Abstract
Rab11a, myosin Vb, and the Rab11-family interacting protein 2 (FIP2) regulate plasma membrane recycling in epithelial cells. This study sought to characterize more fully Rab11-FIP2 function by identifying kinase activities modifying Rab11-FIP2. We have found that gastric microsomal membrane extracts phosphorylate Rab11-FIP2 on serine 227. We identified the kinase that phosphorylated Rab11-FIP2 as MARK2/EMK1/Par-1Bα (MARK2), and recombinant MARK2 phosphorylated Rab11-FIP2 only on serine 227. We created stable Madin-Darby canine kidney (MDCK) cell lines expressing enhanced green fluorescent protein-Rab11-FIP2 wild type or a nonphosphorylatable mutant [Rab11-FIP2(S227A)]. Analysis of these cell lines demonstrates a new role for Rab11-FIP2 in addition to that in the plasma membrane recycling system. In calcium switch assays, cells expressing Rab11-FIP2(S227A) showed a defect in the timely reestablishment of p120-containing junctional complexes. However, Rab11-FIP2(S227A) did not affect localization with recycling system components or the normal function of apical recycling and transcytosis pathways. These results indicate that phosphorylation of Rab11-FIP2 on serine 227 by MARK2 regulates an alternative pathway modulating the establishment of epithelial polarity.
INTRODUCTION
The establishment of polarity is an intricately regulated process in epithelial cells. The apical and basolateral domains must remain separated by the tight junctions to segregate membrane proteins. For example, although the apical domain of Madin-Darby canine kidney (MDCK) cells contains GP-135 (Ojakian and Schwimmer, 1988), the basolateral domain expresses Na+/K+-ATPase (Louvard, 1980). The tight junction serves as a physical barrier between the two protein pools and is characterized by the expression of zonula occludens (ZO)-1, claudins, and occludin in an epithelial monolayer. An adherens junction facilitates cell–cell contact, which is regulated in part by E-cadherin and p120 (reviewed in Miyoshi and Takai, 2005). Each of these proteins must be trafficked to the correct domain of the cell for the epithelial monolayer to function appropriately. These diverse destinations require intricate trafficking pathways to ensure their accuracy. Recently, Rab11a has been implicated in the trafficking of E-cadherin to the adherens junction (Lock and Stow, 2005), suggesting that the Rab11a pathway may be important in the establishment of polarized domains.
Rab11a, a member of the Rab11 subfamily of small GTPases, is well-established as a participant in the regulation of recycling endosomal trafficking. Rab11a is associated with vesicles in the apical portion of epithelial cells near the centrosome and beneath the apical plasma membrane (Casanova et al., 1999). Plasma membrane recycling is critical in maintaining proper membrane protein expression in response to stimuli for such diverse events as nutrient internalization and the recycling of ion channels and receptors (Takei and Haucke, 2001; Volpicelli et al., 2002; Fan et al., 2003; Fan et al., 2004). However, recent work has begun to connect the recycling system to the trafficking pathways of junctional proteins (Le et al., 1999; Lock and Stow, 2005).
The family of small GTPases, including Rab11a, interacts with and is regulated by specific interacting proteins. Numerous binding partners have been elucidated for Rab11a, including 1) myosin Vb (Lapierre et al., 2001); 2) rabphillin11/Rab11-binding protein (Mammoto et al., 1999; Zeng et al., 1999); and 3) a family of Rab11-interacting proteins: Rab11-FIP1, Rab11-FIP2, Rab11-FIP3 (Hales et al., 2001), Rab11-FIP4 (Wallace et al., 2002), Rab11-FIP5 (pp75/Rip11) (Prekeris et al., 2000), and RCP (Lindsay et al., 2002). The Rab11-FIP proteins each interact with Rab11-family members (Rab11a, Rab11b, and Rab25) at their carboxy termini through predicted coiled-coil regions containing an amphipathic α-helical Rab binding domain (Hales et al., 2001; Prekeris et al., 2001). The diversity of multiple Rab11-FIP proteins, all of which bind to Rab11 with similar helices, suggests that each Rab11-FIP may be important in a spatially restricted manner or in separate trafficking processes.
In particular, Rab11-FIP2 seems to form a ternary complex with both Rab11a and myosin Vb (Hales et al., 2002). A truncation of Rab11-FIP2 lacking its amino terminal C2 domain [Rab11-FIP2(129–512)] strongly inhibits plasma membrane recycling (Hales et al., 2002). Nevertheless, studies in nonpolarized cells have also implicated Rab11-FIP2 in the regulation of endocytosis through interaction with the early endosomal protein Reps1 (Cullis et al., 2002).
In this study, we describe the involvement of Rab11-FIP2 in the establishment of polarity. We have biochemically purified a kinase activity that phosphorylated Rab11-FIP2 on serine 227 and that was identified by mass spectrometry as MARK2/EMK1/Par-1Bα (MARK2). Previous studies have associated MARK2 with changes in aspects of polarity (Cohen et al., 2004). Disruption of the junctional integrity with low calcium followed by reestablishment of normal extracellular calcium (calcium switch) showed that phosphorylation of Rab11-FIP2 is important for the timely reestablishment of polarity. The results indicate that phosphorylation of Rab11-FIP2 by MARK2 serves as an important regulatory mechanism for the establishment of epithelial cell polarity.
MATERIALS AND METHODS
Materials
Rabbit anti-Rab11a (VU57) antibodies were developed against the amino terminus of human Rab11a and were specific for Rab11a versus Rab11b and Rab25 (Lapierre, unpublished data). The other antibodies used were rat anti-ZO-1 (Chemicon International, Temecula, CA), mouse anti-ZO-1 (Zymed Laboratories, South San Francisco, CA), mouse pan anti-cadherin (Sigma-Aldrich, St. Louis, MO), mouse anti-p120 catenin (BD Biosciences, San Jose, CA), mouse monoclonal anti-ezrin (4A5; Chemicon International), rabbit anti-occludin (8 μg/ml; Zymed Laboratories), anti-green fluorescent protein (GFP) mouse monoclonal (8362–1; BD Biosciences), and anti-GFP rabbit (AB290; Abcam, Cambridge, MA). All secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Dr. Roy Zent (Department of Medicine, Vanderbilt University, Nashville, TN) kindly provided mouse monoclonal anti-GP135.
Database Searches and Alignment
Rab11-FIP2 homologues were identified through GenBank searches using the Rab11 binding domain. FlyBase, Joint Genome Institute Xenopus, and UCSC Genscan were also used to identify homologues. Alignments were performed using ClustalW (http://www.ebi.ac.uk/clustalw/).
Site-directed Mutagenesis
All site-directed mutagenesis of Rab11-FIP2 was performed using Pfu Turbo polymerase according to the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with a 16-min extension time. Primers were synthesized (Invitrogen, Carlsbad, CA) with one nucleotide change per oligonucleotide sequence. The TCA encoding for amino acid 227 was changed to GCA for the S227A mutant. All constructs were created in pEGFP-C2 (Clontech, Mountain View, CA) and subsequently recloned into pET-30a (Novagen, Madison, WI) with EcoRI and SalI restriction sites.
Protein Production
For recombinant protein production constructs in pET-30a vectors were retransformed into BL21(DE3)pLysS bacteria. Bacteria were grown to log phase and then induced with 400 ng/ml isopropyl β-d-thiogalactoside for 3 h at 37°C. To harvest protein, bacteria were pelleted at 2000 × g and then resuspended in lysis buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl with protease inhibitors [protein buffer], and 10 mM imidazole). Protein was harvested according to the manufacturer’s protocol (Novagen). Briefly, the bacteria were then sonicated four times for 20 s at maximum potency on ice. The lysate was extracted with 0.1% Triton X-100 for 5 min on ice. The extracted lysate was cleared by centrifugation at 15,000 × g, and the resulting supernatant was incubated with nickel-affinity resin at 4°C (His-Bind; Novagen). The beads and protein were washed in protein buffer with 20 mM imidazole. The bound protein was eluted overnight at 4°C with elution buffer (protein buffer with 250 mM imidazole). Recombinat MARK2 was purchased from Upstate Biotechnology (Lake Placid, NY) (14-544).
Rabbit Gastric Tubulovesicle Preparation
Fractions of rabbit gastric mucosal microsomes were prepared as described previously from the fundic mucosa of New Zealand White rabbits (Basson et al., 1991). The rabbit gastric mucosa tissue was homogenized in 5 volumes of 15 mM HEPES, 300 mM sucrose buffer, pH 7.4, with protease inhibitors (Sigma-Aldrich) with a Potter homogenizer at 1000 rpm. The homogenate was sequentially centrifuged at 500 × g for 10 min, 5000 × g for 10 min, 17,000 × g for 20 min, and 100,000 × g for 60 min, and the 100,000 × g pellet was resuspended in the homogenization buffer and frozen at −80°C until use.
Kinase Identification
The 100,000 × g microsomal pellet from rabbit gastric mucosa was thawed on ice and then extracted for 30 min with 1% Triton X-100. The solubilized microsomes were centrifuged at 100,000 × g for 1 h at 4°C. The supernatant from this spin was diluted 1:10 with buffer A (5 mM sodium phosphate, pH 7.2, and 0.1% Triton X-100) for protein purification. The diluted homogenate was loaded on a HiTrap Q FF column (2 ml; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) preequilibrated in buffer A at 1 ml/min. The Rab11-FIP2 kinase activity, which voided the column, was collected and then further purified over a ceramic hydroxylapatite column (Econo-Pac CHT-I 1-ml cartridge; Bio-Rad, Hercules, CA) preequilibrated in buffer A. The void fraction was collected and the bound protein was eluted in a gradient from 0 to 500 mM sodium phosphate, pH 7.2, 0.1% Triton X-100. The Rab11-FIP2 kinase activity eluted at ∼250 mM sodium phosphate. The fractions with the highest activity were pooled, diluted 1:1 in buffer A, and chromatographed over MONO-S resin (5 ml) (GE Healthcare). The bound protein was eluted with a continuous salt gradient from 0 to 1 M NaCl in buffer A. The Rab11-FIP2 kinase activity eluted at 400 mM NaCl. The fractions with the highest activity were pooled and further purified over a Cibachrome blue affinity column (HiTrap Blue, 1 ml; Amersham). The proteins were eluted with a continuous gradient to 2 M NaCl in buffer A. Kinase activity eluted at ∼500 mM NaCl. Finally, the fractions with the highest activity were loaded onto a 10 to 40% glycerol gradient and centrifuged for 24 h at 160,000 × g. The Rab11-FIP2 kinase activity peaked at ∼17% glycerol. Each step was monitored by the in vitro kinase activity assay.
The fraction from the glycerol gradient that contained the greatest amount of kinase activity was subjected to trypsin digestion, and the resulting peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) for protein identification. Before trypsin digestion, the samples were cleaned up using a 10-kDa Ultrafree MC regenerated cellulose filter (Millipore, Billerica, MA), and the proteins were subsequently digested directly off of the filter as described previously (Manza et al., 2005).
LC-MS analysis of the resulting peptides was performed using a Thermo Finnigan (Waltham, MA) LTQ ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Thermo Surveyor HPLC pump, Nanospray source, and Xcalibur 1.4 instrument control. The peptides were separated on a packed capillary tip, 100 μm × 11 cm, with C18 resin (Monitor C18, 5 μm, 100 Å; Column Engineering, Ontario, CA) using an inline solid phase extraction column that was 100 μm × 6 cm packed with the same C18 resin (using a frit generated with liquid silicate Kasil 1; Cortes et al., 1987) similar to that described previously (Licklider et al., 2002), except the flow from the HPLC pump was split before the injection valve. The flow rate during the solid phase extraction phase of the gradient was 1 μl/min and during the separation phase was 700 nl/min. Mobile phase A was 0.1% formic acid, mobile phase B was acetonitrile with 0.1% formic acid. A 95-min gradient was performed with a 15-min washing period (100% A for the first 10 min followed by a gradient to 98% A at 15 min) to allow for solid phase extraction and removal of any residual salts. After the initial washing period, a 60-min gradient was performed where the first 35 min was a slow, linear gradient from 98% A to 75% A, followed by a faster gradient to 10% A at 65 min and an isocratic phase at 10% A to 75 min. The MS/MS spectra of the peptides was performed using data-dependent scanning in which one full MS spectra, using a full mass range of 400-2000 amu, was followed by three MS/MS spectra. Proteins were identified using the SEQUEST algorithm (Yates et al., 1995) and the SEQUEST Browser software in the BioWorks 3.1 software package (Thermo Electron, San Jose, CA), using the nonredundant database from National Center for Biotechnology Information (Bethesda, MD).
For phosphorylation mapping experiments, the Rab11-FIP2 band was excised from a one-dimensional-SDS-PAGE gel and either trypsin or chymotrysin digestion was performed in-gel. The samples were then analyzed using data-dependent analysis similar to that described above with the addition of a neutral loss scan to scan for neutral loss of phosphoric acid (loss of 98) in the top three ions. If a neutral loss ion was found, it was fragmented and an MS/MS/MS spectrum was collected. In addition to using the SEQUEST algorithm to search for phosphorylations on serines, threonines or tyrosines, the data were searched for modifications using the PMOD algorithm (Hansen et al., 2005).
In Vitro Kinase Activity Assay during Purification
The chromatographic fractions were added to the substrate [Rab11-FIP2(190-383)] in a 50 mM Tris buffer containing 5 mM MgCl2, 1 mM EGTA, protease inhibitors (1:100), and 25 μM dithiothreitol on ice. [γ-32P]ATP was added, and the reaction was incubated at 35°C for 10 min. The reactions were terminated with the addition of SDS buffer (final concentrations, 300 mM Tris, pH 7.5, 1% SDS, 20 mM EDTA, and 17.5 mM sucrose) and incubated at 70°C for 10 min. The samples were resolved on 12% SDS-PAGE gels, stained with colloidal Coomassie (GelCode blue; Pierce Chemical, Rockford, IL) for protein detection, dried under vacuum, and analyzed with phosphorimaging (Molecular Dynamics) for [32P]phosphate incorporation.
In Situ Phosphorylation
We used [32P]orthophosphate incorporation to assess phosphorylation in situ. MDCK cells were plated on 60-mm Transwell filters (Corning Life Sciences, Acton, MA) and allowed to polarize at confluence for 3 d. MDCK cells stably expressing enhanced green fluorescent protein (EGFP)-Rab11-FIP2 or EGFP-Rab11-FIP2(S227A) constructs were incubated with [32P]orthophosphate in phosphate-free DMEM supplemented with 2 mg/ml bovine serum albumin for 2 h. The cells were solubilized in lysis buffer [30 mm Tris, pH 8.5, 150 mM NaCl, 20 mM magnesium acetate, 1% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) with protease inhibitors, and phosphatase inhibitors], extracted on ice for 20 min, and centrifuged for 20 min at 15,000 × g to pellet the insoluble material.
For immunoprecipitation, anti-rabbit IgG Dynabeads (Dynal Biotech, Lake Success, NY) were loaded with either 5 μl of anti-GFP antibody AB290 serum (Abcam) or control rabbit serum for 2 h at 4°C. Beads were washed three times with Tris-buffered saline (TBS). Lysate was diluted in immunoprecipitation buffer (final concentration 30 mM Tris, pH 7.5, 150 mM, NaCl, 20 mM magnesium acetate, protease inhibitors, and phosphatase inhibitors) and incubated with the beads overnight at 4°C. The beads were washed twice for 20 min with immunoprecipitation buffer with 0.1% CHAPS and then once with 30 mM Tris, pH 7.5. The beads were eluted in 1% SDS buffer, and proteins were resolved on 10% SDS-PAGE gels, which were either dried and visualized by a phosphorimaging screen (Molecular Dynamics) for 32P-phosphoproteins or transferred to nitrocellulose for anti-GFP immunoblotting.
Immunoblotting
Protein samples were resolved on 10% SDS-polyacrylamide gels following a standard Laemmli protocol (Laemmli, 1970). All incubations were performed at room temperature. Proteins were transferred to nitrocellulose. Blots were blocked for 1 h with 5% dry milk powder (DMP)/TBS and 0.05% Tween 20 (TBS-T). The blots were incubated with primary antibody diluted in 2.5% DMP/TBS-T for 1.5 h (mouse monoclonal anti-GFP 1:2000), washed three times for 10 min in TBS-T, and incubated for 1 h with horseradish peroxidase-labeled anti-mouse secondary; Jackson ImmunoResearch Laboratories) diluted in 1% DMP/TBS-T. The blots were then washed three times with TBS-T followed by one time with TBS, and specific labeling was detected by enhanced chemiluminescence (SuperSignal; Pierce Chemical) with autoradiography using Kodak BioMax ML film (Eastman Kodak, Rochester, NY).
Cell Culture
Parental T23 MDCKs (Barth et al., 1997) as well as the stably transfected cell lines were grown in DMEM supplemented with 10% fetal bovine serum (FBS) (Invitrogen), penicillin-streptomycin, 2 mM l-glutamine, and 0.1 mM minimal essential medium (MEM) nonessential amino acids (Invitrogen). Media for cell lines also contained 0.5 mg/ml G418 sulfate (Cellgro; Mediatech, Herndon, VA), and 0.25 ng/ml hygromycin (Invitrogen). In the stable cell lines, expression of the EGFP chimeras was inhibited with 20 ng/ml doxycycline (Calbiochem, San Diego, CA). To examine EGFP protein expression, cells were grown on 0.4-μm Transwell filters (Corning Life Sciences) without doxycycline in tetracycline screened FBS (Hyclone Laboratories, Logan, Utah) media for 2–4 d.
GFP Constructs and Transfections
Doxycycline-inhibitable expression vectors were generated by excising the EGFP-Rab11-FIP2 wild-type and mutant sequences from pEGFP-vectors with NheI and SmaI and ligating into a pTRE2hyg vector (Clontech) cut with NheI and EcoRV. Transfection was performed using Effectene (QIAGEN, Valenica, CA) following the manufacturer’s protocol. One microgram of vector was transfected into a 60-mm plate of T23 MDCK cells in normal media. The following day, the cells were trypsinized and replated in serial dilutions, including 0.25 ng/ml hygromycin for selection and 20 ng/ml doxycycline for suppression of EGFP expression. Multiple colonies were selected, expanded for 10 d and then screened for EGFP expression in media with tetracycline-screened serum. Multiple clones were initially characterized, all of which showed the same expression pattern and level of the EGFP construct. One clone was selected for each construct to use as the tetracycline-repressible stable cell lines [expressing EGFP-Rab11-FIP2 wild type or EGFP-Rab11-FIP2(S227A)].
Polymeric Immunoglobin A (pIgA) Trafficking in MDCK Cells
Labeling of pIgA and trafficking experiments were done as described previously (Hales et al., 2002) except that we used the cell culture media described above. Cells were loaded from the apical side and fixed at time 0 after a 30-min loading.
Analysis of 125I-IgA Postendocytotic Fate
125I-IgA was iodinated using the ICl method to a specific activity of 1.0–2.0 times] 107 cpm/μg (Breitfeld et al., 1989). The postendocytotic fate of a preinternalized cohort of 125I-IgA (at 5–10 μg/ml) was analyzed as previously described (Apodaca et al., 1994). In brief, filter-grown MDCK cells expressing the various FIP2 constructs and wild-type pIgR were cultured in the presence or absence of doxycycline, and 125I-IgA internalized from the basolateral cell surface of the cells for 10 min at 37°C. The basal surface of the cells was rapidly washed three times, the apical and basolateral medium was aspirated and replaced with fresh medium. The cells were then incubated for 3 min at 37°C. This wash procedure takes 5 min at 37°C. Fresh medium was added to the cells, and they were chased for up to 2 h at 37°C. At the designated time points, the apical and basolateral media (0.5 ml) were collected and replaced with fresh media. After the final time point, filters were cut out of the insert, and the amount of 125I-IgA quantified with a gamma counter. The media samples were precipitated with 10% trichloroacetic acid (TCA) for 30 min on ice and then centrifuged in a microfuge for 15 min at 4°C. The amount of 125I-IgA in the TCA-soluble (degraded) and insoluble fractions (intact) was quantified with a gamma counter.
Calcium Switch
Cells were grown to confluence on filters in regular media with or without doxycycline. Cells were washed with low calcium media (MEM [Cellgro 15–015-CV], 10% dialyzed FBS, penicillin-streptomycin, 2 mM l-glutamine, and 0.1 mM MEM nonessential amino acids; Invitrogen). The cells were incubated in low calcium media overnight with or without doxycycline. Calcium was added to the top and bottom of the filter to a final concentration of 1.8 mM. The cells were collected at the indicated time points.
Laminin Replating Assay
Cells were trypsinized and replated on 24-well plate 0.45-μm laminin-coated filters (BD Biosciences). The cells were collected at the indicated time points.
Immunofluorescence for Calcium Switch and Replating Experiments
Cells were washed one time with phosphate-buffered saline (PBS) and then preextracted on ice in 0.2% Triton X-100 in PBS. Cells were fixed for 30 min in 3% paraformaldehyde at room temperature. Cells were permeabilized with 0.05% Triton X-100 in PBS for 5 min on ice. Cells were incubated with anti-p120 and ZO-1 antibodies in PBS for 1 h on ice. Cells were washed one time with PBS. Cells were incubated with species-specific Cy3-anti mouse IgG and Cy5-anti-rat IgG in PBS for 30 min at room temperature. Cells were washed first with PBS and then with 50 mM sodium phosphate. Finally, cells were stained with 4,6-diamidino-2-phenylindole (DAPI) in sodium phosphate. Filters were cut out of the transwells and mounted with Prolong Anitfade solution (Molecular Probes). Cells were imaged on a Zeiss LSM510 confocal microscope using a 100× lens. The z-sections were 0.3 μm.
Additional Immunofluorescence
Cells were washed three times with PBS and then fixed in 4% paraformaldehyde for 15 min at room temperature. The cells were washed twice with PBS and stored at 4° in PBS until staining. Cells were blocked with extraction buffer (10% normal donkey serum, 150 mM NaCl, 20 mM sodium phosphate, pH 7.4, and 0.3% Triton X-10) for 20 min. Primary antibody was immediately added in antibody buffer (10% normal donkey serum, 150 mM NaCl, 20 mM sodium phosphate, pH 7.4, and 0.05% Tween 20) for 2 h. The cells were washed with PBS three times. Secondary species-specific Cy dye-labeled anti-IgGs were added for 1 h in antibody buffer. After washing with PBS two times, the cells were washed with 50 mM sodium phosphate once and then stained with DAPI in sodium phosphate. Filters were cut out of the transwells and mounted with Prolong Antifade solution (Molecular Probes). Cells were imaged on a Zeiss LSM510 confocal microscope (Carl Zeiss, Thornwood, NY) using a 100× lens. Z-sections were 0.5 μm.
RESULTS
Phosphorylation of Rab11-FIP2
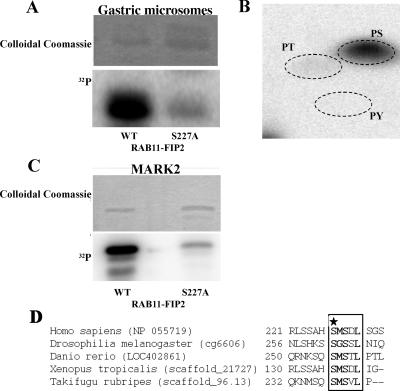
Although previous investigations have noted potential phosphorylation sites on Rab11-FIP2, no discrete kinase activities phosphorylating the protein have been identified. We sought to identify potential phosphorylating activities for Rab11-FIP2 by assaying phosphorylation of recombinant Rab11-FIP2 in vitro using extracts from rabbit 100,000 × g gastric microsomes that are enriched in parietal cell tubulovesicles (H+/K+-ATPase containing apical recycling membranes). Extracts of rabbit gastric microsomes solubilized in 1% Triton X-100 strongly phosphorylated recombinant Rab11-FIP2 (Figure 1A). Phospho-amino acid analysis indicated that Rab11-FIP2 was primarily phosphorylated on serine residues (Figure 1B). Two-dimensional tryptic phosphopeptide mapping revealed that Rab11-FIP2 was phosphorylated on a major site (80%) and a minor site (20%) (our unpublished data). Carboxy- and amino-terminal recombinant truncations narrowed the region of phosphorylation to amino acids 187–356. A further truncation to amino acids 227–356 was not phosphorylated. We therefore performed site-directed mutagenesis on high-probability serine residues between 187 and 230. Mutation of serine 227 to alanine led to a >80% reduction in the in vitro phosphorylation by gastric microsomal extracts (Figure 1A). Serine-to-alanine mutations at serines 223, 224, and 229 had no effect on phosphorylation (our unpublished data).
Figure 1.
Rab11-FIP2 kinase activities. (A) A kinase activity associated with rabbit gastric microsomes phosphorylates Rab11-FIP2 on serine 227. Top, Coomassie stain for protein levels. Bottom, phosphorimage. (B) Phospho-amino acid analysis demonstrates phosphorylation of Rab11-FIP2 is on serine residues. Positions of phospho-amino acid standards are shown by dotted lines. PS, phosphoserine; PT, phosphothreonine; PY, phosphotyrosine. (C) Rab11-FIP2 is phosphorylated by recombinant activated MARK2 in vitro on serine 227 of Rab11-FIP2 wild type by not Rab11-FIP2(S227A). Top blot, Coomassie stain for protein levels. Bottom blot, phosphorimage. (D) Amino acid alignment of the putative Rab11-FIP2 sequences. The conserved phosphorylation site is boxed and in bold. The phosphorylated serine is marked by a star.
Because we had no evidence for the phosphorylation of Rab11-FIP2 by the more common characterized protein kinases, and because serine 227 did not fall into any canonical consensus sites for known kinases, we sought to isolate the Rab11-FIP2 kinase activity from rabbit gastric microsomes. The 100,000 × g microsomal membrane fraction from rabbit gastric mucosa was used to isolate the kinase activity that phosphorylated Rab11-FIP2 on serine 227. Microsomal proteins were extracted with 1% Triton X-100, and proteins were resolved sequentially on Mono-Q, hydroxylapatite, Mono-S, and Cibachrome blue resins followed by resolution on continuous glycerol gradients (see Materials and Methods). The fractions with the highest activity from column chromatography and the final glycerol gradient fraction containing serine 227 Rab11-FIP2 kinase activity were submitted for proteomic analysis by total trypsin digestion and analysis of peptides by LC-MS/MS (Vanderbilt University Proteomics Laboratory, Mass Spectrometry Research Center). Only one kinase was identified consistently in these fractions from two different sample preparations: Par-1Bα/MARK2/EMK1 (MARK2) (5.5–14.7% coverage by amino acid). Seven separate peptides were identified matching either rat or human sequences (Table 1).
Table 1.
Peptides obtained from mass spectrometry for Rab11-FIP2 kinase identification
| Peptide match | Xcorr | z |
|---|---|---|
| AENLLLDADMNIK | 4.54 | 2 |
| KYDGPEVDVWSLGVILYTLVSGSLPFDGQNLK | 3.73 | 3 |
| LFEVIETEK | 2.59 | 2 |
| PSADLTNSSAPSPSHK | 2.44 | 3 |
| TQLNSSSLQK | 2.59 | 2 |
| FLILNPSK | 1.94 | 2 |
| VLNHPNIVK | 1.42 | 2 |
Xcorr, cross-correlation score as identified by SEQUEST (Link et al., 1999); z, charge state of peptide.
Peptide sequences of MARK2 identified in Rab11-FIP2 kinase preparations by mass spectrometry.
To verify MARK2 as a Rab11-FIP2 kinase, we assessed the ability of recombinant activated MARK2 to phosphorylate recombinant Rab11-FIP2 in vitro. Although recombinant MARK2 strongly phosphorylated recombinant Rab11-FIP2, we observed little phosphorylation of Rab11-FIP2(S227A) (Figure 1C). In addition, we examined by LC/MS tandem mass spectrometry phosphorylation sites in recombinant Rab11-FIP2 when the protein was phosphorylated by MARK2 and found that serine 227 was phosphorylated in peptides produced from both trypsin and chymotrypsin digestions. In addition, spectra of the peptides from both digests contained a neutral loss of 98 (NL98) ion, indicative of a phosphorylation site, and an MS/MS/MS spectrum of the NL98 ion in the tryptic peptide confirmed that the ion resulted from neutral loss of phosphoric acid resulting from the phosphorylation.
On inspection of the available genomic databases using the Rab11 binding domain, we found that the serine 227 site is an evolutionarily conserved residue in putative Rab11-FIPs from numerous species, including Drosophila melanogaster and Danio rerio (Figure 1D). Interestingly, this phosphorylation site, the first serine residue of SMSxL, is a noncanoncial site for MARK2. Previous studies have suggested that MARK2 phosphorylates KxGS sites (Drewes et al., 1997). This noncanonical site is also present in two other human Rab11-interacting proteins, RCP and Rab11-FIP1. Activated recombinant MARK2 phosphorylated both RCP and Rab11-FIP1 in vitro (our unpublished data).
Rab11-FIP2 Is Phosphorylated In Situ
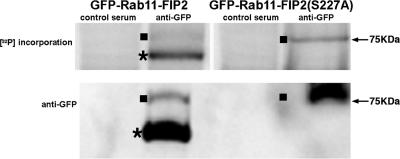
To assess the importance of Rab11-FIP2 phosphorylation in situ, we generated MDCK T23 cell lines with tetracycline-repressible expression of EGFP-Rab11-FIP2 wild type and nonphosphorylatable EGFP-Rab11-FIP2(S227A). EGFP-Rab11-FIP2– and EGFP-Rab11-FIP2(S227A)–expressing cells were grown on permeable filters and labeled for 2 h with [32P]orthophosphate, followed by lysis and immunoprecipitiation of GFP-Rab11-FIP2 proteins with anti-GFP antibodies. An immunoblot using anti-GFP showed that the amount of protein immunoprecipitated was similar between the two conditions. However, although the wild-type Rab11-FIP2 was strongly phosphorylated in situ, Rab11-FIP2(S227A) demonstrated far less in situ phosphorylation (Figure 2). We consistently observed a phosphorylated breakdown product in the EGFP-Rab11-FIP2 wild-type cell line that was not present in the EGFP-Rab11-FIP2(S227A) line. This breakdown was also apparent in recombinant protein preparations, suggesting that Rab11-FIP2 is readily degraded by an as yet unknown mechanism. Recently, PEST sequences have been implicated in the degradation of RCP, a member of the Rab11-FIPs (Marie et al., 2005). Although Rab11-FIP2 does not have PEST sequences, degradation may be a prominent regulatory mechanism in this family. Although mutation of the phosphorylation site seems to stabilize the protein, we have no evidence that phosphorylated Rab11-FIP2 binds 14-3-3 proteins (our unpublished data). These results establish the presence of Rab11-FIP2 phosphorylation on serine 227 in polarized MDCK cells.
Figure 2.
EGFP-Rab11-FIP2 is phosphorylated in situ. Top blot, Phosphorimage of phosphorylation of immunoprecipitated EGFP-Rab11-FIP2 from MDCK cell lines stably expressing either EGFP-Rab11-FIP2 or EGFP-Rab11-FIP2(S227A). Cells were loaded with [32P]orthophosphate, lysed, and immunoprecipitated with an anti-GFP antibody, and proteins were resolved on SDS-PAGE gels. Bottom blot, Immunoblot probing for GFP of immunoprecipitated lysates. Full-length EGFP-Rab11-FIP2 is 75 kDa (denoted with a solid square), but it is consistently broken down to a lower molecular mass species (indicated with a star) by an as yet unknown mechanism.
Overexpression of Cargo Alters Rab11-FIP2 Morphology
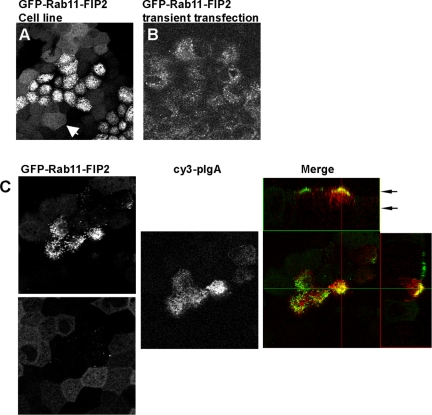
During the characterization of the tetracycline-repressible stable MDCK T23 cell lines, we noticed two morphologies apparent in each of 10 lines cloned of EGFP-Rab11-FIP2 wild type. One morphology was similar to the EGFP-Rab11-FIP2 distribution previously reported by our laboratory in transiently transfected MDCK cells (Hales et al., 2001) and others in A431 cells (Lindsay and McCaffrey, 2004), maintaining an apical vesicular appearance traditionally attributed to the apical recycling endosome. In the second morphology, EGFP-Rab11-FIP2 localized on smaller vesicles and along the lateral membrane. One publication has reported this alternate morphology in A431 cells in response to treatment with EGF (Lindsay and McCaffrey, 2004). Because we isolated multiple individual clones that exhibited both morphologies, we sought to understand the implications of this diversity. One difference between this and our previous study (Hales et al., 2001) was the use of T23 MDCK cells as the parental line (Barth et al., 1997) instead of MDCK II cells. The T23 clone incorporates both the stable expression of polymeric IgA receptor (pIgR) and the tetracycline-repressible transactivator tTA into MDCK II cells. When we compared MDCK II cells transiently expressing EGFP-Rab11-FIP2 with T23 cells expressing EGFP-Rab11-FIP2, the difference in distribution was apparent. The transiently transfected MDCK II cells displayed our previously published distribution for EGFP-Rab11-FIP2, whereas the T23 cells stably expressing EGFP-Rab11-FIP2 displayed both morphologies (Figure 3A). On initiation of pIgA trafficking, the cells changed to the more traditional apical recycling endosomal morphology (Figure 3B). These dynamic localizations suggest an additional new role for Rab11-FIP2 beyond the recycling pathway.
Figure 3.
Distribution of EGFP-Rab11-FIP2 in stably overexpressing cell lines. (A) T23 cells stably expressing EGFP-Rab11-FIP2 exhibit two different morphologies. White arrowhead indicates a cell with a more diffuse distribution of EGFP-Rab11-FIP2. (B) Transiently transfected MDCK II cells show only a punctuate subapical distribution of EGFP-Rab11-FIP2. (C) T23 cells expressing EGFP-Rab11-FIP2 in the presence or absence of pIgA (red in merge). On induction of trafficking, the EGFP-Rab11-FIP2 exhibits dynamic relocalization to the more traditional recycling system. EGFP images are from two different z-positions in the same confocal stack, as indicated in the x-y orthogonal view, to enhance the visualization of both morphologies.
Rab11-FIP2 Proteins Localize to the Subapical Domain
Rab11-FIP2 was initially characterized as an interacting protein for Rab11a (Hales et al., 2001). Rab11a has traditionally been associated with trafficking through the plasma membrane recycling system (Casanova et al., 1999). Therefore, we examined the general effect of overexpression of EGFP-Rab11-FIP2 and it nonphosphorylatable mutant on the distribution of the recycling system components in our inducible cell lines. The phenotype observed in each of these lines was most prominent in polarized cells grown on filters and was difficult to discern in nonpolarized cells grown on glass (our unpublished data).
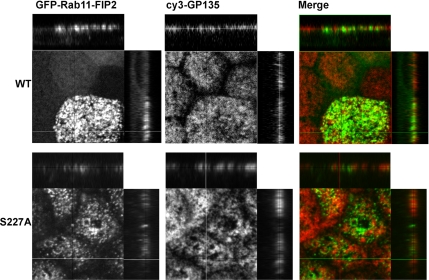
In both cell lines, EGFP-Rab11-FIP2 and EGFP-Rab11-FIP2(S227A) showed overlapping localization with endogenous Rab11a (our unpublished data). The EGFP-Rab11-FIP2 in the wild-type cell line localized near the apical membrane as shown by comparison with the apical marker GP135 (Figure 4). A portion of the EGFP-Rab11-FIP2 was interspersed with the GP135 staining, while the remainder was immediately subapical to the membrane. The nonphosphorylatable EGFP-Rab11-FIP2(S227A) population of vesicles were also located in the subapical region in comparison with GP135 (Figure 4A). Similar results were seen comparing the distribution of EGFP-Rab11-FIP2 proteins with that of ezrin (our unpublished data).
Figure 4.
Subapical distribution of EGFP-Rab11-FIP2 in stably expressing cell lines. Localization of the apical maker GP135 with EGFP-Rab11-FIP2 and EGFP-Rab11-FIP2(S227A) was compared in stable cell lines. Confocal immunofluorescence imaging of the EGFP-Rab11-FIP2 constructs with GP135 showed that EGFP-Rab11-FIP2 partially colocalized and interspersed with the apical marker GP135 (red in merge). EGFP-Rab11-FIP2(S227A) was also partially colocalized with and partially beneath the apical marker GP135 (red in merge). The x-y planes are optical sections taken 2.5 μm apart.
Effects of Phosphorylation Site Mutations on Rab11-FIP2 Recycling System Interactions
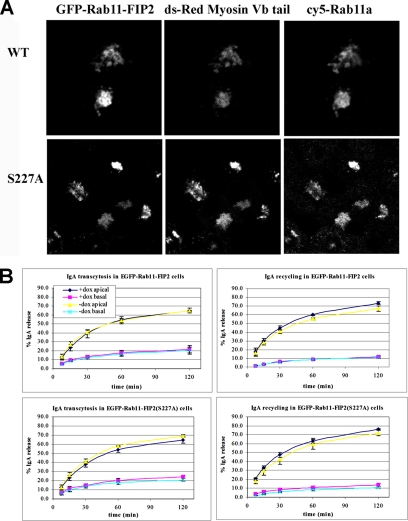
Next, we analyzed the interaction of Rab11-FIP2(S227A) with known apical recycling system components, including association with myosin Vb and Rab11a as well as Rab11-FIP2 dimerization by far Western analysis (Ducharme et al., 2005). All three interactions were maintained regardless of the mutated state of Rab11-FIP2 (our unpublished data). These interactions were confirmed by yeast two-hybrid analyses, which revealed that phosphorylation of S227 did not affect Rab11a or myosin Vb interactions with Rab11-FIP2 or Rab11-FIP2–Rab11-FIP2 dimerization (our unpublished data). We have previously demonstrated that expression of myosin Vb tail causes compaction of the Rab11a-containing apical recycling system and marked inhibition of both apical recycling and transcytosis (Lapierre et al., 2001). Transfection of EGFP-Rab11-FIP2– or EGFP-Rab11-FIP2(S227A)–expressing cell lines with DsRed2-myosin Vb tail elicited colocalization of both EGFP-Rab11-FIP2 proteins with myosin Vb tail and Rab11a in a collapsed recycling system (Figure 5A), further indicating that phosphorylation state does not affect association with myosin Vb. We also examined the effects of overexpression of wild-type EGFP-Rab11-FIP2 or EGFP-Rab11-FIP2(S227A) on trafficking of polymeric IgA in the tetracycline repressible cell lines. Both transcytosis and apical recycling of pIgA, well characterized functions of the Rab11a containing recycling system, were unaffected by the overexpression of wild-type or mutant EGFP-Rab11-FIP2 proteins (Figure 5B).
Figure 5.
Phosphorylation of Rab11-FIP2 does not alter interaction with myosin Vb tail or pIgA trafficking. (A) The phosphorylation state of Rab11-FIP2 does not affect known interactions with myosin Vb. Stable EGFP-RAB11-FIP2 cells lines were transiently transfected with Ds-Red2 myosin Vb tail. Rab11-FIP2 wild type and Rab11-FIP2(S227A) as well as Rab11a were pulled into the myosin Vb tail collapsed recycling system as expected. (B) Recycling functions of Rab11-FIP2 are not affected by phosphorylation on serine 227. Apical recycling and transcytosis of 125I-IgA were assessed in stable transfected cell lines in the presence or absence of doxycycline. Neither apical recycling nor transcytosis were affected by the phosphorylation state of Rab11-FIP2. Top graphs, EGFP-Rab11-FIP2 wild type. Bottom graphs, EGFP-Rab11-FIP2(S227A). Left-hand side shows data from transcytosis assays, whereas the right-hand side shows results from apical recycling assays. Dark blue and pink lines indicate that doxycycline was present in the media (inhibiting expression of the EGFP-chimera). Yellow and light blue lines indicate that the cells were incubated in doxycycline-free media allowing for expression of EGFP-chimera proteins. Dark blue and yellow lines indicate media was collected from the apical side of the transwell. Pink and light blue lines indicate media was collected from the basal side of the transwell. The results show no effects of EGFP-Rab11-FIP2 expression on either transcytosis or apical recycling.
Rab11-FIP2 Phosphorylation Is Important for the Formation of Calcium-dependent Junctions
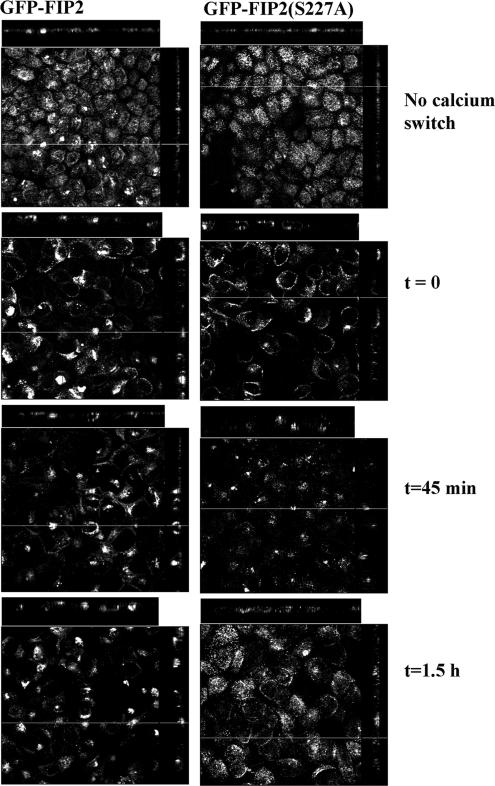
MARK2 has previously been implicated in the proper establishment of polarity in both Caenorhabditis elegans embryos (Guo and Kemphues, 1995) and MDCK cells (Bohm et al., 1997; Cohen and Musch, 2003). Thus, we hypothesized that Rab11-FIP2 phosphorylation may also impact on polarity. We assessed the effect of Rab11-FIP2 phosphorylation mutants on the reestablishment of polarity after calcium switch. Cell polarity was disrupted by switching confluent cells to calcium-free media overnight. Cells were then fixed at indicated time intervals after readdition of 1.8 mM calcium. These cells were stained for ZO-1 and p120 catenin (Figure 6) to assess the reestablishment of tight and adherens junctions, respectively.
Figure 6.
Distribution of EGFP-Rab11-FIP2 during calcium switch. Cell lines expressing EGFP-Rab11-FIP2 or its phosphorylation mutant were subjected to the calcium switch protocol and fixed at the indicated time points after readdition of extracellular calcium. The EGFP-Rab11-FIP2 initially moved to a lateral membrane before accumulating in a tight spot internally. As the junctions reformed, the construct localized to the apical membrane as seen for those cells not subject to the switch. EGFP-Rab11-FIP2(S227A) initially localized in subapical rings. As the junctions reformed, the construct was less compact and became diffusely apical. The x-y planes are compiled stacks of optical sections taken 2.5 μm apart.
EGFP-Rab11-FIP2 wild type exhibited dynamic movements during the reestablishment of polarity. After incubation overnight in calcium-free media, EGFP-Rab11-FIP2 localized along the lateral membrane and in perinuclear pools. After readdition of calcium, Rab11-FIP2 initially moved to perinuclear pools where it colocalized with Rab11a (our unpublished data). Over time, EGFP-Rab11-FIP2 redistributed near the apical membrane, a pattern similar to that in cells that did not undergo a calcium switch (Figure 6). These cells did reform proper junctions as measured by immunofluorescence staining for both p120 catenin and ZO-1 localization by 6 and 3 h, respectively, after reestablishment of normal extracellular calcium (Figure 7, A and B). Similar results were seen with the tight junction marker occludin as well as adherens junction proteins β-catenin and E-cadherin (our unpublished data).
Figure 7.
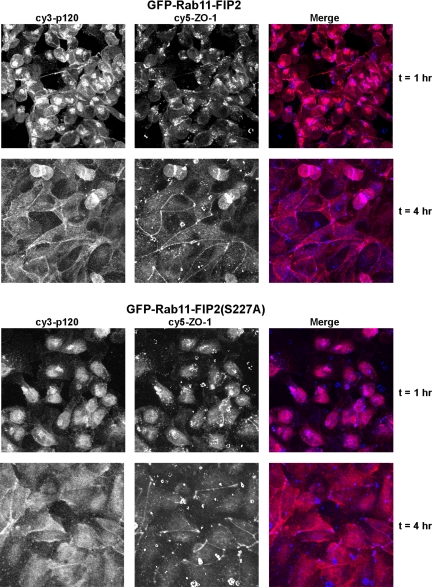
Phosphorylation of Rab11-FIP2 is necessary for the proper reestablishment of p120 at adherens junctions but not ZO-1 at tight junctions. Cell lines expressing EGFP-Rab11-FIP2 or its phosphorylation mutant were subjected to the calcium switch protocol and fixed at the indicated time points. (A) Z sections from cells in the calcium switch experiment stained for p120 catenin. Note the delayed establishment of p120 localization in Rab11-FIP2(S227A) compared with wild type. p120 staining showed a normal reestablishment at the adherens junction in EGFP-Rab11-FIP2 cells grown in the presence of doxycycline (uninduced). (B) Projections of z-stacks from ZO-1 staining during the calcium switch experiment demonstrate the normal reestablishment of ZO-1 at tight junctions in all cell lines.
Interestingly, the EGFP-Rab11-FIP2(S227A) did not exhibit the same dynamic movement during the establishment of polarity. The EGFP-Rab11-FIP2(S227A) initially localized along the apical and lateral membranes after incubation with calcium-free media, often showing up as subapical rings. Moreover, after readdition of calcium, EGFP-Rab11-FIP2(S227A) was less compacted in the internal pools compared with EGFP-Rab11-FIP2. Finally, 1.5 h after the readdition of calcium, the nonphosphorylatable mutant localized diffusely near the apical surface exclusively (Figure 6). EGFP-Rab11-FIP2(S227A)–expressing cells did not reestablish proper adherens junctions as assessed by a lack of p120 catenin localization to the lateral junctions by 6 h after the readdition of calcium (Figure 7A). This cell line did relocalize ZO-1 by 3 h after calcium addition (Figure 7B). When calcium switch was performed in the presence of doxycycline to block expression of EGFP-Rab11-FIP2(S227A), both p120 catenin and ZO-1 relocalized to their junctions after readdition of calcium by 3 h (Figure 7, A and B), as seen previously (Gumbiner et al., 1988; Straight et al., 2004).
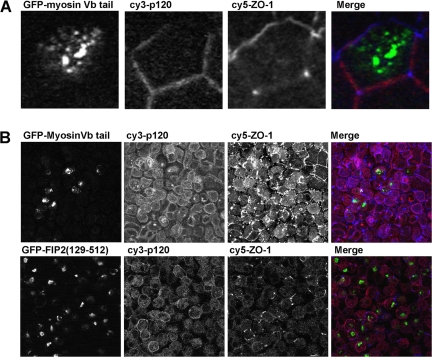
A recent report suggests that the Rab11a pathway is involved in the trafficking of newly synthesized E-cadherin (Lock and Stow, 2005). Therefore, it was possible that the Rab11a pathway was involved in the internalization or recycling of junctional constituents. Nevertheless, we did not observe colocalization of any junctional protein with Rab11-FIP2 during the calcium switch assay. We therefore examined the localization of junctional makers in cells expressing the previously characterized dominant-negative recycling system trafficking mutants EGFP-myosin Vb tail (Figure 8A) and EGFP-Rab11-FIP2(129–512) (our unpublished data). Neither dominant-negative construct showed colocalization with p120 or ZO-1. To confirm that a Rab11a-dependent pathway using Rab11–FIP2 pathway was not involved during trafficking, we examined the effects of these dominant-negative mutants in the calcium switch assay. No p120 was observed in myosin Vb tail or Rab11-FIP2(129–512) containing collapsed recycling systems (Figure 8B). Furthermore, throughout the time course of the calcium add-back, we did not observe localization of any junctional marker within the inhibited recycling system (our unpublished data).
Figure 8.
Dominant-negative Rab11a recycling system trafficking mutants do not colocalize with junctional proteins p120 or ZO-1. (A) X-Y confocal sections showing dual labeling of p120 catenin (Cy 3; pseudocolored red) and ZO-1 (Cy5; pseudocolored blue) in EGFP-myosin Vb tail cells. No colocalization was found between the junctional markers and EGFP-myosin Vb tail. (B) Projections showing dual labeling of p120 catenin (Cy 3; pseudocolored red) and ZO-1 (Cy5; pseudocolored blue) in EGFP-Rab11-FIP2(129-512) and EGFP-myosin Vb tail cells after calcium switch at t = 0. Junctional markers do not colocalize with the dominant-negative inhibitors of the Rab11a apical recycling system. No localization with the collapsed recycling system was seen at any time point after reestablishment of extracellular calcium.
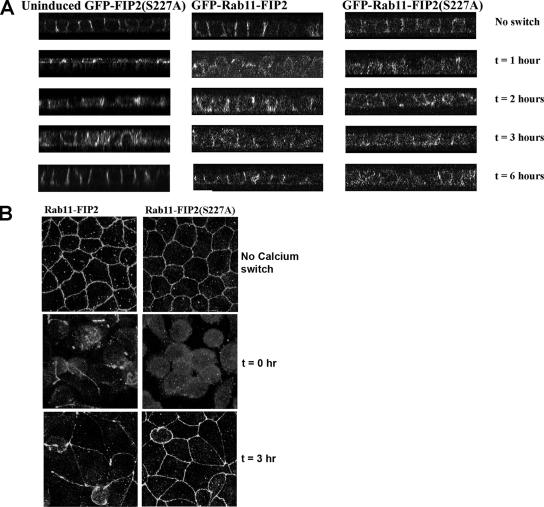
As an independent method of assessing junction formation, we trypsinized cells and replated them at high density on laminin-coated filters. We examined the cells at fixed time points to assess the establishment of junctions by staining for p120 and ZO-1. Cells overexpressing EGFP-Rab11-FIP2 wild type reestablished normal junctions by 4 h after replating, whereas the EGFP-Rab11-FIP2(S227A)–expressing cells did not (Figure 9). ZO-1 localization was reestablished similarly in both lines by 4 h. Thus, the inability to phosphorylate serine 227 of Rab11-FIP2 delays adherens junction formation in multiple assays.
Figure 9.
Phosphorylation of Rab11-FIP2 is necessary for the proper reestablishment of polarity in a replating assay. Cell lines expressing EGFP-Rab11-FIP2 or EGFP-Rab11-FIP2(S227A) were trypsinized and replated on laminin-coated filters. The cells were fixed at the indicated time points and stained with p120 (pseudocolored red) and ZO-1 (pseudocolored blue). These overlays show x-y planes after replating. Although ZO-1 was reestablished at tight junctions in both lines, EGFP-Rab11-FIP2(S227A)–expressing cells demonstrated a deficit in p120 localization 4 h after replating.
DISCUSSION
Traditionally, the apical recycling system is marked by the presence of Rab11a. Rab11a and its known family of interacting proteins (Rab11-FIPs) regulate trafficking through the apical recycling system. For example, removal of the C2 domain from Rab11-FIP2 causes a disruption of the trafficking of pIgA through the apical recycling system (Hales et al., 2002). In addition, we have shown that myosin Vb associates with the Rab11a and Rab11-FIP2 to form a ternary regulatory complex for apical recycling (Hales et al., 2002). However, Rab11-FIP2 also has a role in cellular function beyond apical recycling. We and others have previously shown that a subset of the Rab11-FIP2 is not with the recycling system in quiescent cells (Hales et al., 2001; Cullis et al., 2002). Rab11-FIP2 associates with EH domain proteins such as Reps1, assisting in receptor-mediated endocytosis (Cullis et al., 2002). In the initial characterization of Rab11-FIP2, we found that not all Rab11-FIP2 colocalized with Rab11a. This was apparent particularly when cells were treated with the microtubule-stabilizing drug taxol. Taxol treatment of polarized cells caused Rab11a-containing recycling vesicles to relocate to apical corners near the tight junction complex (Casanova et al., 1999). However, taxol treatment of cells caused Rab11-FIP2 to localize at both the apical corner and laterally (Hales et al., 2001), suggesting that not all of Rab11-FIP2 was involved in the Rab11a-containing recycling pathway. This lateral localization is similar to that seen after the removal of calcium in the EGFP-Rab11-FIP2(S227A) cells. Importantly, in these studies, we have demonstrated that phosphorylation of Rab11-FIP2 on serine 227 by MARK2 is not involved in the traditional apical recycling pathway, but is instead involved in the establishment of epithelial cell polarity.
The recycling system is a dynamic tubular network with multiple regulators and signaling pathways. Present models suggest that the assembly of multiple proteins associating with Rab11a regulates trafficking through the recycling system. Although few of the Rab proteins are directly regulated by protein phosphorylation (van der Sluijs et al., 1992), phosphorylation of Rab-interacting proteins has emerged as an important regulatory mechanism. Thus, the Rab3-interacting protein rabphilin-3 is phosphorylated both by protein kinase A and calmodulin-dependent kinase II (Kato et al., 1994; Numata et al., 1994). Rabphilin-3 phosphorylation reduces its affinity for membranes (Foletti et al., 2001; Lonart and Sudhof, 2001). Rab11-FIP5 (pp75/Rip11) was originally described as an autoantigen phosphoprotein, and as in the case of rabphilin-3, phosphorylation seemed to alter subcellular localization (Wang et al., 1999; Prekeris et al., 2000). The kinase activity responsible for Rab11-FIP5 phosphorylation is unclear, although we have not found phosphorylation of Rab11-FIP5 by MARK2 (our unpublished data). In the present report, we have found that MARK2 can phosphorylate Rab11-FIP2. Nevertheless, mutation of serine 227 does not alter apical membrane recycling or transcytosis. Thus, MARK2 seems to regulate Rab11-FIP2 functions distinct from its role in the recycling system.
MARK2 is a member of the PAR (partitioning-defect) family originally characterized as PAR1 in C. elegans (Kemphues et al., 1988). Since that time, the majority of the work on this kinase has related to phosphorylation of Tau, MAP2C, and MAP4 on their microtubule binding domains, which results in disruption of the microtubule network (Drewes et al., 1995; Drewes et al., 1997; Ebneth et al., 1999). In Drosophila, MARK2 regulates the density, stability, and apicobasal organization of microtubules by regulating the microtubule plus ends (Doerflinger et al., 2003). Although MARK2 phosphorylation of Rab11-FIP2 does not seem to affect the traditional Rab11a trafficking pathway, previous studies have implicated MARK2 in the breakdown of the microtubule network. MARK2 phosphorylation events may also be regulating the movement of vesicles along the microtubule network.
The Schizosaccharomyces pombe family MARK2 member kin1 is necessary to establish the rod-shaped morphology and for progression of cytokinesis (Drewes and Nurse, 2003). This kinase also plays a role in the establishment of polarity. MARK2 is essential for epithelial specific microtubule arrays in polarized cells (Cohen et al., 2004). Overexpression of the kinase inhibits apical/basolateral polarization in MDCK cells partially by altering apical protein transport. Interestingly, cells overexpressing MARK2 form a polarity-axis parallel to the substratum (Cohen and Musch, 2003). Recently, the yeast homologue was reported to interact with components of the exocytic machinery, including the yeast Rab family member Sec4 (Elbert et al., 2005), thus establishing a precedent for the involvement of this kinase family in Rab-related trafficking. The results presented here indicate that phosphorylation of Rab11-FIP2 by MARK2 is necessary for the proper localization of junctional proteins. The importance of the phosphorylation site was most obvious when manipulating the system through a calcium switch assay. The reestablishment of polarity was grossly delayed in cells overexpressing EGFP-Rab11-FIP2(S227A). p120 catenin did not organize into junctions when cells were examined up to 8 h after the addition of normal media. Similar defects in the establishment of polarity in EGFP-Rab11-FIP2(S227A)–expressing cells were also observed in replating assays. Nevertheless, we did not observe any difference in transepithelial resistance (TER) between cells expressing EGFP-Rab11-FIP2, EGFP-Rab11-FIP2(S227A), or uninduced cell lines (our unpublished data). However, cells that have ZO-1 knocked out also do not have reduced TER, suggesting that other junctional proteins such as ZO-2 can compensate for a lack of junctional ZO-1 (Umeda et al., 2004).
Previous work by Parkos and colleagues demonstrated that junctional proteins including cadherin, p120, and ZO-1 are internalized in response to calcium depletion (Ivanov et al., 2004). During Drosophila cellularization, cadherin is initially localized along the entire lateral membrane and then becomes sequestered into both apical lateral and basal lateral compartments; finally, it is restricted to the apically oriented adherens junction (Le Bivic, 2005). This dual localization found during mid-cellularization is reminiscent of the localization of p120 observed in cells overexpressing EGFP-Rab11-FIP2(S227A), suggesting that these cells may never achieve proper adherens junctions. The role of Rab11-FIP2 in adherens junction protein trafficking is supported by a recent study by Stow and colleague demonstrating that newly synthesized cadherin requires functional Rab11 for proper localization (Lock and Stow, 2005). However, our work suggests that establishment of junctions is not blocked by expression of previously characterized inhibitors of the apical recycling system, Rab11-FIP2(129-512) and Myosin Vb tail. Another Rab11-FIP, Rab Coupling Protein (RCP), was also recently found to relocalize with Rab11a to the lateral membrane in the presence of calpeptin, a potent blocker of calcium-mediated calpain actions (Marie et al., 2005). In the presence of calpain inhibitor, RCP relocalized in a pattern similar to that for Rab11-FIP2 during the initial stages of junctional reformation after readdition of calcium. Interestingly, RCP also has the MARK2 phosphorylation site and is phosphorylated by MARK2 in vitro, suggesting a possible common regulatory mechanism for both RCP and Rab11-FIP2 in response to calcium.
In summary, the present study has defined a novel consensus site for phosphorylation by MARK2 of Rab11-FIP2. Manipulation of the serine 227 phosphorylation site in Rab11-FIP2 alters the establishment of polarity in polarized MDCK cells, but it does not affect traditional recycling system trafficking pathways. Previous investigations have suggested that MARK2 plays an important role in the establishment of polarity. Although the exact compendium of phosphorylation substrates for MARK2 is unclear, it seems likely that a cascade of phosphorylation events on a number of proteins may ensue from activation of MARK2. These phosphorylation events may be directly active (e.g., promoting cytoskeletal interactions for proteins destined for the adherens junctions) or inhibitory for functions that compete with polarity (e.g., general nonspecific trafficking to all plasma membrane surfaces). At this time, we cannot determine the exact role of Rab11-FIP2 in these processes, but these studies do define a previously unrecognized class of MARK2 substrates that likely act as components of a coordinated cascade of events that lead to the orderly establishment of polarity. All of these findings demonstrate that Rab11-FIP2 plays an important role in regulating epithelial cell polarity, distinct from its function in the Rab11a-containing recycling pathways.
ACKNOWLEDGMENTS
We thank Dr. Roy Zent for the gift of antibody reagents. Confocal fluorescence imaging was performed in part through the use of the VUMC Cell Imaging Shared Resource, supported by National Institutes of Health (NIH) Grants CA-68485, DK-20593, DK-58404, and HD15052. We thank Vanderbilt University for institutional support of the Proteomics Laboratory in the Mass Spectrometry Research Center through the Academic Venture Capital Fund. We thank Kenya Avant, Cathy Caldwell, Salisha Hill, Dr. Karen Hobdy-Henderson, Dr. Joseph Roland, and Dr. Min Jin for support of these studies. This work was supported by NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grants DK-48370 and DK-43405 (to J.R.G.) and by NIH NIDDK Grant R01 DK-51970 (to G. A.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0736) on June 14, 2006.
REFERENCES
- Apodaca G., Katz L. A., Mostov K. E. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J. Cell Biol. 1994;125:67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A.I.M., Pollack A. L., Altschuler Y., Mostov K. E., Nelson W. J. NH2-terminal deletion of β-catenin results in stable colocalization of mutant β-catenin with adenomatous polyposis coli protein and altered MDCK cell adhesion. J. Cell Biol. 1997;136:693–706. doi: 10.1083/jcb.136.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. D., Goldenring J. R., Tang L. H., Lewis J. J., Padfield P., Jamieson J. D., Modlin I. M. Redistribution of 23 kDa tubulovesicle-associated GTP-binding proteins during parietal cell stimulation. Biochem. J. 1991;279:43–48. doi: 10.1042/bj2790043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm H., Brinkmann V., Drab M., Henske A., Kurzchalia T. V. Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr. Biol. 1997;7:603–606. doi: 10.1016/s0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]
- Breitfeld P. P., Casanova J. E., Harris J. M., Simister N. E., Mostov K. E. Expression and analysis of the polymeric immunoglobulin receptor in Madin-Darby canine kidney cells using retroviral vectors. Methods Cell Biol. 1989;32:329–337. doi: 10.1016/s0091-679x(08)61178-4. [DOI] [PubMed] [Google Scholar]
- Casanova J. E., Wang X., Kumar R., Bhartur S. G., Navarre J., Woodrum J. E., Altschuler Y., Ray G. S., Goldenring J. R. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 1999;10:47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Brennwald P. J., Rodriguez-Boulan E., Musch A. Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J. Cell Biol. 2004;164:717–727. doi: 10.1083/jcb.200308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Musch A. Apical surface formation in MDCK cells: regulation by the serine/threonine kinase EMK1. Methods. 2003;30:269–276. doi: 10.1016/s1046-2023(03)00033-1. [DOI] [PubMed] [Google Scholar]
- Cortes H. J., Pfeiffer C. D., Richter B. E., Stevens T. Porous ceramic bed supports for fused silica packed capillary columns used in liquid chromatography. J. High Resolut. Chromatogr. Chromatogr. Commun. 1987;10:446–448. [Google Scholar]
- Cullis D. N., Philip B., Baleja J. D., Feig L. A. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- Doerflinger H., Benton R., Shulman J. M., St Johnston D. The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development. 2003;130:3965–3975. doi: 10.1242/dev.00616. [DOI] [PubMed] [Google Scholar]
- Drewes G., Ebneth A., Preuss U., Mandelkow E. M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Drewes G., Nurse P. The protein kinase kin1, the fission yeast orthologue of mammalian MARK/PAR-1, localises to new cell ends after mitosis and is important for bipolar growth. FEBS Lett. 2003;554:45–49. doi: 10.1016/s0014-5793(03)01080-9. [DOI] [PubMed] [Google Scholar]
- Drewes G., Trinczek B., Illenberger S., Biernat J., Schmitt-Ulms G., Meyer H. E., Mandelkow E. M., Mandelkow E. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J. Biol. Chem. 1995;270:7679–7688. doi: 10.1074/jbc.270.13.7679. [DOI] [PubMed] [Google Scholar]
- Ducharme N. A., Jin M., Lapierre L. A., Goldenring J. R. Assessment of Rab11-FIP2 interacting proteins in vitro methods in enzymology. In: William C.J.D., Balch E., Hall Alan, editors. GTPases Regulating Membrane Targeting and Fusion. San Diego: Academic Press; 2005. pp. 706–715. [DOI] [PubMed] [Google Scholar]
- Ebneth A., Drewes G., Mandelkow E. M., Mandelkow E. Phosphorylation of MAP2c and MAP4 by MARK kinases leads to the destabilization of microtubules in cells. Cell Motil. Cytoskeleton. 1999;44:209–224. doi: 10.1002/(SICI)1097-0169(199911)44:3<209::AID-CM6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Elbert M., Rossi G., Brennwald P. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol. Biol. Cell. 2005;16:532–549. doi: 10.1091/mbc.E04-07-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G. H., Lapierre L. A., Goldenring J. R., Richmond A. Differential regulation of CXCR2 trafficking by Rab GTPases. Blood. 2003;101:2115–2124. doi: 10.1182/blood-2002-07-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G.-H., Lapierre L. A., Goldenring J. R., Sai J., Richmond A. Rab11-Family Interacting Protein 2 and Myosin Vb are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol. Biol. Cell. 2004;15:2456–2469. doi: 10.1091/mbc.E03-09-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletti D. L., Blitzer J. T., Scheller R. H. Physiological modulation of rabphilin phosphorylation. J. Neurosci. 2001;21:5473–5483. doi: 10.1523/JNEUROSCI.21-15-05473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B., Stevenson B., Grimaldi A. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J. Cell Biol. 1988;107:1575–1587. doi: 10.1083/jcb.107.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hales C. M., Griner R., Hobdy-Henderson K. C., Dorn M. C., Hardy D., Kumar R., Navarre J., Chan E. K., Lapierre L. A., Goldenring J. R. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hales C. M., Vaerman J. P., Goldenring J. R. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- Hansen B. T., Davey S. W., Ham A. J., Liebler D. C. P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. J. Proteome Res. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- Ivanov A. I., Nusrat A., Parkos C. A. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol. Biol. Cell. 2004;15:176–188. doi: 10.1091/mbc.E03-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Sasaki T., Imazumi K., Takahashi K., Araki K., Shirataki H., Matsuura Y., Ishida A., Fujisawa H., Takai Y. Phosphorylation of Rabphilin-3A by calmodulin-dependent protein kinase II. Biochem. Biophys. Res. Commun. 1994;205:1776–1784. doi: 10.1006/bbrc.1994.2875. [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W., Jr., Mercer J. A., Bahler M., Goldenring J. R. Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. L., Yap A. S., Stow J. L. Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 1999;146:219–232. [PMC free article] [PubMed] [Google Scholar]
- Le Bivic A. E-cadherin-mediated adhesion is not the founding event of epithelial cell polarity in Drosophila. Trends Cell Biol. 2005;15:237–240. doi: 10.1016/j.tcb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Licklider L. J., Thoreen C. C., Peng J., Gygi S. P. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal. Chem. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- Lindsay A. J., Hendrick A. G., Cantalupo G., Senic-Matuglia F., Goud B., Bucci C., McCaffrey M. W. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- Lindsay A. J., McCaffrey M. W. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane 10.1242/jcs.01280. J. Cell Sci. 2004;117:4365–4375. doi: 10.1242/jcs.01280. [DOI] [PubMed] [Google Scholar]
- Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Lock J. G., Stow J. L. Rab11 in recycling endosomes regulates the sorting and basolateral transport of E-cadherin. Mol. Biol. Cell. 2005;16:1744–1755. doi: 10.1091/mbc.E04-10-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonart G., Sudhof T. C. Characterization of rabphilin phosphorylation using phospho-specific antibodies. Neuropharmacology. 2001;41:643–649. doi: 10.1016/s0028-3908(01)00126-5. [DOI] [PubMed] [Google Scholar]
- Louvard D. Apical membrane aminopeptidase appears at site of cell-cell contact in cultured kidney epithelial cells. Proc. Natl. Acad. Sci. USA. 1980;77:4132–4136. doi: 10.1073/pnas.77.7.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A., Ohtsuka T., Hotta I., Sasaki T., Takai Y. Rab11BP/Rabphilin-11, a downstream target of rab11 small G protein implicated in vesicle recycling. J. Biol. Chem. 1999;274:25517–25524. doi: 10.1074/jbc.274.36.25517. [DOI] [PubMed] [Google Scholar]
- Manza L. L., Stamer S. L., Ham A.-J. L., Codreanu S. G., Liebler D. C. Sample preparation and digestion for proteomic analysis using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- Marie N., Lindsay A. J., McCaffrey M. W. Rab coupling protein is selectively degraded by calpain in Ca 2+-dependent manner. Biochem. J. 2005;389:223–231. doi: 10.1042/BJ20042116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi J., Takai Y. Molecular perspective on tight-junction assembly and epithelial polarity. Adv. Drug Deliv. Rev. 2005;57:815–855. doi: 10.1016/j.addr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Numata S., Shirataki H., Hagi S., Yamamoto T., Takai Y. Phosphorylation of Rabphilin-3A, a putative target protein for Rab3A, by cyclic AMP-dependent protein kinase. Biochem. Biophys. Res. Commun. 1994;203:1927–1934. doi: 10.1006/bbrc.1994.2413. [DOI] [PubMed] [Google Scholar]
- Ojakian G., Schwimmer R. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 1988;107:2377–2387. doi: 10.1083/jcb.107.6.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R., Davies J. M., Scheller R. H. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001;276:38966–38970. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Scheller R. H. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell. 2000;6:1437–1448. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- Straight S. W., Shin K., Fogg V. C., Fan S., Liu C.-J., Roh M., Margolis B. Loss of PALS1 expression leads to tight junction and polarity defects. Mol. Biol. Cell. 2004;15:1981–1990. doi: 10.1091/mbc.E03-08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K., Haucke V. Clathrin-mediated endocytosis: membrane factors pull the trigger. Trends Cell Biol. 2001;11:385–391. doi: 10.1016/s0962-8924(01)02082-7. [DOI] [PubMed] [Google Scholar]
- Umeda K., Matsui T., Nakayama M., Furuse K., Sasaki H., Furuse M., Tsukita S. Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J. Biol. Chem. 2004;279:44785–44794. doi: 10.1074/jbc.M406563200. [DOI] [PubMed] [Google Scholar]
- van der Sluijs P., Hull M., Huber L. A., Male P., Goud B., Mellman I. Reversible phosphorylation–dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J. 1992;11:4379–4389. doi: 10.1002/j.1460-2075.1992.tb05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli L. A., Lah J. J., Fang G., Goldenring J. R., Levey A. I. Rab11a and myosin Vb regulate recycling of the M4 muscarinic acetylcholine receptor. J. Neurosci. 2002;22:9776–9784. doi: 10.1523/JNEUROSCI.22-22-09776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. M., Lindsay A. J., Hendrick A. G., McCaffrey M. W. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem. Biophys. Res. Commun. 2002;299:770–779. doi: 10.1016/s0006-291x(02)02720-1. [DOI] [PubMed] [Google Scholar]
- Wang D., Buyon J. P., Zhu W., Chan E. K. Defining a novel 75-kDa phosphoprotein associated with SS-A/Ro and identification of distinct human autoantibodies. J. Clin. Investig. 1999;104:1265–1275. doi: 10.1172/JCI8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Zeng J., et al. Identification of a putative effector protein for rab11 that participates in transferrin recycling. Proc. Natl. Acad. Sci. USA. 1999;96:2840–2845. doi: 10.1073/pnas.96.6.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]