Abstract
A major drawback of most studies on how bacteria become resistant to antibiotics is that they concentrate mainly on bacteria that can be cultivated in the laboratory. In the present study, we cloned part of the oral metagenome and isolated a novel tetracycline resistance gene, tet(37), which inactivates tetracycline.
All of the tetracycline resistance genes that have been investigated so far have been isolated from cultivable microflora. A major limitation of this approach is that a large proportion of the microflora cannot be cultivated in the laboratory (4, 5). To begin to tackle this problem, we devised an approach that did not require the bacteria to be cultured: cloning of the oral metagenome. DNA was extracted from the bacteria present in saliva and dental plaque samples and cloned into expression libraries in Escherichia coli. We have detected a new tetracycline resistance gene, demonstrating the effectiveness of this approach in isolating antibiotic resistance genes from oral microflora.
Dental plaque and saliva samples were collected from 20 healthy adults who had not received antibiotics during the previous 3 months.
Bacteria were harvested by centrifugation (3,500 × g, 10 min, 4°C) from the saliva and plaque specimens. The resulting bacterial pellet was washed gently in sterile saline. Two aliquots of the cells were prepared, and DNA was extracted from one of these aliquots by using the Puregene gram-positive DNA isolation protocol and from the other by using the Puregene gram-negative DNA isolation protocol (Gentra Systems) according to the manufacturer's instructions. The extracted DNA was subsequently pooled. To prepare the DNA for library construction, 0.2 ml of DNA (at a concentration of 250 ng/ml) was sonicated for 5 s on ice at 80% power by using an ultrasonic homogenizer (IKA-WERKE). The ends of the DNA were repaired by treating them with 2 U of mung bean nuclease (Promega)/mg in a final volume of 100 μl at 37°C for 1 h to produce blunt ends. The resulting DNA fragments were separated by agarose gel electrophoresis, and fragments between 800 and 3,000 bp were cut from the gel and purified by using the Qiagen agarose purification kit. To generate 3′-A overhangs prior to cloning into a TOPO-XL vector, the DNA fragments were incubated at 75°C for 1 h with 1 U of Taq DNA polymerase (Bioline)/mg in a final volume of 100 μl in the presence of 2 mM dATP in 1× Taq buffer. Prior to ligation into the vector, the DNA was treated by using the Qiagen PCR purification kit. Ligation of DNA into TOPO-XL and subsequent transformation into E. coli TOP10 cells were performed according to the manufacturer's protocol. Plasmid DNA was isolated from E. coli by using the Qiagen Miniprep kit.
The library was screened on Luria-Bertani agar plates containing tetracycline at a concentration of 5 μg/ml. Plasmid DNA from antibiotic-resistant clones was used to retransform E. coli to confirm that antibiotic resistance was encoded on the insert DNA.
To detect the tetracycline resistance genes encoding ribosomal protection proteins [tet(M), tet(O), tetB(P), tet(Q), tet(S), tet(T)], PCRs with primers and cycling conditions previously reported (1) were used. tet(W) tet(37)-specific primers (Tet37F, 5′-AGGGATATTGGTTGGAGA-3′; Tet37R, 5′-ATCAGTCTCATATTTCGACA-3′) were used in standard PCRs.
All PCR products were sequenced with forward primer XLF-2 (5′-CGC CAG TGT GAT GGA TAT-3′) and reverse primer XL-2R (5′-TAG AAT ACT CAA GCT ATG C-3′). The sequencing reactions were performed by using the ABI PRISM BigDye Terminator cycle sequencing protocol on either an Applied Biosystems model 310 genetic analyzer or a model 373 DNA sequencer according to the manufacturer's instructions.
DNA sequences were analyzed with the DNAMAN version 5.2.2 program (Lynnon Biosoft). Similarity analysis was carried out with the Advance Blast program of GenBank (National Center for Biotechnology Information, National Institutes of Health, Washington, D.C.), and alignments were performed by using the CLUSTAL W program service at the European Bioinformatics Institute (http://www.ebi.ac.uk/).
The enzymatic activity of tet(37) was determined as previously described (3).
A total of 450 transformants were obtained from the genomic library. Of these, 18 (4%) were tetracycline resistant. These were screened by PCR for the presence of the previously characterized tetracycline resistance genes, and tet(M) and tet(O) were detected in two clones and tet(Q) was detected in one.
Some of the other clones were not homologous to sequences in the databases. However, one contained a novel tetracycline resistance gene, designated tet(37), which conferred an MIC for E. coli of 30 μg/ml aerobically but less than 1 μg/ml anaerobically. MICs for E. coli carrying only TOPO-XL were 1 μg/ml aerobically. The complete DNA sequence of the 390-bp tet(37) gene was obtained (GenBank accession number AF540889).
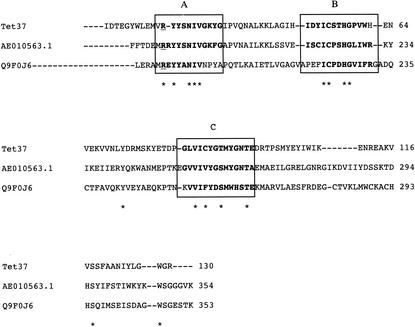
A BLAST search of the deduced amino acid sequence encoded by this gene showed that it is homologous to a number of proteins related to flavoproteins, oxidoreductases, and NAD(P)-requiring enzymes. The closest relative to Tet 37 in the database is from Fusobacterium nucleatum subsp. nucleatum strain ATCC 255 (GenBank accession number AE010563.1), with 44% identity. A multiple alignment of Tet 37 with other oxidoreductases is presented in Fig. 1 and shows conserved motifs between Tet 37 and these oxidoreductases.
FIG. 1.
Multiple sequence alignment of the Tet 37 sequence with other oxidoreductases by using CLUSTAL W (version 1.82). Flavoprotein from F nucleatum subsp. nucleatum ATCC 255 (GenBank accession number AE010563.1) had 44% identity; Rubredoxin-oxygen oxidoreductase from Desulfovibrio gigas (GenBank accession number Q9F0J6) had 29% identity. Amino acid sequence positions are indicated with numbers, and the blocks named A, B, and C show the possible ADP binding sites, metal binding, and the redox center flavodoxin-like domain (I-YGTM-GNTE----S), respectively. The conserved arginine residue is underlined in the sequence, and the conserved active-site residues are marked with an asterisk.
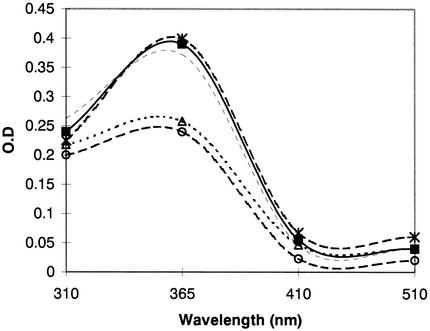
The enzymatic activity of E. coli cell extracts containing tet(37) was determined. There was a marked decrease in the prominent peak in the tetracycline absorbance spectrum at 365 nm after incubation for 90 min (Fig. 2), indicating inactivation of tetracycline. A decrease in absorbance was also observed when cell extracts containing tet(X) were used, confirming the observations of Speer and Salyers (3). There was no such decrease when cells containing the vector alone were used (Fig. 2).
FIG. 2.
Comparison of the visible absorption UV-Vis spectrum. Spectral changes were observed upon adding NADPH (1 mM) to incubation mixtures containing tetracycline (30 μg/ml) and cell extract of E. coli containing tet(X) before incubation (-×-) and after 90 min of incubation (-○-) and containing tet(37) before incubation (—) and after 90 min of incubation (--▵--). E. coli cell extracts containing tetracycline and the vector only showed no change in absorbance after 90 min of incubation (—▪—). O.D., optical density.
When intact cells containing the tet(37) gene were used instead of cell extracts, no reduction in the absorbance at 365 nm occurred. Incubation of NADH alone with disrupted cells resulted in a decrease in the height of the NADPH peak at 290 nm. Similar data were obtained with Tet X, indicating the use of NADPH by these enzymes. In the absence of NADPH, no decrease in the tetracycline peak was observed.
That tetracycline was being inactivated was confirmed by a biological assay in which broth containing tetracycline was incubated with E. coli containing tet(37) or a control E. coli isolate containing tet(M) (a ribosome protection resistance gene) and then incorporated into fresh broth after filtration. Tetracycline-sensitive bacteria could grow in the broth that had previously been incubated with the organisms containing tet(37) but not in the broth that had been grown with the control organism.
The properties of tet(37) are similar to those of tet(X), originally isolated from Bacteroides (2). However there is no homology between the deduced amino acid sequence of Tet 37 and that of Tet X.
In conclusion, we have shown that antibiotic resistance genes can be cloned and expressed from DNA isolated from the whole array (i.e., cultivable and noncultivable) of oral microflora. It should be possible to use this technique to isolate all of the antibiotic resistance genes from a particular ecological niche regardless of whether or not the original host bacteria can be cultivated in the laboratory.
Acknowledgments
This project was supported by Medical Research Council grant G9900875.
We thank Rustam Aminov for supplying the strains carrying characterized tetracycline-resistant genes and A. Salyers for providing the E. coli strain carrying the tet(X) gene used as the control in this study.
REFERENCES
- 1.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protective proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemaker, N. B., H. Vamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speer, B. S., and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spratt, D. A., A. J. Weightman, and W. G. Wade. 1999. Diversity of oral asaccharolytic Eubacterium species in periodontitis-identification of novel phylotypes representing uncultivated taxa. Oral Microbiol. Immunol. 14:56-59. [DOI] [PubMed] [Google Scholar]
- 5.Wade, W. 2002. Unculturable bacteria—the uncharacterized organisms that cause oral infections. J. R. Soc. Med. 95:81-83. [DOI] [PMC free article] [PubMed] [Google Scholar]