DNA Miniprep kit (Sigma-Aldrich), or for most samples, by phenol-chloroform-isoamyl alcohol extraction. In the latter case, cells were first lysed for 10 min in lysis buffer (10 mM Tris-HCl, pH 8, 100 mM EDTA, and 0.5% SDS) and digested with 0.1 mg/ml proteinase K overnight. Bisulfite conversion (Warnecke et al., 2002) was performed using the MethylEasy DNA bisulfite modification kit (Human Genetic Signatures, Sydney, Australia). Converted DNA was used fresh or stored at −20°C. Converted DNA was amplified by PCR using primer sets purchased from Human Genetic Signatures for the LEP, MYOG, CD31 and GAPDH genes. These primers sets are commercially available (www.geneticsignatures.com). We also designed primers using the Methprimer software (www.urogene.org/methprimer/index1.html) for the PPARG2, FABP4, and LPL genes (Table 1). For PPARG2, FABP4, and LPL, PCR conditions were 95°C for 7 min and 40 cycles of 95°C 1 min, 54°C 2 min and 72°C 2 min, followed by 10 min at 72°C. For LEP, MYOG, CD31, and GAPDH, nested PCRs were performed, each as follows: 95°C for 3 min and 30 cycles of 95°C for 1 min, 50°C for 2 min, and 72°C for 2 min, followed by 10 min at 72°C. PCR products were directly sequenced or cloned into bacteria using the TOPO TA cloning kit (Invitrogen, Oslo, Norway). Clones were sequenced using commercial services from MWG Biotech (Ebersberg, Germany).
Table 1.
Bisulfite sequencing primers used in this study
| Gene name | Forward primer (F)Reverse primer (R) | Product size (bp) |
|---|---|---|
| FABP4 | F: GGTAATTTTTGAGATAGGAGTGTTT | 413 |
| R: CCAATTAAAAATAAAATCCAATCATTT | ||
| LPL | F: GGGAGGATTGTAAGTGATAAATAGG | 457 |
| R: CAACTAAAAATAAACAACTTTCCCTT | ||
| PPARG2 | F: GTTGAAGTTTTTAAGAAAGTAAATT | 480 |
| R: AAAAAAAATATTACCACACTATCTC | ||
| CD31 | Seminested primer seta | 386 |
| GAPDH | Seminested primer seta | 217 |
| LEP | Seminested primer seta | 218 |
| MYOG | Seminested primer seta | 217 |
a Purchased from Human Genetic Signatures.
Real-Time Reverse Transcription (RT)-PCR
RT-PCR was carried from 500 ng of total RNA using the Iscript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative (Q)RT-PCR reactions were performed in triplicates on a MyiQ real-time PCR Detection System using IQ SYBR Green (Bio-Rad). Most samples were analyzed in duplicates from two separate cDNA preparations. Primers used are listed in Table 2. SYBR Green PCR conditions were 95°C for 4.5 min and 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, using GAPDH as a normalization control. mRNA levels were calculated as described previously (Pfaffl, 2001).
Table 2.
Real-time RT-PCR primers used in this study
| Gene name | Forward primer (F) 5′→3′Reverse primer (R) 5′→3′ | Product size (bp) |
|---|---|---|
| CD31 | F: AGCAGCATCGTGGTCAACATA | 105 |
| R: GATGGAGCAGGACAGGTTCAG | ||
| FABP4 | F: TCAGTGTGAATGGGGATGTGAT | 310 |
| R: TTCAATGCGAACTTCAGTCCAG | ||
| GAPDH | F: TTGCCATGGGTGGAATCATA | 148 |
| R: TCGGAGTCAACGGATTTGGT | ||
| LEP | F: TTTCACACACGCAGTCAGTCT | 61 |
| R: CCAGGAATGAAGTCCAAACC | ||
| LPL | F: CCTGAAGTTTCCACAAATAAGACC | 321 |
| R: ATGCCGTTCTTTGTTCTGTAGAT | ||
| MYOG | F: ACCGACTTCCTCTTACACACCTTAC | 224 |
| R: TATGAGACATCCCCCTACTTCTACC | ||
| PPARG2 | F: CTTCCATTACGGAGAGATCCAC | 125 |
| R: AAGCGATTCCTTCACTGATACAC |
RESULTS
Adipogenic Gene Promoters Are Hypomethylated in Clonally Cultured ASCs and Exhibit Between-Clone Heterogeneity in 5′-3′ CpG Methylation Pattern
We first examined the DNA methylation status of four adipogenic genes in polyclonal cultures of ASCs. CpG methylation analysis of LEP, PPARG2, FABP4, and LPL promoter regions (Figure 1A) across several populations of cultured undifferentiated ASCs revealed hypomethylated promoters (Figure 1, B–E). The LEP promoter region contained CpGs, which were at most 32% methylated (nos. 1, 19, and 21), whereas the rest of the CpGs displayed 0–25% methylation (Figure 1B). PPARG2 was also hypomethylated (8–23% methylated CpGs; Figure 1C), as were the FABP4 and LPL promoters (Figure 1, D and E). Despite the overall hypomethylation, however, distinct CpGs were clearly more methylated than others in the LEP, FABP4, and LPL promoters.
Figure 1.
CpG-specific methylation level at the LEP, PPARG2, FABP4, and LPL promoters in cultured ASCs. (A) Distribution of CpGs in each promoter region examined. Numbers indicate nucleotide number upstream of the ATG translational start site (see Supplemental Figure S1 for sequences). Tick marks indicate the position of each CpG. (B–E) Percentage of 5′–3′ CpG methylation determined by bisulfite sequencing at indicated promoters in polyclonal populations of cultured undifferentiated ASCs. Number 1 refers to the 5′-most CpG.
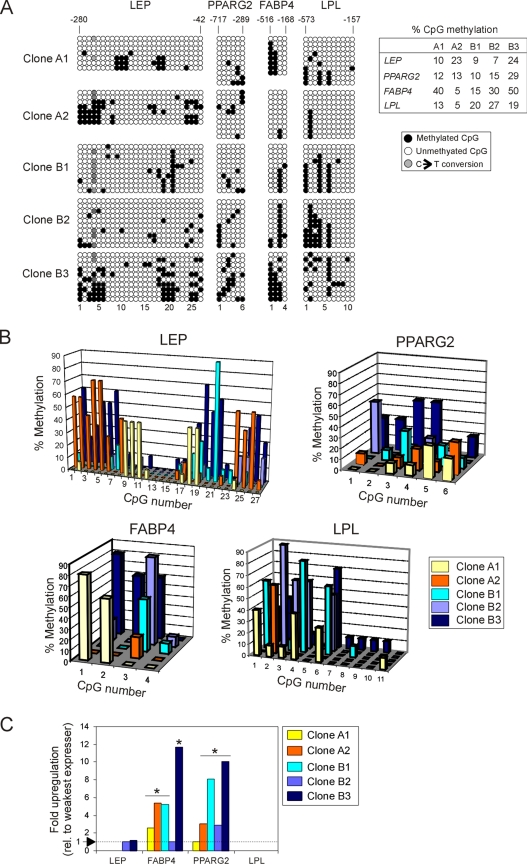
The variation in differentiation potential between clonal cultures of ASCs observed previously (Boquest et al., 2005) prompted the analysis of DNA methylation in five clonal lines of ASCs established from single freshly isolated cells. Clones A1 and A2 were from one donor and clones B1–B3 were from another donor. All clones were analyzed at passage 4, i.e., after ∼20 population doublings from single cells. Figure 2A shows the methylation status of each CpG for each gene, in 7–10 bacterial clones of PCR products. For each gene and in each cell clone, the overall percentage of methylation was under 50% (Figure 2A, right), and no gene was consistently more methylated than any other (Figure 2A, right). Thus, DNA hypomethylation of adipogenic promoters is a common feature of undifferentiated ASC clones.
Figure 2.
Adipogenic loci are hypomethylated in cultured ASC clones, irrespective of gene expression. (A) Bisulfite analysis of LEP, PPARG2, FABP4, and LPL in five ASC clones from two donors (A and B clones, respectively). Table shows the percentage of global CpG methylation (● in A) at each locus for each clone. (B) Proportion of individual methylated CpGs at the LEP, PPARG2, FABP4, and LPL promoter in each clone. (C) QRT-PCR analysis of expression of LEP, PPARG2, FABP4, and LPL in undifferentiated ASC clones, relative to the lowest expressing clone (level 1) for a given gene. Asterisk (*) indicates a statistical difference in expression at the p < 0.01 level (t test) relative to the weakest expressing clone (level 1).
Although no overt differences were detected in the overall proportion of methylation between cell clones, several observations were made from the analysis of each clone. First, despite some heterogeneity (see below), the clones displayed overlapping areas of preferred methylation, as judged by graphic representation of percentages of methylation of each CpG for each clone (Figure 2B). Second, there was no consistency in the methylation status of a given CpG within clones derived from a particular donor. Indeed, we detected as much variation between clones within one donor (i.e., within A or B clones) as between clones derived from different donors. Third, except for clone A1, which surprisingly displayed two main methylation profiles (Figure 2A), there was relative homogeneity in the methylation pattern within a clone. Fourth, there were nevertheless differences in methylation profiles between clones (Figure 2B). This was particularly evident in the LEP promoter: a cluster of six CpGs (nos. 1–6; Supplemental Figure S1A) were 30–70% methylated in clones A2 and B3, but they were essentially unmethylated in the other clones (Figure 2B). Similarly, methylation of two other areas (CpGs nos. 18–21 and 24–27) differed highly between clones. Of note, CpG 21 (nucleotide position −107 in the LEP promoter) showed 90 and 60% methylation in clones B1 and B3, respectively, whereas it was completely unmethylated in the other clones. Another example is CpG 26 (position −48), which was 50% methylated in clones A2 and B3 and unmethylated in the other clones. Clone A1 also displayed nearly 40% methylation in two distinct areas (CpG nos. 9–11 and 18–19), which were unmethylated in the other clones. For PPARG2, similar differences were noted, although to a lesser extent and mostly caused by the overall higher methylation of clone B3 (Figure 2B). FABP4 also displayed methylation variation between clones at CpGs nos. 1–3, CpG no. 4 being largely unmethylated. Clones A1 and B3 were highly methylated at CpGs nos. 1 and 2, in contrast to the others clones, which showed no or little methylation (Figure 2B). Last, the LPL promoter also displayed between-clone methylation differences within methylated areas (CpG nos. 1–4 and 6), whereas CpG nos. 5 and 7–11 were essentially unmethylated in all clones (Figure 2B).
We concluded from these observations that the LEP, PPARG2, FABP4, and LPL promoters are largely hypomethylated in undifferentiated cultured ASCs. Nevertheless, we detected in each locus areas where methylation preferentially occurs. In these areas, however, the extent of methylation of specific CpGs can vary between cell clones, even when derived from a single donor. Furthermore, although global methylation profiles overlap within a clone, CpG methylation is mosaic.
CpG Methylation Pattern Is Unrelated to Gene Expression in Undifferentiated Cultured ASCs
To determine whether there was any correlation between CpG methylation and gene expression in undifferentiated cultured ASCs, expression of LEP, PPARG2, FABP4, and LPL was analyzed by real-time RT-PCR. Consistent with previous cDNA microrray analyses (Boquest et al., 2005), some of the genes were transcribed in undifferentiated cells, albeit at variable levels between clones (Figure 2C). Specifically, LEP expression was only detected in clones B2 and B3 and at similar levels (p > 0.1; t test). FABP4 was expressed in all cell clones, with clone B2 being by far the lowest expresser (p < 0.01) and clone B3 the highest expresser (p < 0.001 compared with all other clones). Similarly, PPARG2 was expressed in all clones at variable levels with clone A1 being the weakest expresser (p < 0.01) and clone B3 the highest expresser (p < 0.01). LPL was not expressed in any of the clones (Figure 2C).
Most significantly, the relatively low CpG methylation level at each locus and in each clone was irrespective of gene expression level (Figure 2, A–C). For example, LEP was notably differentially methylated in clones A1, A2, and B1 (Figure 2A), but not transcribed in any of these clones (Figure 2C). Moreover, clones B2 and B3 expressed LEP at similar levels despite a different methylation pattern and level (Figure 2A). Similarly, FABP4 and PPARG2 expression levels (Figure 2C) were unrelated to DNA methylation profile at these promoters (Figure 2A). Furthermore, the lack of LPL expression in all clones could not strongly be correlated to CpG methylation, because, as for LEP and FABP4, there was marked mosaicism in the methylation state of specific CpGs at this locus (Figure 2A). Therefore, for each region examined in these adipogenic genes, we could not attribute a specific CpG methylation status to respective mRNA levels in undifferentiated ASCs.
DNA Methylation of Adipogenic Genes upon Adipogenic Differentiation In Vitro
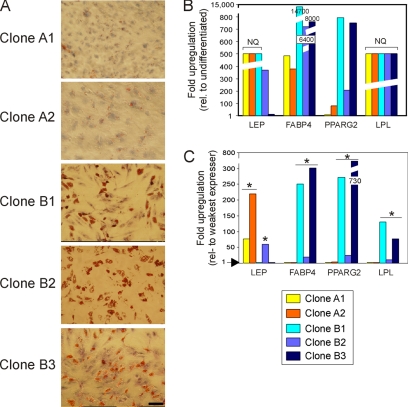
The localization of ASCs in adipose tissue argues that adipogenesis is a natural differentiation pathway for these cells. To determine whether methylation of LEP, PPARG2, FABP4, and LPL was altered upon adipogenic differentiation, the five ASC clones were stimulated for 3 wk toward the adipogenic pathway. Each clone responded to stimulation, with various efficiencies, with clones B1–B3 being more efficient than clones A1 and A2 on the basis of Oil Red-O staining (Figure 3A). QRT-PCR analysis of differentiated cells with respect to undifferentiated counterparts established the induction of expression of LEP and LPL, and strong up-regulation of PPARG2 and FABP4, confirming adipogenic differentiation (Figure 3B). Note that in Figure 3B, NQ refers to nonquantified LEP and LPL mRNA levels due to the lack of expression of these genes in undifferentiated cells (Figure 2C); these levels were arbitrarily set on the graph.
Figure 3.
Adipogenic differentiation of ASC clones. (A) Morphological evidence of differentiation after 3 wk of adipogenic stimulation (Oil Red-O staining). Bar, 50 μm. (B) QRT-PCR analysis of expression of indicated genes in each ASC clone after 3 wk of differentiation, relative to expression level in the same but undifferentiated clone. (C) Gene expression analysis as in B, but expressed relative to the lowest expressing clone for a given gene. All samples were analyzed in triplicates. NQ indicates gene expression, but level was not quantified due the absence of expression in undifferentiated ASCs (value was arbitrarily set on graph). *p < 0.005 (t test) for all transcripts with an expression level >10-fold relative to the weakest expressing clone (level 1), and p < 0.001 for genes indicated in the text.
Expression of each gene relative to the lowest expressing clone indicated that mRNA levels varied between clones after differentiation, likely as a result of variations in differentiation efficiency (Figure 3C). The t test analysis of expression levels relative to the weakest expressing clones within each gene revealed highly significant differences in expression (p < 0.005 to < 0.0001 for transcripts up-regulated >10-fold compared with the lowest expressing clone; Figure 3C). Furthermore, we found a correlation between the low expression levels of FABP4, PPARG2, and LPL and weak Oil-Red-O staining in clones A1 and A2 (Figure 3, A and C). In contrast, LEP was most strongly expressed in clones A1 and A2 (p < 0.001 compared with clone B1), and LEP expression was inversely proportional to that of FABP4, PPARG2, and LPL (Figure 3C). This suggests that peak LEP expression is temporally distinct from that of FABP4, PPARG2, and LPL. Thus, although all clones were induced to differentiate, we detected considerable variation in the relative expression levels of individual genes. Additionally, strong up-regulation of FABP4, PPARG2, and LPL expression in clones B1 and B3 (p < 0.001) seemed to correlate with strong phenotypic changes elicited by adipogenic stimulation.
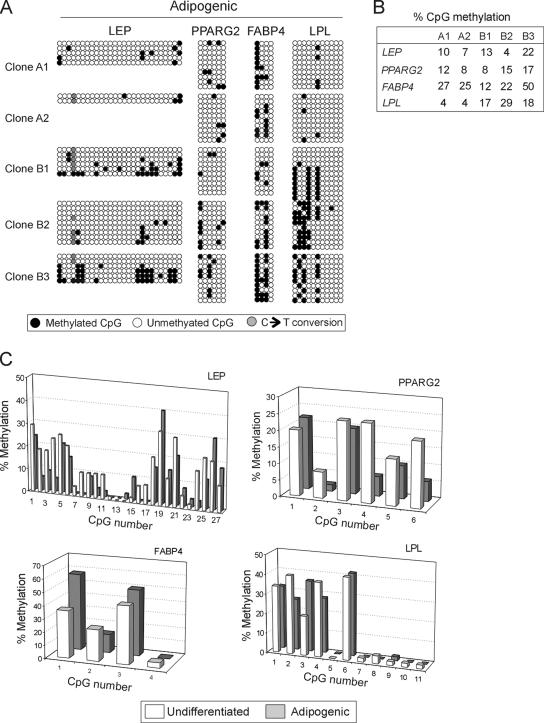
With a few exceptions (see below), the global DNA methylation pattern of all genes examined remained unexpectedly stable upon adipogenic differentiation. Methylation of each CpG is shown in Figure 4A, and transitions in CpG methylation after adipogenic induction across all clones are illustrated in Figure 4C. Transitions for each individual clones are shown in Supplemental Figure S2. Global methylation over the regions examined in the LEP, PPARG2, FABP4, and LPL promoters remained unchanged after differentiation (p > 0.1, t tests; see Supplemental Table S1; Figure 4, A and B; compare with Figure 2A). Thus, upon adipogenic differentiation, each clone globally maintains its methylation profile. An average of the percentages of methylation at individual CpGs across all clones supported this observation; however, t test analysis of (de)methylation of individual CpGs revealed some noticeable changes (Figure 4C). Specifically, in the LEP promoter, CpG nos. 2, 3, 4, 21, 24, and 25 displayed significant (albeit not complete) demethylation upon adipogenic differentiation (p < 0.001), whereas all other cytosines remained unaffected (p > 0.05). In the PPARG2 promoter, CpG nos. 4 and 6 underwent demethylation (p < 0.001 and < 0.01, respectively), whereas CpGs no. 1 in the FABP4 promoter and CpG no. 3 in the LPL promoter underwent methylation (p < 0.01; Figure 4C).
Figure 4.
Bisulfite sequencing analysis of DNA methylation of LEP, PPARG2, FABP4, and LPL promoters in ASC clones after adipogenic differentiation. (A) Bisulfite analysis. (B) Percentage of global CpG methylation (◇ in A) at each promoter for each clone. (C) Average percentage of individual CpG methylation, across all clones, in undifferentiated ASCs and after adipogenic differentiation. Statistical analysis (paired t tests) of differences in percentage methylation is provided in the text.
A few alterations in CpG methylation were also observed, which were specific for individual clones. In the LEP promoter, the most noticeable change was complete CpG nos. 9–11 and 18–19 demethylation in clone A1 (Figure 4A and Supplemental Figure S2A), which was accompanied by induction of LEP expression. Cytosines 1–5 and 24–25 also apparently underwent demethylation in clone A2; however, the data are based on only two sequenced PCR product clones after differentiation (Figure 4A and Supplemental Figure S2A) due to extreme cloning difficulty. Furthermore, CpG 21 was clearly demethylated upon adipogenic differentiation in clone B1 (Figure 4A and Supplemental Figure S2A), but this was not indicative of strong expression, because clone B1 was the weakest LEP expresser (Figure 3B). In all other clones, DNA methylation profiles were maintained (Figure 4C and Supplemental Figure S2A) regardless of LEP expression levels. In the PPARG2 promoter, DNA methylation patterns remained unaltered such as the same between-clone variation was observed as in undifferentiated cells (Figure 4, A and C and Supplemental Figure S2B). FAPB4 promoter methylation also remained stable, with the exception of CpG No. 2 in clones A1 and B3, which underwent demethylation (∼60–20–30% methylation; Figure 4A and Supplemental Figure S2C). This demethylation, however, did not relate to particularly strong expression of FABP4, because whereas FABP4 was strongly induced in clone B3, it was barely up-regulated in clone A1 (Figure 3C). The LPL promoter retained its undifferentiated methylation pattern in clones B1, B2, and B3 despite induction of expression; however, specific CpGs displayed alterations in other clones (Figure 4, A and C, and Supplemental Figure S2D). In clone A1 (40% methylated CpG nos. 1 and 4) underwent complete demethylation, and in clone A2, the 60% methylated CpG no. 2 was completely demethylated. Either of these changes correlated with induction of LPL transcription (Figure 3B) but not with strong expression compared with other clones (Figure 3C).
These results indicate that globally, average methylation of LEP, PPARG2, FABP4, and LPL promoters across ASC clones remain stable upon adipogenic differentiation. Nevertheless, methylation and demethylation events are identified at specific CpGs in all promoters, but there is no consistent response to differentiation induction between clones.
To assess the physiological relevance of methylation changes, or lack thereof, detected in the LEP promoter upon in vitro ASC differentiation, we examined LEP promoter methylation in fully differentiated cultured Simpson–Golabi–Behmel syndrome (SGBS) human adipocytes (Wabitsch et al., 2001). Figure 5indicates that the LEP promoter in mature adipocytes was also hypomethylated (11% methylation; Figure 5A), and the 5′-3′ CpG methylation profile was nearly identical to that of adipogenic differentiated ASCs (Figure 5B; p = 0.79; t test). LEP was expressed in SGBS cells, as expected from this cell type (Figure 5C). Thus, the methylation pattern of adipogenic differentiated ASCs reflects that of other differentiated human adipocytes.
Figure 5.
DNA methylation analysis of the LEP promoter in differentiated human SGBS adipocytes. (A) Bisulfite sequencing analysis. (B) Percentage of individual CpG methylation in adipogenic-differentiated ASCs (pool of all clones shown in A) and in SGBS adipocytes. (C) Endpoint RT-PCR analysis of LEP and GAPDH expression in differentiated SGBS adipocytes.
Adipogenic Loci Are Also Hypomethylated in Freshly Isolated, Uncultured ASCs
Our results indicate so far that the adipogenic genes LEP, PPARG2, FABP4, and LPL are largely hypomethylated in cultured ASCs. ASCs in passage 4 of clonal culture have undergone ∼20 population doublings. This is due to the time required for single cells to initiate replication and for obtaining cell numbers compatible with these analyses. Thus, we hypothesized that DNA hypomethylation of adipogenic loci in ASCs might be a result of culture, because global DNA demethylation is known to occur upon long-term culture of other cell types (Catania and Fairweather, 1991; Hornsby et al., 1992; Zheng et al., 2006).
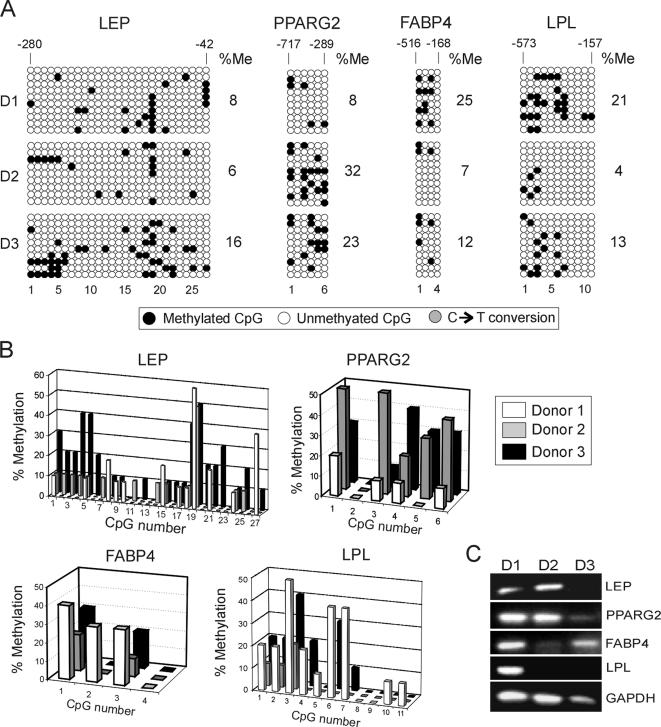
To test this hypothesis, we examined CpG methylation of LEP, PPARG2, FABP4 and LPL in ASCs immediately after isolation from three healthy women of comparable age. The data are shown in Figure 6, A and B. First, all loci were globally hypomethylated in ASCs purified from each donor. The global percentage of methylation ranged from 4% (LPL; donor 2) to 32% (PPARG2; donor 2) (Figure 6A) and was consistent with data obtained from cultured cells (Figure 2A). Thus, freshly isolated ASCs display hypomethylated adipogenic promoters, and little change occurs globally upon culture, when the methylation percentage of all CpGs is taken into account (Figure 7; p > 0.16; and Supplemental Table S1).
Figure 6.
DNA methylation analysis of LEP, PPARG2, FABP4, and LPL in freshly isolated, uncultured ASCs. (A) Bisulfite analysis of CpG methylation in ASCs from three donors (D1–D3). Percentage of overall CpG methylation (%Me; ●) is shown. (B) Proportions of individual methylated CpGs at each promoter and or each donor. CpG numbers are indicated, no. 1 being the 5′ most CpG. Note that analysis of 10 bacterial clones for each donor was barely sufficient for statistical comparisons. (C) Endpoint RT-PCR analysis of expression of indicated genes in ASCs purified from donors D1, D2, and D3.
Figure 7.
Comparison of CpG methylation profiles in uncultured versus cultured ASCs. Average percentages of methylation of individual CpGs in the LEP, PPARG2, FABP4, and LPL promoters in freshly isolated ASCs from all three donors (uncultured) and across all five undifferentiated ASC clones (cultured) are shown.
Second, however, some alterations were noted at specific CpGs upon culture. Chi-square analysis of CpG methylation percentages indicated enhanced methylation upon culture of cytosines C21 or LEP (p < 0.001), C3 of FAPB4 (p < 0.0001), and C1, C4, and C6 of LPL (p < 0.001), whereas cytosines C19 of LEP (p < 0.001) and C1 of PPARG2 (p = 0.006) underwent hypermethylation upon culture. All other CpGs remained altered (p > 0.05). Third, there was minor heterogeneity in the 5′-3′ CpG methylation profile between donors and overall methylation profiles largely overlapped (Figure 6B). Variation was gene-specific, because no one donor displayed consistent methylation across all loci relative to any other donor. More specifically, a between-donor comparison of methylation frequencies for individual CpGs revealed no significant variation in CpG methylation at each locus examined (p = 0.06–0.968; Supplemental Table S1), with three exceptions: for LEP (donor 2 versus donor 3; p = 0.006), PPARG2 (donor 1 versus donor 2; p = 0.02), and LPL (donor 1 versus donor 2; p = 0.007). Fourth, we also detected some mosaicism with donors, although again, the methylated areas were conserved. Fifth, RT-PCR analysis of uncultured ASCs from each donor indicated that all genes were expressed; however, not all three donors expressed all genes (Figure 6C). This is consistent with previous cDNA microarray analyses of freshly isolated ASCs (Boquest et al., 2005). Last, there was no correlation between gene expression in one given donor and global or pattern-specific methylation.
We concluded that adipogenic loci are hypomethylated in freshly isolated ASCs and that methylation profiles are rather homogenous between donors despite some mosaicism. Areas of higher methylation within each locus are consistent with those detected in cultured undifferentiated cells, despite a few specific differences. This observation was consistent regardless of whether all donors and all clonal cultures were pooled to provide average methylation levels at each CpG (Figure 7) or whether individual donors and clones were examined (compare Figure 2A with 6A). DNA hypomethylation of adipogenic loci in ASCs, therefore, is a characteristic of these stem cells and does not arise as a result of culture.
Lineage-specific, Nonadipogenic Loci Are Methylated in ASCs
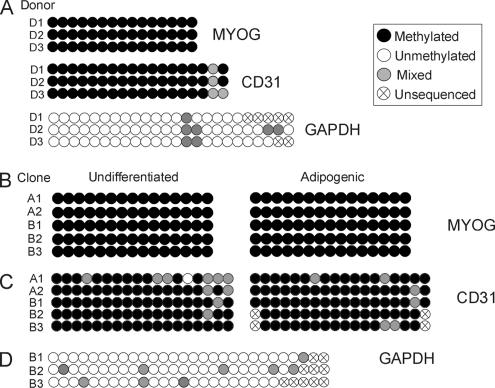
The overall DNA hypomethylation reported for adipogenic genes in ASCs was not generalized to all multilineage priming genes. In contrast to LEP, PPARG2, FABP4, or LPL, the myogenic locus MYOG (Supplemental Figure S1E) revealed methylation at all CpGs examined in ASCs from three donors (Figure 8A). MYOG methylation was maintained upon clonal culture as well as upon adipogenic differentiation (Figure 8B). MYOG methylation was evident even without cloning PCR products generated from bisulfite-converted DNA (Figure 8, A and B, and Supplemental Figure S3A). Of note, however, MYOG was methylated despite its expression in undifferentiated ASCs (Boquest et al., 2005). Therefore, methylation of MYOG in ASCs does not correlate with its expression.
Figure 8.
DNA methylation analysis of MYOG, CD31, and GAPDH in uncultured and cultured ASCs. (A) Analysis of ASCs purified from donors D1–D3 resulting from direct sequencing of PCR products after bisulfite conversion. Representative sequences are shown in Supplemental Figure S3. (B and C) Analysis of MYOG (B) and CD31 (C) methylation in undifferentiated and adipogenic-differentiated ASC clones. (D) Analysis of GAPDH methylation in undifferentiated ASC clones B1–B3. The “mixed” methylation pattern is due to the analysis of mixed cell populations because PCR products resulting from bisulfite conversion were not cloned.
Similar observations were made for the endothelial cell-specific CD31 gene promoter. CD31 was heavily methylated both in freshly isolated and in cultured ASCs (Figure 8, A and C, and Supplemental Figure S3B). This was in agreement with selection of ASCs against the CD31 surface antigen upon isolation (Boquest et al., 2005). In contrast, the CD31 promoter region examined was unmethylated in CD31− endothelial precursor cells (our unpublished data; Boquest, Noer, Sørensen, Vekterud, and Collas, manuscript in preparation). Nevertheless, methylation of MYOG and CD31 in undifferentiated ASCs did not preclude its expression in undifferentiated ASCs and in vitro differentiation toward myogenic and endothelial pathways (Boquest, Noer, Sørensen, Vekterud, and Collas, unpublished data). Last, as expected from its ubiquitous expression, the GAPDH promoter was largely unmethylated in ASCs purified from each donor (Figure 8A) and in undifferentiated cultured cells (Figure 8D and Supplemental Figure S3C).
These results suggest that DNA hypomethylation in both freshly isolated and cultured ASCs is restricted to adipogenic and housekeeping gene promoters. Genes apparently not involved in adipogenesis, such as MYOG or CD31, are highly methylated. This suggests tissue type specificity in the extent of methylation of multilineage priming genes in ASCs within their tissue of residence as well as upon culture in undifferentiated state. For any of those genes, however, methylation profile does not correlate with expression.
The Leptin Promoter Is Methylated in Nonadipose Differentiated Somatic Cells
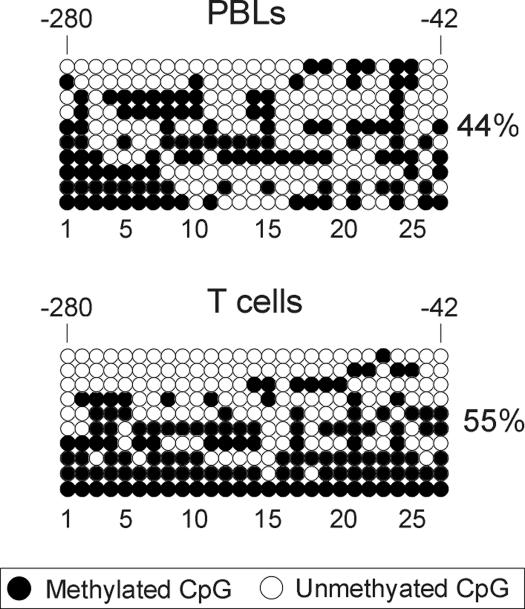
To determine whether DNA hypomethylation of adipogenic gene loci was restricted to stem cells or was a constitutive property, we examined the DNA methylation status of the LEP promoter in primary human cells, either isolated from donors or cultured. In purified human (uncultured) peripheral blood lymphocytes (PBLs) and T-cells, the LEP promoter was hypermethylated, albeit not totally methylated, compared with ASCs (Figure 9A; p < 0.0001). We concluded from these observations that DNA hypomethylation of the LEP promoter is a property of ASCs, regardless of their differentiation state, and of differentiated adipocytes.
Figure 9.
Bisulfite sequencing analysis of CpG methylation of LEP in primary human nonadipocytic cells. Analysis of uncultured PBLs and T-cells. Percentage of global methylation (●) is shown. CpG numbers are shown, with no. 1 being the 5′-most CpG.
DISCUSSION
This study presents the first assessment, to our knowledge, of promoter DNA methylation at the nucleotide level in relation to gene expression in freshly isolated, cultured, and differentiated human MSCs. Several features seem to epigenetically characterize stem cells from lipoaspirates at the DNA methylation level. 1) Freshly isolated cells display hypomethylated adipogenic promoters, in contrast to myogenic or endothelial genes. 2) ASCs exhibit a mosaic CpG methylation profile, on the basis of heterogeneous methylation patterns between individual cells, and of variations in the percentage of methylation of a given CpG between donors and between cell clones. 3) DNA methylation profiles reflect neither the transcriptional status in undifferentiated cells nor the potential for gene expression upon in vitro differentiation. 4) Clonal culture of ASCs established from single isolated cells preserves the overall hypomethylation of adipogenic promoters; nevertheless, between-clone mosaicism at specific CpGs occurs. 5) Within-clone mosaicism is also detectable despite the overall overlap of methylated areas in a given locus. 6) In vitro differentiation toward the adipogenic pathway maintains global methylation patterns at the loci examined despite the induction or up-regulation of gene expression. Nevertheless, specific CpGs undergo demethylation, particularly in the LEP promoter. 7) Adipogenic genes are more methylated in primary differentiated cells unrelated to adipogenesis, arguing for ASC specificity of the hypomethylated state of these loci. Mosaic hypomethylation of adipogenic promoters in ASCs may therefore constitute a molecular signature of ASCs.
Hypomethylation of Adipogenic Loci in Undifferentiated ASCs
Bisulfite sequencing analysis of ASCs reveals the overall hypomethylation of undifferentiated, freshly isolated, or cultured stem cells. The average percentage of CpG methylation in the promoter regions examined in the LEP, PPARG2, FABP4, and LPL promoters in cells from three donors ranged from 10 to 15%. These values agree with the hypomethylation reported for human colon (endodermal) crypt stem cells (Yatabe et al., 2001; Kim et al., 2005). Nevertheless, the LEP promoter of preadipocytes cultured from adipose tissue were found in a separate study (Melzner et al., 2002) to be highly methylated (73%), a surprising finding for a CpG island. However, in contrast to ASCs characterized in this study, these preadipocytes were found not to express LEP (Melzner et al., 2002), possibly reflecting a less committed cell type or a result of culture. CpG methylation in ASCs was heterogeneous across adipogenic promoters examined. Both in uncultured and cultured ASCs, we identified areas of preferred methylation, but these areas did not exceed 5–40% methylation. Analysis of cells from individual donors and of clonal ASC lines, however, reveals a broader range of methylation frequencies at specific CpGs. Nevertheless, although adipogenic promoters are hypomethylated, DNA hypomethylation is not a ubiquitous feature of ASCs, because MYOG and CD31, myogenic and endothelial markers, respectively, are highly methylated.
Hypomethylation of adipogenic loci in undifferentiated cells may reflect a commitment of these cells to a specific lineage. In vivo, the very location of ASCs in the stromal vascular fraction of adipose tissue predicts a preferred commitment toward adipogenic differentiation. To support this view, we found, in agreement data of Melzner et al. (2002) in differentiated adipocytes, consistent unmethylation of SP1-binding sites (covering CpGs nos. 11–12 and 15–16 in our study) and of a C/EBP-binding site (covering CpG no. 21; clones A1, A2, and B2) in the LEP promoter in undifferentiated ASCs. Similarly, the PPARγ-response element between CpGs 7 and 8, and the sterol response element (between CpGs 9 and 10) in the LPL promoter, are also consistently unmethylated (Merkel et al., 2002). Unmethylation of these sites likely ensures accessibility to these transcription factors. Lineage commitment is also supported by the overall hypomethylated state of DNA in ESCs in early passage cultures, when they retain pluripotency (Hoffman and Carpenter, 2005; Maitra et al., 2005; Zvetkova et al., 2005). Adipogenic lineage-specific promoter hypomethylation may, therefore, constitute a molecular signature of ASCs. An implication, then, is that although similar to ASCs at the transcriptome and immune phenotype levels (Kern et al., 2006), MSCs from nonadipogenic tissues may display a different extent of methylation at adipogenic loci. Conversely, promoters of other lineage-specific genes may in turn be undermethylated in such MSCs, relative to stem cells from adipose tissue. Our results raise the hypothesis, therefore, that MSCs of different tissues may be marked by lineage-specific promoter hypomethylation.
Mosaic Methylation in Adipose Stem Cell Populations
Despite the overall hypomethylation of ASCs, we consistently observed heterogeneous methylation patterns at adipogenic loci in freshly isolated cells. There was minor variation in the percentage of methylation of specific cytosines between donors, despite the sequence overlap between the methylated areas. Furthermore, within individuals, we detected mosaicism between cells, both in the number of methylated cytosines and in the methylation pattern. This is in agreement with heterogeneity in 5′-to-3′ CpG methylation patterns reported in stem cells from single intestinal crypts (Yatabe et al., 2001; Kim et al., 2005). Mosaic methylation may result from stochastic methylation, which accumulates independently in different cells (CpG-rich sites are unmethylated at birth; Bird, 2002) as a result of exposure to environmental, aging, and health factors (Esteller, 2005; Hoffman and Carpenter, 2005; Laird, 2005; Ushijima, 2005; Zardo et al., 2005), in combination with a propensity for specific CpGs to be more methylated than others (Pfeifer et al., 1990; Silva et al., 1993). Thus, by analogy to the genetic diversity generated during evolution, stochastic methylation may reflect an epigenetic drift arising within stem cell reservoirs in somatic tissues.
Depending on the level of analysis, heterogeneous methylation profiles of ASCs are maintained or enhanced upon culture of undifferentiated cells. Averaging of methylation percentages at each CpG examined across all donors (uncultured cells) and across all cell clones shows a stable methylation profile and frequency in all adipogenic loci, in addition to GAPDH, MYOG, and CD31. Thus, polyclonal stem cell populations can display stable DNA methylation profiles. Nonetheless, we detected enhanced mosaicism at all adipogenic loci between clones of ASCs compared with that identified between stem cell donors. Clones from single isolated ASCs have been cultured for ∼1 wk before first division and then for ∼10 population doublings to reach sufficient cell numbers for first passaging, followed by another ∼10 population doublings by the time of analysis (passage 4). Twenty rounds of DNA replication are expected to elicit fidelity errors in maintenance methylation. A nonexclusive alternative accounting for enhanced heterogeneous methylation patterns is that different cells in the starting stem cell population display mosaic CpG methylation. Furthermore, asymmetric cell division, a characteristic of pluripotent stem cells (Clevers, 2005; Giebel et al., 2006), would also be expected to generate a differential epigenetic pattern in each daughter cell within a clonal cell line. It should be noted, however, that heterogeneous CpG methylation profiles are not specific for pluripotent cells, because mosaic methylation has also been reported in other clonal primary cell cultures (Zhu et al., 1999), tumor-derived clones (Silva et al., 1993; Graff et al., 2000), or uncultured PBLs and T-cells (Figure 9; this study).
DNA Methylation May Not Be a Determinant of Gene Expression or Potential for Expression in ASCs
The relationship between DNA methylation and gene expression or expression potential in undifferentiated ASCs remains complex (Jones and Takai, 2001). A typical observation in our study is the LEP promoter, which shows 9–23% methylation in three nonexpressing clones (A1, A2, and B1) and 7 and 24% methylation in two expressing clones (B1 and B2). Nevertheless, CpG no. 21 in the LEP promoter (which notably is contained within an C/EBP-binding site) is 60–90% methylated in clones B1 and B3, in which LEP up-regulation is significantly weaker (p < 0.001) than in any of the other clones in which CpG no. 21 is unmethylated. Furthermore, heavily methylated loci do not preclude expression. For example, the CD31 gene is highly methylated (this study) but nonetheless transcribed in ASCs with a CD31− immunophenotype (Boquest et al., 2005). Therefore, gene expression in undifferentiated ASCs does not correlate with a specific methylation pattern at any of the loci examined. Evidence against a direct role of DNA methylation as the primary determinant of gene expression has been addressed previously (Jones and Takai, 2001), and it is becoming clear that the lack of correlation between DNA methylation and transcription is not necessarily restricted to pluripotent cells (Kaneko et al., 2004).
DNA methylation does not seem to be a predictor of differentiation potential of ASCs. The adipogenic genes examined were hypomethylated, yet transcriptional up-regulation upon differentiation varied from 2- to >700-fold with respect to the lowest expressing clone. Furthermore, we found no correlation between any pattern of CpG methylation and gene expression or differentiation potential. Because ASCs can differentiate toward myogenic and endothelial lineages despite complete methylation of MYOG and CD31 in undifferentiated cells, this contention seems to also hold true for nonadipogenic genes. Nevertheless, differentiation toward nonadipogenic lineages may be more challenging due to the more methylated state of the DNA at key control elements. We are currently testing this hypothesis. In contrast to genes required for differentiation to nonadipogenic lineages, adipogenic gene promoters in undifferentiated ASCs may be maintained in a transcriptionally poised state by a mechanism that relies on DNA hypomethylation.
What, then, controls expression potential of lineage-specific genes in pluripotent cells? Recent evidence that neuronal differentiation of (hypomethylated) ESCs is regulated by the removal of a repressor complex (Ballas et al., 2005) argues that determinants of differentiation potential in other stem cell types may involve additional levels of regulation. As recently illustrated for pluripotent ESCs (Azuara et al., 2006), it is possible that a key transcriptional brake in undifferentiated ASCs involves histone H3 lysine 27 methylation, controlled by polycomb-group proteins (Pasini et al., 2004; Ringrose et al., 2004; Montgomery et al., 2005). The known interplay between DNA methylation and transcriptionally repressive histone modifications is also likely to operate in mesenchymal stem cells (Ayyanathan et al., 2003; Fujita et al., 2003; Lehnertz et al., 2003).
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Millar and J. Melki for T-cell DNA, H. K. Blomhoff for PBLs, and C. Drevon for SGBS adipocytes. We also thank L. Stijac for technical assistance. This work was supported by the Research Council of Norway, the Norwegian Cancer Society, The Norwegian Stem Cell Network, and The University of Oslo.
Abbreviations used:
- ASC
adipose stem cell
- ESC
embryonic stem cell
- FBS
fetal bovine serum
- HBSS
Hank’s balanced salt solution
- QRT
quantitative reverse transcription.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0322) on June 7, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Antequera F. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., Yamada Y., Tanaka K., Torigoe K., Rauscher F. J., III, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bey L., Etienne J., Tse C., Brault D., Noe L., Raisonnier A., Arnault F., Hamilton M. T., Galibert F. Cloning, sequencing and structural analysis of 976 base pairs of the promoter sequence for the rat lipoprotein lipase gene. Comparison with the mouse and human sequences. Gene. 1998;209:31–38. doi: 10.1016/s0378-1119(98)00003-1. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Boquest A. C., Shahdadfar A., Fronsdal K., Sigurjonsson O., Tunheim S. H., Collas P., Brinchmann J. E. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression following in vitro cell culture. Mol. Biol. Cell. 2005;16:1131–1141. doi: 10.1091/mbc.E04-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brero A., Easwaran H. P., Nowak D., Grunewald I., Cremer T., Leonhardt H., Cardoso M. C. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., O’Brien C. D., Zhou Z., Sanders S. M., Greenbaum J. N., Makrigiannakis A., DeLisser H. M. Involvement of human PECAM-1 in angiogenesis and in vitro endothelial cell migration. Am. J. Physiol. 2002;282:C1181–C1190. doi: 10.1152/ajpcell.00524.2001. [DOI] [PubMed] [Google Scholar]
- Catania J., Fairweather D. S. DNA methylation and cellular ageing. Mutat. Res. 1991;256:283–293. doi: 10.1016/0921-8734(91)90019-8. [DOI] [PubMed] [Google Scholar]
- Chi J. T., et al. Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. USA. 2003;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Stem cells, asymmetric division and cancer. Nat. Genet. 2005;37:1027–1028. doi: 10.1038/ng1005-1027. [DOI] [PubMed] [Google Scholar]
- Cousin B., Andre M., Arnaud E., Penicaud L., Casteilla L. Reconstitution of lethally irradiated mice by cells isolated from adipose tissue. Biochem. Biophys. Res. Commun. 2003;301:1016–1022. doi: 10.1016/s0006-291x(03)00061-5. [DOI] [PubMed] [Google Scholar]
- Cowan C. M., Shi Y. Y., Aalami O. O., Chou Y. F., Mari C., Thomas R., Quarto N., Contag C. H., Wu B., Longaker M. T. Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat. Biotechnol. 2004;22:560–567. doi: 10.1038/nbt958. [DOI] [PubMed] [Google Scholar]
- Deb-Rinker P., Ly D., Jezierski A., Sikorska M., Walker P. R. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J. Biol. Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- Fajas L., et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J. Biol. Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- Fraser J. K., Wulur I., Alfonso Z., Hedrick M. H. Fat tissue: an underappreciated source of stem cells for biotechnology. Trends Biotechnol. 2006;24:150–154. doi: 10.1016/j.tibtech.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Fujita N., Watanabe S., Ichimura T., Tsuruzoe S., Shinkai Y., Tachibana M., Chiba T., Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 2003;278:24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- Giebel B., Zhang T., Beckmann J., Spanholtz J., Wernet P., Ho A. D., Punzel M. Primitive human hematopoietic cells give rise to differentially specified daughter cells upon their initial cell division. Blood. 2006;107:2146–2152. doi: 10.1182/blood-2005-08-3139. [DOI] [PubMed] [Google Scholar]
- Graff J. R., Gabrielson E., Fujii H., Baylin S. B., Herman J. G. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J. Biol. Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Tontonoz P., Platt K. A., Ross S. R., Spiegelman B. M. Identification of a fat cell enhancer: analysis of requirements for adipose tissue-specific gene expression. J. Cell. Biochem. 1992;49:219–224. doi: 10.1002/jcb.240490303. [DOI] [PubMed] [Google Scholar]
- Gronthos S., Zannettino A. C., Hay S. J., Shi S., Graves S. E., Kortesidis A., Simmons P. J. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J. Cell Sci. 2003;116:1827–1835. doi: 10.1242/jcs.00369. [DOI] [PubMed] [Google Scholar]
- Grunau C., Clark S. J., Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L. M., Carpenter M. K. Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- Hornsby P. J., Yang L., Raju S. G., Maghsoudlou S. S., Lala D. S., Nallaseth F. S. Demethylation of specific sites in the 5′-flanking region of the CYP17 genes when bovine adrenocortical cells are placed in culture. DNA Cell Biol. 1992;11:385–395. doi: 10.1089/dna.1992.11.385. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Vaessen B., Lenvik T., Blackstad M., Reyes M., Verfaillie C. M. Multipotent progenitor cells can be isolated from postnatal murine bone marrow, muscle, and brain. Exp. Hematol. 2002;30:896–904. doi: 10.1016/s0301-472x(02)00869-x. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kaneko K. J., Rein T., Guo Z. S., Latham K., Depamphilis M. L. DNA methylation may restrict but does not determine differential gene expression at the Sgy/Tead2 locus during mouse development. Mol. Cell. Biol. 2004;24:1968–1982. doi: 10.1128/MCB.24.5.1968-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. K., Lee D. H., Bae Y. C., Kim H. K., Baik S. Y., Jung J. S. Improvement of neurological deficits by intracerebral transplantation of human adipose tissue-derived stromal cells after cerebral ischemia in rats. Exp. Neurol. 2003;183:355–366. doi: 10.1016/s0014-4886(03)00089-x. [DOI] [PubMed] [Google Scholar]
- Katz A. J., Tholpady A., Tholpady S. S., Shang H., Ogle R. C. Cell surface and transcriptional characterization of human adipose-derived adherent stromal (hADAS) cells. Stem Cells. 2005;23:412–423. doi: 10.1634/stemcells.2004-0021. [DOI] [PubMed] [Google Scholar]
- Kern S., Eichler H., Stoeve J., Kluter H., Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Je C. M., Sin J. Y., Jung J. S. Effect of partial hepatectomy on in vivo engraftment after intravenous administration of human adipose tissue stromal cells in mouse. Microsurgery. 2003;23:424–431. doi: 10.1002/micr.10178. [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Siegmund K. D., Tavare S., Shibata D. Age-related human small intestine methylation: evidence for stem cell niches. BMC Med. 2005;3:10. doi: 10.1186/1741-7015-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Laird P. W. Cancer epigenetics. Hum. Mol. Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role of DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Maitra A., et al. Genomic alterations in cultured human embryonic stem cells. Nat. Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- Mason M. M., He Y., Chen H., Quon M. J., Reitman M. Regulation of leptin promoter function by Sp1, C/EBP, and a novel factor. Endocrinology. 1998;139:1013–1022. doi: 10.1210/endo.139.3.5792. [DOI] [PubMed] [Google Scholar]
- Massari M. E., Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzner I., Scott V., Dorsch K., Fischer P., Wabitsch M., Bruderlein S., Hasel C., Moller P. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J. Biol. Chem. 2002;277:45420–45427. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- Merkel M., Eckel R. H., Goldberg I. J. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- Montgomery N. D., Yee D., Chen A., Kalantry S., Chamberlain S. J., Otte A. P., Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Otto T. C., Lane M. D. Adipose development: from stem cell to adipocyte. Crit. Rev. Biochem. Mol. Biol. 2005;40:229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- Panning B., Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A. P., Jensen M. R., Lazzerini D. E., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:E45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Hansen R. S., Gartler S. M., Riggs A. D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc. Natl. Acad. Sci. USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Reseland J. E., Syversen U., Bakke I., Qvigstad G., Eide L. G., Hjertner O., Gordeladze J. O., Drevon C. A. Leptin is expressed in and secreted from primary cultures of human osteoblasts and promotes bone mineralization. J. Bone Miner. Res. 2001;16:1426–1433. doi: 10.1359/jbmr.2001.16.8.1426. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Ehret H., Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Rodic N., Oka M., Hamazaki T., Murawski M. R., Jorgensen M., Maatouk D. M., Resnick J. L., Li E., Terada N. DNA methylation is required for silencing of ant4, an adenine nucleotide translocase selectively expressed in mouse embryonic stem cells and germ cells. Stem Cells. 2005;23:1314–1323. doi: 10.1634/stemcells.2005-0119. [DOI] [PubMed] [Google Scholar]
- Rodriguez A. M., et al. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem. Biophys. Res. Commun. 2004;315:255–263. doi: 10.1016/j.bbrc.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Ross S. R., Graves R. A., Greenstein A., Platt K. A., Shyu H. L., Mellovitz B., Spiegelman B. M. A fat-specific enhancer is the primary determinant of gene expression for adipocyte P2 in vivo. Proc. Natl. Acad. Sci. USA. 1990;87:9590–9594. doi: 10.1073/pnas.87.24.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J., Ward K., White R. Mosaic methylation in clonal tissue. Dev. Biol. 1993;156:391–398. doi: 10.1006/dbio.1993.1086. [DOI] [PubMed] [Google Scholar]
- Timper K., Seboek D., Eberhardt M., Linscheid P., Christ-Crain M., Keller U., Muller B., Zulewski H. Human adipose tissue-derived mesenchymal stem cells differentiate into insulin, somatostatin, and glucagon expressing cells. Biochem. Biophys. Res. Commun. 2006;341:1135–1140. doi: 10.1016/j.bbrc.2006.01.072. [DOI] [PubMed] [Google Scholar]
- Urs S., Smith C., Campbell B., Saxton A. M., Taylor J., Zhang B., Snoddy J., Jones V. B., Moustaid-Moussa N. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J. Nutr. 2004;134:762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat. Rev. Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Wabitsch M., Brenner R. E., Melzner I., Braun M., Moller P., Heinze E., Debatin K. M., Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- Warnecke P. M., Stirzaker C., Song J., Grunau C., Melki J. R., Clark S. J. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- Yatabe Y., Tavare S., Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl. Acad. Sci. USA. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardo G., Fazi F., Travaglini L., Nervi C. Dynamic and reversibility of heterochromatic gene silencing in human disease. Cell Res. 2005;15:679–690. doi: 10.1038/sj.cr.7290337. [DOI] [PubMed] [Google Scholar]
- Zheng Q. H., Ma L. W., Zhu W. G., Zhang Z. Y., Tong T. J. p21(Waf1/Cip1) plays a critical role in modulating senescence through changes of DNA methylation. J. Cell. Biochem. 2006 doi: 10.1002/jcb.20838. Mar 2; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Zhu X., Deng C., Kuick R., Yung R., Lamb B., Neel J. V., Richardson B., Hanash S. Analysis of human peripheral blood T cells and single-cell-derived T cell clones uncovers extensive clonal CpG island methylation heterogeneity throughout the genome. Proc. Natl. Acad. Sci. USA. 1999;96:8058–8063. doi: 10.1073/pnas.96.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk P. A., Zhu M., Mizuno H., Huang J., Futrell J. W., Katz A. J., Benhaim P., Lorenz H. P., Hedrick M. H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- Zvetkova I., Apedaile A., Ramsahoye B., Mermoud J. E., Crompton L. A., John R., Feil R., Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.