Abstract
RNA interference (RNAi) is a conserved gene-silencing phenomenon that can be triggered by delivery of double-stranded RNA (dsRNA) to cells and is a widely exploited technology in analyses of gene function. Although a number of proteins that facilitate RNAi have been identified, current descriptions of RNAi and interrelated mechanisms are far from complete. Here, we report that the Caenorhabditis elegans gene haf-6 is required for efficient RNAi. HAF-6 is a member of the ATP-binding cassette (ABC) transporter gene superfamily. ABC transporters use ATP to translocate small molecule substrates across the membranes in which they reside, often against a steep concentration gradient. Collectively, ABC transporters are involved in a variety of activities, including protective or barrier mechanisms that export drugs or toxins from cells, organellar biogenesis, and mechanisms that protect against viral infection. HAF-6 is expressed predominantly in the intestine and germline and is localized to intracellular reticular organelles. We further demonstrate that eight additional ABC genes from diverse subfamilies are each required for efficient RNAi in C. elegans. Thus, the ability to mount a robust RNAi response to dsRNA depends upon the deployment of two ancient systems that respond to environmental assaults: RNAi mechanisms and membrane transport systems that use ABC proteins.
INTRODUCTION
When introduced into eukaryotic cells, double-stranded RNA (dsRNA) elicits a foreign genome response that is orchestrated by RNA interference (RNAi) mechanisms (Fire et al., 1998). RNAi results in the directed decay of mRNAs with sequences related to the input dsRNA (Montgomery et al., 1998), and in some cases, transcriptional-level silencing of genes with similar sequence (Bernstein and Allis, 2005; Grishok, 2005). RNAi mechanisms are conserved in eukaryotes and protect organisms from potentially deleterious nucleic acids such as those found in transposons and viruses (Robert et al., 2004; Vastenhouw and Plasterk, 2004; Li and Ding, 2005). Many of the components of the RNAi mechanism are shared with related RNA-directed silencing pathways, and the collective silencing mechanisms have vital functions in cells and in development. For example, RNAi and related pathways participate in the establishment and proper functioning of heterochromatin, in regulation of gene expression, and in posttranscriptional regulation of cellular mRNAs by microRNAs (miRNAs) and endogenous small interfering RNAs (siRNAs) (Grewal and Rice, 2004; Noma et al., 2004; Du and Zamore, 2005; Lee et al., 2006). Some of the different pathways use common proteins that may be limiting in availability, and emerging evidence suggests competition between RNA-directed silencing mechanisms under certain conditions (Duchaine et al., 2006; Lee et al., 2006). The interrelatedness of RNAi mechanisms and the number of RNAi components with essential roles in development necessitate a better and broader understanding of RNAi mechanisms and pathways.
A number of conserved proteins that effect gene silencing in response to dsRNA have been uncovered through analyses of Caenorhabditis elegans mutants that are defective in RNAi. Altogether, the genes function in the processing of exogenous dsRNA into siRNAs; in processing of endogenous precursor RNAs into siRNAs, miRNAs, and tiny noncoding RNAs; in RNA-dependent RNA polymerase-dependent activities associated with RNAi responses; in inhibition of RNAi; in cellular mechanisms that allow passive entry of small dsRNA molecules into cells; in mechanisms that presumably use endocytosis machinery to allow cellular entry of dsRNAs; and in mechanisms that modify chromatin (Ketting et al., 1999; Tabara et al., 1999; Smardon et al., 2000; Knight and Bass, 2001; Sijen et al., 2001; Dudley et al., 2002; Simmer et al., 2002; Tabara et al., 2002; Tijsterman et al., 2002a,b, 2004; Winston et al., 2002; Caudy et al., 2003; Timmons et al., 2003; Kennedy et al., 2004; Grishok et al., 2005; Kim et al., 2005; Tops et al., 2005; Wang et al., 2005). Although a number of genes that function in RNAi have been identified using C. elegans, the list is far from complete.
C. elegans remains an important genetics tool for the elucidation of RNAi mechanisms in multicellular animals. Genetic screening is facilitated by a number of protocols that are available for dsRNA delivery. dsRNA can be introduced to C. elegans via feeding (allowing animals to ingest bacteria engineered to express dsRNA) (Timmons et al., 2001), by soaking animals in solutions containing dsRNA (Tabara et al., 1998), and by injecting animals with dsRNA (Fire, 1999), and all methods can elicit a systemic RNAi response in treated animals and their progeny. dsRNA delivered to animals using bacterial “food” is introduced at a relatively lower dosage, allowing for the identification of mutants with subtle or tissue-specific RNAi defects, or allowing for the identification of hypomorphic mutations in otherwise essential genes.
Here, we demonstrate that the Caenorhabditis elegans ATP-binding cassette (ABC) transporter gene haf-6 is required for efficient RNAi and that eight additional ABC transporters have similar roles in RNAi.
MATERIALS AND METHODS
C. elegans Strains
Worm husbandry and genetic crosses were performed according to standard protocols (Brenner, 1974). The following strains, used in mapping, were generous gifts from Dr. Andrew Fire (Stanford University, Stanford, MA): LGI: dpy-5(e61) unc-54(e1092); LGIV: unc-17(e245) dpy-4(e1166); LGV: dpy-11(e224) unc-60(e723); LGII: dpy-10(e128) unc-52(e669); and LGIII: dpy-18(e449) unc-32(e189).
The haf-6(ne335) strain was a generous gift from Craig Mello (University of Massachusetts Medical School, Worcester, MA) (Tabara et al., 1999). The following mutants were used in phenotypic analyses and were obtained from the C. elegans Genetics Center (University of Minnesota, Minneapolis, MN) or the National Bioresources Project in Japan: abt-1(tm703); abt-2(ok669); abt-4(ok633); C05D10.3(tm688); C56E6.1(ok865); cft-1(ok1180); C16C10.12(2 alleles: ok927, ok962); ced-7(5 alleles: n2690, n1892, n2094, n1996, n1997); F02E11.1(ok1007); F19B6.4(ok806); haf-1(ok705); haf-2(gk13); haf-3(ok1086); haf-4(2 alleles: gk240, ok1042); haf-5(2 alleles: gk155, gk161); haf-7(gk46); haf-8(gk12); haf-9(gk23); mrp-1(pk89); mrp-3(ok955); mrp-4(ok1095); mrp-6(ok1027); mrp-8(ok1360); NL130: pgp-1(pk17), pgp-3(pk18); NL132: pgp-1(pk17); NL152: pgp-1(pk17), pgp-3(pk18), mrp-1(pk89); pgp-10(ok991); pgp-11(tm333); pgp-12(gk19); pgp-13(ok747); pgp-15(ok987); pgp-2(gk114); NL131: pgp-3(pk18); pgp-4(gk16); pgp-5(ok856); pgp-7(ok528); pmp-1(ok773); pmp-3(ok1087); pmp-4(ok396); RB1047: deletion in pgp-6 and pgp-7(ok994); T26A5.1(ok882); T27E9.7(ok771); F18E2.2(ok830); Y42g9a.6(ok812); and Y49e10.9(ok1044).
Feeding Strains and Feeding-based Assays for RNAi Defects
“Feeding plates” harboring bacteria engineered to express dsRNA were prepared using HT115(DE3) host bacteria as described previously (Timmons et al., 2001; Hull and Timmons, 2004). Plasmid L4440 was the cloning vector for plasmids transformed into this bacterial strain (Timmons and Fire, 1998). HT115(DE3) strains harboring a pop-1 cDNA insert in L4440 (pop-1 food) targets the TCF/LEF1 transcription factor and produces sterility in young animals reared on this food. unc-22 food (Timmons and Fire, 1998) targets the muscle-specific unc-22 transcript and induces a twitching phenotype. Other feeding strains were obtained from MRC/Geneservice RNAi library (Kamath et al., 2003), or they were generated by inserting PCR-generated fragments of cDNAs into L4440.
RNAi was induced using the feeding protocol by placing animals as L1/L2 larvae onto fresh plates prepared as described previously (Timmons et al., 2001; Hull and Timmons, 2004). In each experiment, control feeding plates with wild-type worms and OP50 plates with mutant worms were used to monitor for effectiveness of the delivery protocol and for potential environmental contributions to the phenocopy, respectively. Assays were performed at 15, 20, and 25°C. pop-1(RNAi) was assayed by tabulating the number of viable progeny produced per adult on the feeding plate. This value can be affected by temperature, the time frame of the experiment, and the staging of the animals at the time of placement onto the feeding plate; therefore, the value is comparative, not absolute. In each experiment, RNAi phenocopies in mutants were closely compared with similarly treated wild-type worms. Comparative values for unc-22(RNAi) activity were obtained by calculating the percentage of F1 animals that twitch.
Positional Cloning of haf-6(ne335), Transgene Rescue of RNAi Defects, and Complementation Tests with smg-2
Ingestion of pos-1 food by wild-type adults can induce lethality in resulting embryos—the presence of viable progeny is evidence of an RNAi defect. haf-6(ne335) mutants were isolated in a screen for RNAi-defective animals using pos-1 food (Tabara et al., 1999). haf-6(ne335) is recessive, and ne335 is the only haf-6 allele to date. Many RNAi-defective strains have no other phenotypes, and this raises the potential that a second uncharacterized mutation might contribute to RNAi defects in some strains. To assay for, or guard against, this possibility, we produced different versions of haf-6(ne335) strains by outcrossing ne335 to various wild-type stocks from different origins (e.g., N2 stocks obtained from the C. elegans Genetics Center versus N2 stocks maintained for several years in the laboratory of Andrew Fire). We selected for robust RNAi activity, viability, and fertility at 25°C, and for normal brood sizes in wild-type animals before using them in outcrosses. In initial stages of mutant characterization, feeding assays were used to select for ne335 homozygotes; later, a PCR strategy was used to identify mutant homozygotes from additional outcrosses. Primers 683 GCCATCCTCTCAGCCTAC and 684 CCACCCACGCTCTTACATG were used in genotyping. The differently outcrossed ne335 strains behaved identically in our assays.
A rough genetic map was obtained using two- and three-factor mapping strategies and genetically marked strains. Heterozygous F1 cross-progeny were set aside and allowed to produce F2 progeny. Single F2 progeny were placed at L2/L3 stage onto pop-1 feeding plates. Homozygous haf-6 animals gave rise to F3 progeny, and the presence of the marker phenotype was noted for estimations of linkage or recombination frequency. The ne335 mutation mapped to the left end of chromosome I. A series of overlapping yeast artificial chromosomes (YACs) corresponding to this region were obtained from the C. elegans Sequencing Consortium (Sanger Institute, Cambridge, United Kingdom). YAC DNA was obtained by preparing total genomic DNA from yeast. Purified DNA was injected into ne335 mutants and rescue of RNAi defects was assayed using pop-1 and elt-2 food.
For complementation testing, crosses were performed using one hermaphrodite per mating on standard OP50 plates. The presence of 50% males in the progeny indicated a successful cross. Complementarity was assessed in reciprocal crosses: 1) haf-6(ne335) hermaphrodites were mated with smg-2(e2008); him-5(e1417) males, and 2) smg-2(e2008) hermaphrodites were mated with haf-6(ne335); him-5(e1417); ccIs8160 [rpL28::gfp] males. The green fluorescent protein (GFP)-expressing transgene (generously provided by Dr. A. Fire) (Timmons et al., 2003) facilitated detection of cross-progeny. Cross-progeny were transferred as L1/L2 larvae onto pop-1 feeding plates; unc-22 food elicited similar results. Complementation between smg-2 and haf-6 was tested at 20 and 25°C, with similar results.
Plasmids and Transgenic Strains
Transgene lines were established as complex arrays to better ensure expression in the germline (Kelly et al., 1997). DNA sequences were linearized before injecting. A plasmid harboring a dominant mutation in the rol-6 gene was used as transformation marker; bacteriophage lambda DNA or C. elegans genomic DNA was used as carrier.
DNA Segments Used in Plasmid Cloning.
1) The haf-6 cDNA extends from the initiator ATG, lacks a 5′-untranslated region (UTR), includes its 3′-UTR, and was generated by reverse transcription (RT)-PCR using wild-type RNA as template and primers 472 ATGTCAATTCTATCTAAACTATCCC and 473 CTTTAATATTTCTACTAAAATTTAAA. 2) A smg-2 cDNA, lacking in 5′-UTR sequences, harbors an initiator ATG and 3′-UTR and was amplified by RT-PCR using wild-type RNA as template and primers 488 ATGGACGATTCGGACGACGAATATTCGAGAAGCCACGGGG and 489 AATTTTTCTAAAAATTTCGGATTTTCGAGAGAAAATCAAGC. 3) The ends of the haf-6/smg-2 intergenic region are bounded by the 5′-UTRs corresponding to each gene. This fragment was obtained by PCR amplification using wild-type DNA as template and primers 501 TTTACTAGTGATATCTAAAAACCAGGAAAAATCAATACAAATCAA and 502 TTTTTCTAGAGCTAGCTACTGGAAGAAAAGTGCGTTTTTGAGGGGT. 4) Five hundred and forty-nine base pairs of let-858 promoter (including its 5′-UTR) were used to drive ubiquitous expression (Kelly et al., 1997). Primers 343 TTTTTGCATGCGTCGACGCTCTGAAAAACGAAAGTGGA and 344 TTTGGATCCCTTACTATAAAAAAGTTTGAATAC were used to amplify this segment from wild-type DNA. 5) The smg-2 frameshift (insertion) was generated by first assembling a plasmid with tripartite sequences composed of the smg-2 and haf-6 cDNA sequences flanking the smg-2/haf-6 intergenic region configured in the chromosomal context with cDNAs in opposite orientation. The tripartite plasmid was then digested with AgeI, which cuts once within the smg-2 cDNA (1923 base pairs downstream from ATG), and AgeI ends were filled in with T4 DNA polymerase and ligated to regenerate a circular plasmid with a four- base pair insertion in the smg-2 coding region. 6) The smg-2 deletion was generated from the tripartite plasmid by digesting the smg-2 cDNA region with Nde I and religating, resulting in the removal of base pairs 391-2866 (with ATG at position 1). 7) 2385 base pairs of myo-3 promoter sequence, including its 5′-UTR were used to drive expression in muscle (Fire et al., 1998). Primers 497 ttagcggatccgctagcAAGCTTGGGCTGCA GGTCGGC and 498 ttagcggatccgcggccgcTCTAGATGGATCTAGTGGTCGTGG were used to amplify the promoter. DNA sequencing confirmed each fragment was error-free. Fragments were cloned into standard vectors and combined to produce the plasmids described using standard cloning techniques. Transgene lines were established as described above.
Transgene-mediated Rescue of RNAi.
RNAi activity in transgenic animals was assessed by placing animals as L2/L3 larvae on pop-1 or elt-2 food. Absence of live progeny is indicative of an intact RNAi response.
Transgene-mediated Rescue of Nonsense-mediated Decay (NMD).
Plasmids were injected as described above into smg-2(r908), unc-54(r293) hosts, and phenotypes in the resulting transgenic strains were noted. unc-54(r293) carries a premature stop codon that produces a paralyzed phenotype in wild type; the phenotype is suppressed in animals defective for smg-2 (Hodgkin et al., 1989). (unc-54 mRNA is not degraded in smg-2 mutants, allowing for expression of a near full-length UNC-54 protein.) Functional SMG-2 activity is evidenced by a paralyzed phenotype.
HAF-6::GFP Reporters.
Seven plasmids were designed for assays of protein localization and mutant rescue. GFP sequences were derived from plasmids pPD119.45, pPD118.37, pPD96.02, pPD119.16, and pPD102.33 (obtained from FireLab Vector kit; Andrew Fire). Plasmids were coinjected with carrier DNA and transformation marker sequences (dominant rol-6) into wild-type animals and were established as extrachromosomal, complex arrays. GFP fluorescence patterns were monitored in live animals, and the subcellular patterns were similar to those observed using our anti-HAF-6 polyclonal antibody. GFP expression was also monitored using an anti-GFP monoclonal (Sigma-Aldrich, St. Louis, MO) to detect expression in the germline.
Antibodies and Immunofluorescence Microscopy
A peptide corresponding to amino acids 69–82 (IDHLRTTEDQNASMC) was cross-linked to keyhole limpet hemocyanin as hapten [coupled using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride] and injected into rabbits (Harlan Bioproducts for Science, Indianapolis, IN) (Supplemental Figure 1). Immune serum was affinity purified using a column coupled with HAF-6::glutathione S-transferase (GST) fusion protein derived from a pGEX-4T-2 vector (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The HAF-6::GST fusion harbored the first 100 amino acids of HAF-6. The fusion protein was covalently coupled to CNBr-activated Sepharose 4B (Sigma-Aldrich). Remaining antibodies that might cross-react to bacterial proteins were adsorbed onto a second column coupled with bacterial proteins derived from a similar purification protocol as HAF-6::GST. Tissue was prepared for immunofluorescence using a number of different fixation and permeabilization conditions and solutions, including methanol, formaldehyde, or paraformaldehyde fixatives. Animals were permeabilized by freeze-cracking on glass slides using fixative solutions containing either methanol, 4% formaldehyde, or 4% paraformaldehyde in phosphate-buffered saline. Not surprisingly, methanol fixation removed some signal from membranes, confirming the specificity of the antibody preparation; paraformaldehyde fixation resulted in a more pronounced nuclear envelope staining; and formaldehyde fixative or formaldehyde/paraformaldehyde mixes produced consistent staining with patterns similar to reporter GFP patterns in live animals. We also permeabilized animals en masse in Eppendorf tubes (Finney and Ruvkun, 1990) with variations on the protocol that included omission of methanol and/or detergents. Additionally, animals were dissected in fixative using syringe needles to achieve permeabilization. The anti-Complex IV subunit I antibody was purchased from MitoSciences (Eugene, OR); anti-calreticulin antibodies were a gift from Joohong Ahnn (Gwangju Institute of Technology, Gwangju, Korea) (Park et al., 2001). Fluorescent images were obtained using a Zeiss LSM510 Meta confocal microscope system (Carl Zeiss Microimaging, Thornwood, NY). Goat anti-mouse and goat anti-rabbit secondary antibodies were coupled to Alexa 488 or Alexa 594 (Invitrogen, Carlsbad, CA).
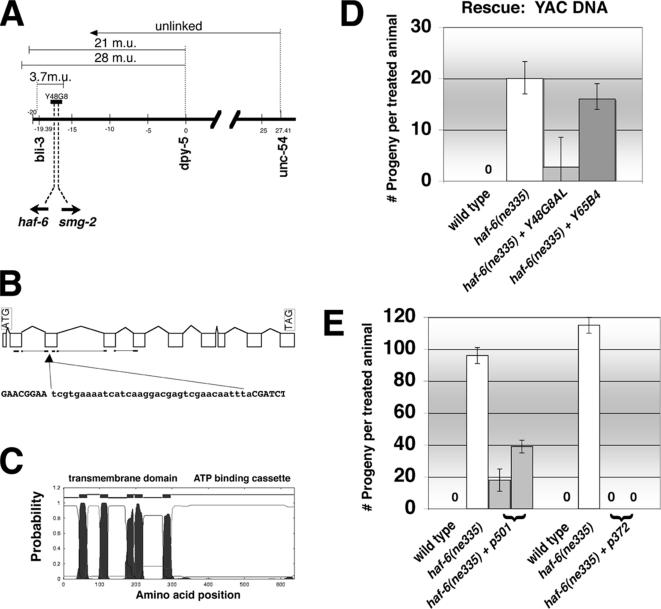
Figure 1.
ne335 is an allele of haf-6. (A) haf-6 maps to the left arm of chromosome I. We scored 300–500 F2 progeny animals in each mapping experiment. YAC Y48G8, which rescues the RNAi defect in haf-6(ne335) mutants, is indicated. The haf-6 gene is approximately 700 bp upstream of smg-2 and is oppositely transcribed, an indication that the genes may have regulatory elements that are shared or that compete. Both genes reside in Y48G8. (B) Revised intron/exon structure of haf-6. We found an additional 303 base pairs of coding region not predicted in Wormbase (release WS146). The 35-base pair deletion in haf-6(ne335) is indicated by a filled triangle. The deleted region is lowercase. The five horizontal lines at the bottom of diagram highlight the relative positions of the five predicted transmembrane helices. The thick lines represent exons; thin lines indicate introns. (C) Predicted transmembrane topology of HAF-6 (Krogh et al., 2001); the ABC is C-terminal. The set of five transmembrane helices constitutes a transmembrane domain (TM). (D) Rescue of RNAi defects in haf-6(ne335) mutants by Y48G8 but not from an unrelated YAC. RNAi defects were assessed on pop-1 feeding plates at 20°C. Bars represent the average of five experiments; error bars represent SD. In total, seven overlapping YACs were tested; data from Y65B4 are included in the figure as a representative negative control. Only Y48G8 provided rescue, demonstrating that specific sequences, as opposed to the presence of a transgene, provide rescue. (E) Rescue of RNAi defects in haf-6 mutants. The pLT501 and pLT372 plasmids harbor a full-length haf-6 cDNA driven from haf-6 upstream/promoter sequences (see Figure 2, construct 5). The HAF-6 sequence in pLT501 harbors a G562R amino acid substitution; plasmid pLT372 is wild type for HAF-6. Bars represent an average of two experiments; error bars denote the range. Gray bars represent different transgenic lines derived independently from the same DNA injection mix.
RESULTS
ne335 Is an Allele of haf-6
The ne335 allele was isolated based on its “method-of-delivery-dependent” RNAi defects (Tabara et al., 1999). ne335 mutants elicit RNAi when dsRNA is introduced by injection (Fire et al., 1998), but they are RNAi defective when worms are reared on “dsRNA food” (bacteria that express dsRNA corresponding to a particular worm gene) (Timmons et al., 2001). We mapped the ne335 mutation to the left end of chromosome I using standard methodology and pop-1 or unc-22 food to identify homozygotes and recombinants (Figure 1A). The single gene Y48G8AL.11 rescued the RNAi defects in ne335 (Figure 1, A, B, D, and E). Y48G8AL.11 is named haf-6 based on its homology to half-molecule ABC transporters. Although full ABC transporter proteins have two sets of transmembrane-spanning domains and two domains with homology to ATP-binding cassettes, half-transporters have only one of each domain (Figure 1C and Supplemental Figure 1). The molecular lesion in the ne335 allele is a 35-base pair deletion in exon 3 of the haf-6 gene (Figure 1B), and the mutated gene is predicted to produce a partial protein with only the first two transmembrane-spanning helices but no ATPase domain.
The Neighboring Gene, smg-2, Does Not Contribute to the RNAi Defect in haf-6(ne335)
Although robust RNAi phenocopies are observed in response to delivery of dsRNA, the effect is not permanent. Faster recovery of RNAi has been observed in smg-2 mutants injected with dsRNA in comparison with similarly injected wild-type animals (Domeier et al., 2000). The smg-2 gene is located ∼700 base pairs upstream of haf-6, and the genes are oppositely transcribed (Figure 1A). Because the two genes are juxtaposed and may have overlapping regulatory elements, we considered that smg-2 activity might be impaired in ne335 mutants, contributing to the RNAi defects we observe. SMG-2 activity is required for some aspects of mRNA surveillance, a mechanism that degrades defective mRNAs, in particular those mRNAs harboring a premature stop codon. smg-2 mutants act as allele-specific suppressors of NMD (Hodgkin et al., 1989; Page et al., 1999; Anders et al., 2003; Grimson et al., 2004), and we used this property in functional assays for SMG-2 activity.
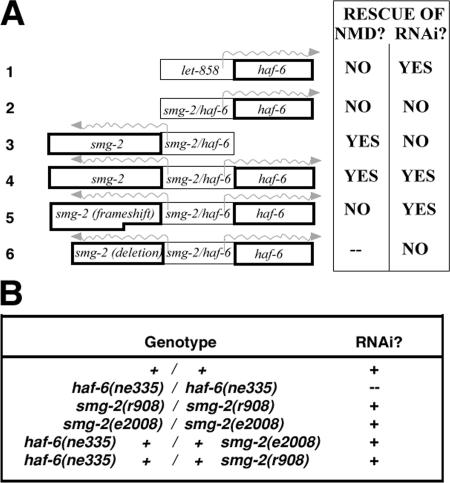
We took a transgenic approach to determine whether smg-2 activity might be affected by the mutation in haf-6 (Figure 2A). cDNA sequences, and not genomic regions, for haf-6 and smg-2 were used due to the presence of unclonable inverted repeats in several introns of both genes; furthermore, no cosmids were subcloned from this region by the C. elegans DNA Sequencing Consortium. The haf-6 cDNA alone rescued the RNAi defects in haf-6(ne335) when driven ubiquitously from a let-858 promoter (Figure 2A, construct 1). The smg-2 cDNA alone did not rescue the RNAi defects in haf-6(ne335) mutants, but it did rescue NMD defects in smg-2 mutants (Figure 2A, construct 3). A single construct harboring both cDNAs flanking the intergenic region was able to rescue both the NMD defect in smg-2 and the RNAi defect in haf-6(ne335) mutants, as expected (Figure 2A, construct 4). Plasmids with this tripartite configuration of DNA segments that harbored a smg-2 cDNA with a frameshift mutation (Figure 2A, construct 5) or with a deletion (Figure 2A, construct 5) were generated. These plasmids did not allow rescue of NMD, as expected; furthermore, construct 5 did rescue RNAi defects in haf-6(ne335). Extra copies of smg-2 in the haf-6(ne335) mutant background did not improve the RNAi defects in our transgene-based assays; thus, the ne335 mutation has no apparent effect on smg-2. By contrast, we observed that part of the smg-2 coding region is required for full haf-6 expression (Figure 2A, constructs 2 and 6 compared with constructs 4 and 5), possibly acting as a transcriptional enhancer.
Figure 2.
smg-2 does not contribute to the RNAi defects in haf-6(ne335). (A) DNA constructs injected into mutants were configured as indicated. Transgenic lines for each plasmid were established in both haf-6(ne335) and smg-2(r908), unc-54(r293) mutant backgrounds. Each haf-6(ne335) transgenic strain was tested for RNAi activity; each smg-2(r908), unc-54(r293) transgenic strain was tested for NMD as described in Materials and Methods. haf-6 mutant animals without transgenes did not display RNAi activity in these assays; likewise, the nontransgenic smg-2, unc-54 mutant strain did not display NMD. Rescue of RNAi defects in haf-6 mutants was dependent upon proper expression of haf-6 sequences; rescue of NMD in smg-2 mutants was dependent on intact SMG-2 activity. The design of constructs 3–6 reflects the chromosomal, oppositely oriented, arrangement of the genes. A smg-2 coding segment is required for full haf-6 expression (compare 2 and 4 with 6 and 5). (B) No RNAi defects are observed in smg-2 mutants using feeding assays, and the haf-6 and smg-2 alleles do not complement. (Both pop-1 and unc-22 foods were used in these experiments.)
Additional data support the conclusion that smg-2 activity is not affected by the 35-base pair deletion in haf-6(ne335) and that the RNAi defects we observe are solely due to defects in haf-6: 1) The smg-2 sequence is wild type in haf-6 mutants, ruling out the possibility that mutations in two genes coexist in this strain. 2) We easily obtained smg-2 cDNA by RT-PCR using RNA from haf-6 mutants as template, suggesting the smg-2 gene is expressed (our unpublished data). 3) The RNAi defects in haf-6 are complemented by smg-2 mutants (Figure 2B). 4) When our feeding assays (pop-1 and unc-22 foods) are used to test the RNAi capability of smg-2(r908) and smg-2(e2008) mutants, we observed no RNAi defects at any growth temperature (15, 20, or 25°C; Figure 2B). The proximity of the genes, the presence of haf-6 regulatory elements in the smg-2 coding region, and the potential for other forms of shared regulation raise the possibility that some smg-2 mutants, especially those with large deletions, might also have disruptions in haf-6, complicating analyses of RNAi defects in these strains.
An Intact ABC Domain Is Required for HAF-6 Activity in RNAi
We verified our subcloned fragments by DNA sequencing and found that the haf-6 cDNA in plasmid pLT501 (Figure 1E) had acquired a point mutation during PCR amplification. [This mutation is not present in the genome of wild type or haf-6(ne335).] The mutation is predicted to result in an amino acid substitution of Arg for Gly at amino acid position 562 (Supplemental Figure 1). Gly562 is strictly conserved and resides only three amino acids upstream from the signature conserved motif (LSGGQ) specific to ABC transporters (Supplemental Figure 1), and G562 likely plays an important role in nucleotide binding. The G562R mutation rescued RNAi defects in haf-6(ne335); however, the relative ability of the R562G form to rescue RNAi was notably reduced in comparison with the ability of intact haf-6 sequences to rescue (Figure 1E, plasmid 501 has the G562R mutation and plasmid 372 is wild type). Our interpretation of the results is that the G562R substitution produces a HAF-6 protein with a weakened ability to bind and/or hydrolyze nucleotides with the result that the RNAi activity is also weakened. Thus the RNAi role for HAF-6 likely requires substrate transport.
Phenotypic Analysis of haf-6(ne335)
haf-6 mutant animals are viable and fertile, yet display method-of-delivery–dependent RNAi defects. We used the full spectrum of dsRNA delivery methods available in C. elegans to better understand the nature of the RNAi defects in haf-6(ne335). We report two main observations.
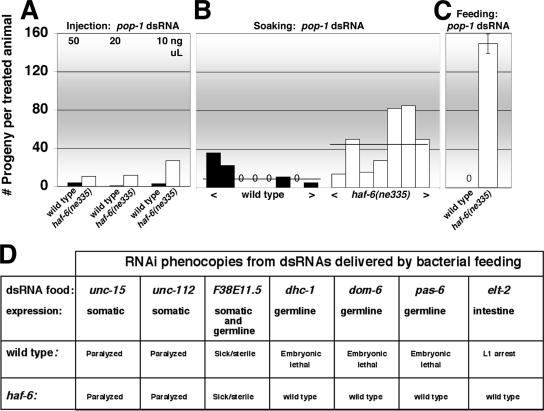
First, the RNAi defect in haf-6(ne335) mutants is dosage sensitive with respect to the amount of dsRNA. An RNAi response is observed when relatively high concentrations of dsRNA are injected; lower concentrations are less effective in eliciting an RNAi response in mutants (Figure 3A). Experiments using soaking methods also reveal a concentration-dependent RNAi response (Figure 2B)—treatment of mutants with a relatively high concentration of dsRNA was required for a robust RNAi phenocopy in the population. With soaking, wide variations in penetrance of RNAi phenocopies can be attributed to individual variations in the amount ingested. Although soaking and injection can elicit RNAi in mutants, feeding is most effective in revealing their RNAi defects (Figure 2C).
Figure 3.
haf-6 mutants display dose-dependent and tissue-specific RNAi defects. Injection (A), soaking (B), and feeding (C) methods were used to deliver dsRNA. (A–C) pop-1(RNAi) phenocopies (sterility) were assessed by scoring the number of F1 progeny per treated animal for a defined time frame. (A) Various dilutions (from 10 to 50 ng/μl) of pop-1 dsRNA were injected into the body cavity of L3/L4 animals that were subsequently cultured on standard plates at 20°C and also at 25°C (our unpublished data). No temperature-dependent differences in RNAi activity were observed. Ten to 18 animals (average 12.2) were injected in each set. (B) Animals were soaked in 50 ng/μl pop-1 dsRNA as L1/L2 at 20°C, and each animal was recovered onto individual culture plates. Bars represent the number of progeny produced over a defined time period from treated individuals. The horizontal lines above each set of bars represent the average number of progeny produced for the particular strain: 3.3 progeny/adult in wild type; 44.6 for haf-6(ne335). Individual soaked mutants displayed a more variable phenocopy penetrance than did wild type; however, average overall RNAi response in mutants was lower than wild type. The variation in response is likely correlated to the amount of material ingested by individuals and hence to the dosage of dsRNA. (C) Animals were placed onto pop-1 food as L1/L2 and monitored for sterility at 20°C over a defined time, identical conditions were used in each set of experiments. (D) haf-6 mutants are RNAi defective in germline and intestine. The feeding protocol was used to deliver dsRNAs corresponding to the genes indicated. Animals were placed at L1/L2 stage onto feeding plates. All feeding strains elicited an RNAi phenocopy in wild-type. Experiments were performed at 15, 20, and 25°C, with similar results.
Second, we observe RNAi defects in haf-6(ne335) for genes that are expressed in germline and intestine but not other tissues (Figure 3D). We initially considered that haf-6, a method-of-delivery–dependent mutant, might be defective in the dissemination of feeding-derived dsRNAs from the intestine to other tissues. However, some systemic RNAi responses are intact in haf-6 mutants, as evidenced by the appearance of RNAi phenocopies 1) in somatic tissue when dsRNA is introduced by feeding and 2) in treated animals and their progeny from dsRNA introduced by soaking and injection. Thus, an efficient RNAi response to dsRNA, in particular to dsRNA delivered by bacterial feeding, is sensitive to the relative levels of HAF-6 activity as well as to the dosage of dsRNA.
Subcellular Localization of HAF-6
Most organisms harbor multiple ABC protein genes (Holland et al., 2003)—60 ABC transporter genes have been identified in C. elegans (Sheps et al., 2004). Collectively, the different ABC transporters localize to most membranes in the cell, including the plasma membrane, endoplasmic reticulum, and mitochondria. Determining the subcellular localization of HAF-6 is key to understanding the role of HAF-6 in RNAi and to the identification of transportable substrates that are relevant to its RNAi function. HAF-6 is a half-transporter member of the B subfamily, and an additional eight half-transporter genes in subfamily B are encoded by the C. elegans genome (Sheps et al., 2004). The precise functions, substrates, and subcellular localizations of half-transporters in C. elegans have not been fully investigated. Mammalian half-transporters from subgroup B function as heterodimers, and possibly homodimers in the endoplasmic reticulum; others localize to mitochondria or lysosomes (Monaco et al., 1990; Spies et al., 1990; Allikmets et al., 1996; Csere et al., 1998; Hogue et al., 1999; Yamaguchi et al., 1999; Mitsuhashi et al., 2000; Zhang et al., 2000a,b). Because not all members of the same subfamily are similarly localized, subfamily classification does not allow predictions of subcellular localization. Additionally, localization domains have been identified for a relatively few transporter proteins, and the degree of evolutionary conservation of such motifs has yet to be determined.
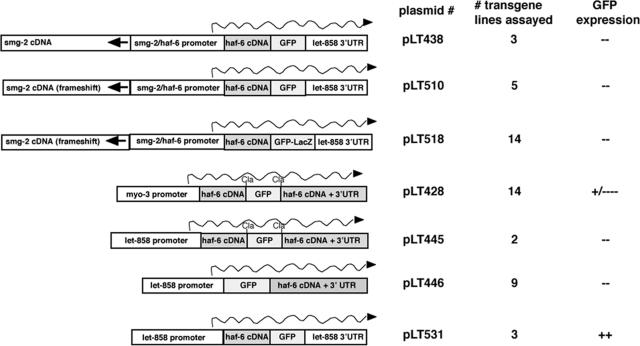
We first built plasmids to express HAF-6::GFP fusion proteins in C. elegans (Figure 4). The different constructs produced three variations of GFP fusions: an N-terminal GFP::HAF-6 protein, a C-terminal HAF-6:: GFP protein, and an internal GFP sequence in-frame within the entire HAF-6 protein. Expression of GFP fusion proteins was driven from haf-6/smg-2 promoter segments that provided rescue in previous experiments (Figure 2, construct 5), from a myo-3 promoter segment that drives expression in muscle, or from a let-858 promoter that drives expression in all cells. Each construct was injected into wild-type and haf-6 mutant animals as described in Materials and Methods, and transgenic lines were established. GFP accumulation was observed in only two plasmid configurations (Figure 4): 1) Muscle cells in transgenic lines harboring plasmid pLT428 expressed GFP; however, the expression was weak and was observed only in a few anterior muscle cells in each animal. Additionally, not all transgenic animals (with the Roller transformation marker phenotype) displayed GFP fluorescence, and GFP expression was not meiotically stable—no GFP was observed in these lines past the F3 generation. However, in GFP-expressing muscle cells, we observed intracellular fluorescence that was clearly reticular and not mitochondrial (our unpublished data). 2) The second construct, pLT531, displayed a more robust GFP fluorescence (Figure 5A). HAF-6::GFP expression in this plasmid was regulated by a let-858 promoter that drove ubiquitous expression. Fluorescence from the same HAF-6::GFP construct did not accumulate to visible levels when driven from an endogenous haf-6/smg-2 promoter (pLT438 and pLT510). Because the let-858 promoter drives ubiquitous expression, we anticipated that GFP would be observed in all cells of transgenic lines harboring pLT531. However, we observed GFP expression mostly in the germline and intestine, those tissues that display RNAi defects (Figure 3D). (Germline expression of GFP was observed using anti-GFP antibodies and immunofluorescent microscopy.) The more limited tissue expression pattern may reflect the absence of a heterodimeric binding partner in the nonfluorescent tissues.
Figure 4.
GFP reporter plasmid configurations used to build transgenes. Plasmids were configured as indicated and injected into wild type and haf-6 mutants. The number of independently established lines is indicated. GFP fusion proteins expressed in muscle from plasmid pLT428 displayed weak fluorescence, and the ability of the transgenic animals to express GFP was lost over generational time. We observed a clear ER localization pattern for GFP in the few muscle cells with fluorescence. Only plasmid pLT531 elicited stable GFP fluorescence. The pLT531 plasmid was injected into haf-6(ne335) mutants to generate strain XX915. XX915 animals displayed GFP fluorescence in a pattern similar to that observed in wild type, and the RNAi phenotypes were rescued.
Figure 5.
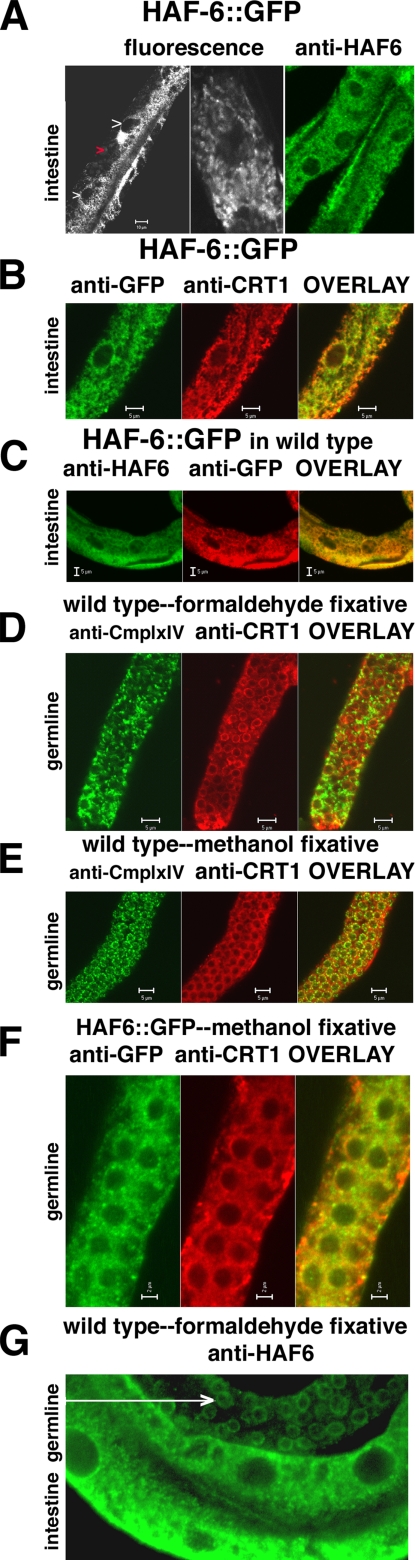
HAF-6 is expressed in intestine and germline and localizes to intracellular, reticular membranes. Endogenous GFP fluorescence from a rescuing HAF-6::GFP fusion protein (Figure 4, plasmid pLT531) and immunolocalizations using anti-HAF-6 and control antibodies were used to investigate subcellular distribution patterns. (A) HAF-6::GFP fluorescence patterns in live animals (left and center) are similar to the patterns observed in fixed animals decorated with an anti-HAF-6 antibody (right). White arrows, intestinal nuclei; red arrows, autofluorescent granules. (B) The HAF-6::GFP protein (left) is localized to the same compartment as calreticulin (CRT-1), a primarily ER resident with a C-terminal KDEL sequence. Similar results were obtained in cohybridization experiments using the anti-HAF-6 antibody with anti-CRT-1. HAF-6 is a transmembrane protein, and 100% spectral overlap with CRT-1 is not anticipated; nonetheless, co-compartmentalizaion is observed. (C) Cohybridization experiments were performed in transgenic animals expressing both endogenous HAF-6 and a HAF-6::GFP fusion protein. The signals from anti-GFP and anti-HAF-6 overlapped, indicating that the fusion protein is expressed in all the compartments where the endogenous protein is located. (D) A control experiment demonstrating that subcellular compartments can be distinguished in germline tissue fixed with formaldehyde. Mouse anti-Complex IV subunit I antibodies recognize mitochondrial proteins. Rabbit anti-CRT-1 antibodies recognize proteins localized mostly to the endoplasmic reticulum. (E) A control experiment demonstrating that distinct compartmentalization patterns are also observed using methanol as fixative. (F) HAF-6 localization patterns do not resemble mitochondrial patterns in germline. HAF-6 patterns are reticular in germline. (G) HAF-6 protein is perinuclear in germline tissue; perinuclear staining is less prevalent in intestinal cells. Both tissues shown are from the same animal. Confocal images were captured using a Zeiss LSM 510 META. Bars, 5 μm on all panels except A (10 μm) and F and G (2 μm).
The RNAi defect in mutant animals was rescued by the HAF-6::GFP fusion protein; however, this does not preclude the possibility that some GFP fluorescence might originate from misfolded or nonfunctional proteins trapped in the endoplasmic reticulum. We therefore validated our GFP fluorescence observations using immunolocalization techniques and an antibody specific to HAF-6 protein.
ABC transporter proteins have a high degree of amino acid similarity. The N-terminal region of HAF-6 has little similarity to other ABC proteins in C. elegans, so we generated an anti-peptide polyclonal antibody targeting this region (Supplemental Figure 1). The deletion in the haf-6(n335) is not predicted to affect the region that encodes the epitope. Indeed, we do observe antibody staining in the mutant, indicating that HAF-6 protein, probably a truncated version, is expressed. [We were not able to isolate a full-length cDNA from haf-6(ne335) RNA by RT-PCR and anticipated that the RNA might be more susceptible to degradation.] The specificity of the antibody is demonstrated by the observation of similar staining patterns in experiments using anti-HAF-6 antibody versus experiments using GFP reporters from a construct that provides rescue of the RNAi defects. Furthermore, we have developed antibodies against other HAF proteins, and we observe patterns that are distinct from HAF-6 (our unpublished data).
Immunofluorescence staining revealed that HAF-6 was predominantly expressed in intestinal and germline tissue (Figure 5), a pattern that mirrors the RNAi defects observed in mutants (Figure 3D). Thus haf-6 RNAi defects are apparently limited to the tissues where HAF-6 is expressed. In double-labeling experiments, the anti-HAF-6 antibody did not decorate the same cellular compartment as a mitochondrial antibody (anti-Complex IV). We did observe co-compartmentalization using anti-HAF-6 and anti-Calreticulin antibodies, indicating that HAF-6 likely resides in the endoplasmic reticulum and not in mitochondria, as was expected from comparisons of sequence similarity to human half-transporters. Interestingly, HAF-6 localization extended to perinuclear regions in germline tissue, which is consistent with an endoplasmic reticulum (ER) localization (Figure 5G). However, localization of HAF-6 to the nuclear periphery was not prominent in intestinal tissue. Furthermore, the subcellular patterns observed in experiments using anti-HAF-6 antibody were similar to the patterns of GFP expression observed in transgenic animals expressing the HAF-6::GFP fusion protein (Figure 5A).
Additional ABC Transporters Facilitate RNAi
The C. elegans genome encodes 61 genes with sequence homology to ABC transporters (Sheps et al., 2004). Strains that harbor defects in 43 of these genes were tested for RNAi activity using pop-1 and unc-22 food (see Materials and Methods). We observed strong defects in eight additional strains (Table 1)—this set includes full-transporter as well as half-transporter molecules from a wide distribution of subfamilies. Some transporter genes have overlapping patterns of expression in somatic tissue, whereas others have nonoverlapping and more restricted patterns of expression (Zhao et al., 2004). Given the high degree of sequence homology between ABC transporters, the diversity of expression patterns, and the variety of configurations of transmembrane and ABC motifs in this set of genes, we cannot yet ascribe an RNAi function to a common sequence-related feature. Because all the strains in this set are RNAi-defective for germline targets, we predict a germline function for these transporters. Most surprising is the observation that nine ABC proteins are nonredundantly required for efficient RNAi; however, because the RNAi defect in any single mutants is rather weak, their effects may be additive.
Table 1.
Additional ABC transporters are required for efficient RNAi in C. elegans
| ABC subfamily | Strain | RNAi activity on |
Predicted topology | |
|---|---|---|---|---|
| pop-1 food | unc-22 food | |||
| A | abt-1(tm0703) | Defective | Wild type | (6TM-ABC)2 |
| B | pgp-4(gk16) | Defective | Wild type | (6TM-ABC)2 |
| B | pgp-11(tm33) | Defective | Wild type | (6TM-ABC)2 |
| B | haf-2(gk13) | Defective | Defective | 8TM-ABC |
| B | haf-6(ne335) | Defective | Defective, heat sensitive | 5TM-NFB |
| C | mrp-1(pk89) | Defective, cold sensitive | Wild type | (12/6TM-ABC)2 |
| D | pmp-1(ok773) | Defective | Wild type | 4TM-ABC |
| G | C16C10.12(ok962) | Defective | Wild type | ABC-4TM |
| G | C05D10.3(tm688) | Defective | Wild type | ABC-4TM |
Deletion strains of ABC transporters were reared on pop-1 and unc-22 feeding plates at 15, 20, 25, and 26°C. pop-1 RNAi phenocopies (sterility) and unc-22 phenocopies (twitching) were assessed and compared with those of wild type. Notations on the predicted topology and ABC classification systems were derived from Sheps et al. (2004). Strains testing negative in these assays harbored mutations in the following genes (see Materials and Materials): abt-2; abt-4; C56E6.1; ced-7; cft-1; F02E11.1; F19B6.4; haf-1; haf-3; haf-4; haf-5; haf-7; haf-8; haf-9; mrp-3; mrp-4; mrp-6; mrp-8; pgp-1; pgp-2; pgp-3; pgp-5; pgp-6; pgp-7; pgp-10; pgp-12; pgp-13; pmp-3; pmp-4; pmp-15; T26A5.1; T27E9.7; TAG-167; Y42g9a.6; Y49e10.9.
DISCUSSION
The haf-6 gene encodes a half-molecule transporter with homology to members of the B subclass of ABC transporters. Most organisms harbor many different ABC transporter genes, and the C. elegans genome has at least 60 (Sheps et al., 2004). Transporters are classified into subgroups based on the overall level of homology and the numbers and arrangement of the two conserved domains: the transmembrane domain, made up of multiple transmembrane-spanning helices, and the ABCs. ABC transporters can be found in virtually all membranes of the cell, including those of subcellular organelles, although some ABC proteins lack transmembrane-spanning domains. The small molecule substrates transported by the collective ABC transporters include lipids, peptides, ions, nucleotides, carbohydrates, and drugs. Substrates can be transported into or out of cells or between intracellular compartments, and substrate transport is involved in various essential and nonessential cellular activities.
Generally, the amino acid sequence of an ABC transporter allows few conclusions to be drawn regarding the substrates that might be transported. We first anticipated that the homology between HAF-6 and the better-studied human half-transporter molecules would provide clues to function, or perhaps subcellular localization. HAF-6 has the most overall sequence similarity to human transporters that localize to mitochondria. However, HAF-6 seems to localize to reticular membranes rather than mitochondrial membranes in our observations of live animals expressing GFP and of antibody-stained fixed tissue. Thus, the overall amount of homology between transporters from different species does not allow predictions regarding subcellular localization.
We observed RNAi defects in eight additional ABC transporter-defective strains. Transporters are often expressed in a tissue-specific manner in multicellular organisms, and expression patterns for most of the nine transporters in C. elegans have been reported (Zhao et al., 2004). pgp-4 and pgp-11 expression was observed in the excretory cell; pgp-11 is also expressed in intestinal tissue, as is mrp-1 and pmp-1; additional mrp-1 expression was observed in the pharynx and vulva, and C05D10.3 gene expression was observed in embryos and the gut of males (Zhao et al., 2004). The expression patterns were determined using transgenes configured to drive expression of GFP in live animals from promoter (upstream) sequences of an ABC transporter gene. However, it is generally the case that transgenes are silenced in the germline and in young embryos (Kelly et al., 1997), so this method is not a reliable indicator of gene expression in these tissues. The RNAi defects we observed using pop-1 food (Table 1) indicate a role in the germline for these genes.
Of the nine transporters that facilitate RNAi, haf-2 is the only gene expressed in muscle (Zhao et al., 2004), and haf-2(gk13) is the only mutant we tested with a strong muscle-specific RNAi defect. Because the remaining eight mutant strains did not display unc-22(RNAi) phenocopies in response to ingestion of unc-22 food, the RNAi defects are likely only observed in tissues where the ABC transporter in question is normally expressed. None of the mutants is likely to be strongly defective in the dissemination of silencing signals derived from ingestion of bacterial food, because RNAi is active in some somatic tissues (muscle). However, a fully systemic RNAi phenocopy in response to ingested dsRNAs may depend upon the proper function and expression of multiple ABC transporters in multiple tissues. We anticipate that tissue-specific RNAi defects may be observed in additional ABC transporter mutants. We are currently testing the remaining mutants using bacterial feeding strains to target genes with expression patterns that overlap with that of the ABC transporter in question.
Some ABC transporters export heavy metals, drugs, and other toxins from cells, thereby providing protection from harsh environments. A developmental program that results in arrested dauer larvae, which are better adapted for survival in harsh conditions, is initiated when C. elegans encounters environmental stress. The ABC transporter mrp-1 allows animals to survive in environments laden with heavy metals (Broeks et al., 1996) and has been demonstrated to function in preventing dauer formation (Yabe et al., 2005). Here, we report that mrp-1 mutants harbor RNAi defects (Table 1). Thus, an unappreciated role for ABC proteins during stress may be to influence RNAi mechanisms and thereby affect developmental reprogramming.
ABC transporters are required for various physiological activities, including protective or barrier mechanisms that export drugs or toxins from cells (Broeks et al., 1996; Bauer et al., 1999; Begley, 2004; Fromm, 2004) and mechanisms that protect against viral infection (Trowsdale et al., 1990; Abele and Tampe, 2004). A variety of human pleiotropic disorders are linked to mutations in ABC transporters (Dean, 2005), and transporters are up-regulated in some drug-resistant cancers (Gottesman and Ambudkar, 2001; Gottesman and Ling, 2006). Because alterations in ABC transporter function contribute to a number of pathologies, understanding how ABC transporters influence endogenous RNAi mechanisms will prove relevant to disease etiology as well as environmental adaptation. Furthermore, an ability to influence RNAi may help explain the patterns of evolutionary conservation of this diverse group of genes. We propose several models to reason how ABC protein function might influence RNAi: 1) ABC transporters may provide an appropriate intracellular environment for proper RNAi biochemistry, 2) ABC proteins may actively signal RNAi mechanisms through the transport of specific substrates; or 3) ABC protein activity may redirect the activity of associated proteins toward an RNAi function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hiroaki Tabara, Craig Mello, Andrew Fire, and the C. elegans Genetics Center for strains and reagents; Joohong Ahnn for the anti-CRT-1 antibody; the C. elegans Gene Knockout Consortium (Oklahoma Medical Research Foundation, Oklahoma City, OK; University of British Columbia, Vancouver, British Columbia, Canada) and the National Bioresources Project in Japan (Tokyo) for ABC gene deletion mutants; David Moore-Nichols for microscopy assistance; the Sanger Institute and C. elegans Sequencing Consortium for YACS and cosmids; and William G. (Bill) Kelly and Lishia Chen for review of the manuscript. This work was supported by National Institutes of Health Grants P20RR015563 and P20RR016475 and funds from the American Cancer Society.
Abbreviations used:
- ABC
ATP-binding cassette
- dsRNA
double-stranded RNA
- RNAi
RNA interference.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0192) on May 24, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abele R., Tampe R. The ABCs of immunology: structure and function of TAP, the transporter associated with antigen processing. Physiology. 2004;19:216–224. doi: 10.1152/physiol.00002.2004. [DOI] [PubMed] [Google Scholar]
- Allikmets R., Gerrard B., Hutchinson A., Dean M. Characterization of the human ABC superfamily: isolation and mapping of 21 new genes using the expressed sequence tags database. Hum. Mol. Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]
- Anders K. R., Grimson A., Anderson P. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg P. Adrenoleukodystrophy and other peroxisomal diseases. Curr. Opin. Genet. Dev. 1994;4:407–411. doi: 10.1016/0959-437x(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Bauer B. E., Wolfger H., Kuchler K. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta. 1999;1461:217–236. doi: 10.1016/s0005-2736(99)00160-1. [DOI] [PubMed] [Google Scholar]
- Begley D. J. ABC transporters and the blood-brain barrier. Curr. Pharm. Des. 2004;10:1295–1312. doi: 10.2174/1381612043384844. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Allis C. D. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A., Gerrard B., Allikmets R., Dean M., Plasterk R. H. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J. 1996;15:6132–6143. [PMC free article] [PubMed] [Google Scholar]
- Caudy A. A., Ketting R. F., Hammond S. M., Denli A. M., Bathoorn A. M., Tops B. B., Silva J. M., Myers M. M., Hannon G. J., Plasterk R. H. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Csere P., Lill R., Kispal G. Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Lett. 1998;441:266–270. doi: 10.1016/s0014-5793(98)01560-9. [DOI] [PubMed] [Google Scholar]
- Dean M. The genetics of ATP-binding cassette transporters. Methods Enzymol. 2005;400:409–429. doi: 10.1016/S0076-6879(05)00024-8. [DOI] [PubMed] [Google Scholar]
- Domeier M. E., Morse D. P., Knight S. W., Portereiko M., Bass B. L., Mango S. E. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science. 2000;289:1928–1931. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- Du T., Zamore P. D. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Duchaine T. F., et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Dudley N. R., Labbe J. C., Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fromm M. F. Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol. Sci. 2004;25:423–429. doi: 10.1016/j.tips.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Ambudkar S. V. Overview: ABC transporters and human disease. J. Bioenerg. Biomembr. 2001;33:453–458. doi: 10.1023/a:1012866803188. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580:998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Rice J. C. Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Grimson A., O’Connor S., Newman C. L., Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol. Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 2005;579:5932–5939. doi: 10.1016/j.febslet.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Grishok A., Sinskey J. L., Sharp P. A. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue D. L., Liu L., Ling V. Identification and characterization of a mammalian mitochondrial ATP-binding cassette membrane protein. J. Mol. Biol. 1999;285:379–389. doi: 10.1006/jmbi.1998.2259. [DOI] [PubMed] [Google Scholar]
- Holland I. B., Cole S.P.C., Kuchler K., Higgins C. F. ABC Proteins: From Bacteria to Man. London: Academic Press; 2003. [Google Scholar]
- Hull D., Timmons L. Methods for delivery of double-stranded RNA into Caenorhabditis elegans. Methods Mol. Biol. 2004;265:23–58. doi: 10.1385/1-59259-775-0:023. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Haverkamp T. H., van Luenen H. G., Plasterk R. H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kim J. K., et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Knight S. W., Bass B. L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lee R. C., Hammell C. M., Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. W., Ding S. W. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N., et al. MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis. J. Biol. Chem. 2000;275:17536–17540. doi: 10.1074/jbc.275.23.17536. [DOI] [PubMed] [Google Scholar]
- Monaco J. J., Cho S., Attaya M. Transport protein genes in the murine MHC: possible implications for antigen processing. Science. 1990;250:1723–1726. doi: 10.1126/science.2270487. [DOI] [PubMed] [Google Scholar]
- Montgomery M. K., Xu S., Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S. I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Page M. F., Carr B., Anders K. R., Grimson A., Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. J., et al. Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2835–2845. doi: 10.1091/mbc.12.9.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., Vastenhouw N. L., Plasterk R. H. RNA interference, transposon silencing, and cosuppression in the Caenorhabditis elegans germ line: similarities and differences. Cold Spring Harb. Symp. Quant. Biol. 2004;69:397–402. doi: 10.1101/sqb.2004.69.397. [DOI] [PubMed] [Google Scholar]
- Sheps J. A., Ralph S., Zhao Z., Baillie D. L., Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R. H., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J., Plasterk R. H. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke J. M., Stacey S. C., Klein M. E., Mackin N., Maine E. M. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Spies T., Bresnahan M., Bahram S., Arnold D., Blanck G., Mellins E., Pious D., DeMars R. A gene in the human major histocompatibility complex class II region controlling the class I antigen presentation pathway. Nature. 1990;348:744–747. doi: 10.1038/348744a0. [DOI] [PubMed] [Google Scholar]
- Tabara H., Grishok A., Mello C. C. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W. G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C. C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit E., Siomi H., Mello C. C. The dsRNA binding Protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tijsterman M., Ketting R. F., Okihara K. L., Sijen T., Plasterk R. H. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 2002a;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- Tijsterman M., May R. C., Simmer F., Okihara K. L., Plasterk R. H. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol. 2004;14:111–116. doi: 10.1016/j.cub.2003.12.029. [DOI] [PubMed] [Google Scholar]
- Tijsterman M., Okihara K. L., Thijssen K., Plasterk R. H. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr. Biol. 2002b;12:1535–1540. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Timmons L., Tabara H., Mello C., Fire A. Inducible Systemic RNA Silencing in Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:2972–2983. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tops B. B., Tabara H., Sijen T., Simmer F., Mello C. C., Plasterk R. H., Ketting R. F. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 2005;33:347–355. doi: 10.1093/nar/gki183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J., Hanson I., Mockridge I., Beck S., Townsend A., Kelly A. Sequences encoded in the class II region of the MHC related to the ‘ABC’ superfamily of transporters. Nature. 1990;348:741–744. doi: 10.1038/348741a0. [DOI] [PubMed] [Google Scholar]
- Vastenhouw N. L., Plasterk R. H. RNAi protects the Caenorhabditis elegans germline against transposition. Trends Genet. 2004;20:314–319. doi: 10.1016/j.tig.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D., Jr., Kim J. K., Gabel H. W., Kamath R. S., Mello C. C., Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Winston W. M., Molodowitch C., Hunter C. P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Yabe T., Suzuki N., Furukawa T., Ishihara T., Katsura I. Multidrug resistance-associated protein MRP-1 regulates dauer diapause by its export activity in Caenorhabditis elegans. Development. 2005;132:3197–3207. doi: 10.1242/dev.01909. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Kasano M., Terada T., Sato R., Maeda M. An ABC transporter homologous to TAP proteins. FEBS Lett. 1999;457:231–236. doi: 10.1016/s0014-5793(99)01042-x. [DOI] [PubMed] [Google Scholar]
- Zhang F., Hogue D. L., Liu L., Fisher C. L., Hui D., Childs S., Ling V. M-ABC2, a new human mitochondrial ATP-binding cassette membrane protein. FEBS Lett. 2000a;478:89–94. doi: 10.1016/s0014-5793(00)01823-8. [DOI] [PubMed] [Google Scholar]
- Zhang F., Zhang W., Liu L., Fisher C. L., Hui D., Childs S., Dorovini-Zis K., Ling V. Characterization of ABCB9, an ATP binding cassette protein associated with lysosomes. J. Biol. Chem. 2000b;275:23287–23294. doi: 10.1074/jbc.M001819200. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Sheps J. A., Ling V., Fang L. L., Baillie D. L. Expression analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J. Mol. Biol. 2004;344:409–417. doi: 10.1016/j.jmb.2004.09.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.