SYNOPSIS
Objective
A prospective space-time scan statistic was applied to Chicago's 2002 shigellosis surveillance data to evaluate its utility in objectively describing clusters and assisting in the prioritization of investigations.
Methods
The prospective space-time module of SaTScan, a free software available online, was used to identify “live” clusters of disease, meaning cases that were current as of the date of the analysis and strongly associated in place and time. Fifty-two separate space-time analyses were run, one simulation for each week of 2002. Identified clusters were described in terms of space, time, risk factors reported by involved case-patients, and cases' links to venue-associated outbreaks.
Results
Twelve live clusters were detected at the p<0.05 significance level: two single-household clusters and 10 community clusters. The community clusters ranged in size from 194 to 367 census tracts (median=294), and in disease burden from 21 to 41 cases (median=29). Geographically, all of the community clusters were located in the west-central part of the city and had a temporal span of 28 days. Within the 10 community clusters, 15 different day care centers were identified as potential exposure settings for case-patients or their close contacts.
Conclusions
The prospective space-time scan statistic offers local health departments an objective way of describing clusters of shigellosis cases. The method used in this study could help prioritize the assignment and investigation of cases, particularly when overall shigellosis incidence exceeds expected numbers or when an agency's resources are stressed by other events, such as outbreaks.
Shigella infections cause an estimated 448,000 gastrointestinal illnesses in the United States each year.1 One serotype, Shigella sonnei, accounts for over two-thirds of reported cases, and has frequently been associated with epidemic transmission.2 Day care centers,3–5 recreational water venues,6 and summer camps for mentally retarded individuals7 have been implicated as transmission settings in large outbreaks of Shigella sonnei infection, and community-wide outbreaks have occurred among members of a religious group8 and men who have sex with men.9 Although most cases of Shigella sonnei infection are acquired through close contact with an infected person, when food contamination occurs, resulting foodborne outbreaks may cause large numbers of cases in multiple jurisdictions.10
Many public health departments conduct surveillance for shigellosis to identify cases, investigate potential sources of infection, and intervene to prevent further transmission. Geographic information systems (GIS) offer a variety of tools to assist with these functions. The uses of GIS include the geocoding of cases to postal codes or census tracts for calculation of counts and rates by geographic area, and the visualization or modeling of spatial and temporal patterns of disease occurrence.11–13 In community outbreaks of Shigella sonnei infection, both statistical and mapping techniques have been implemented to assess clustering of cases and prioritize geographic areas for intensive investigation and intervention.14
In Chicago from August to October of 2002, the number of human disease case reports registered by the Chicago Department of Public Health's (CDPH) Communicable Disease Surveillance System (CDSS) exceeded the expected number for 12 consecutive weeks, with the report burden in some weeks reaching as high as five times the number expected. Surveillance for cases of central nervous system disease during the large West Nile virus disease outbreak occurring locally and nationally contributed to the increase,15 but CDPH also noted elevated numbers of shigellosis cases.
During this period, shigellosis clusters were delineated by periodically plotting the case-patients' reported residences on a city map using ArcView software16 and subjectively deciding which cases should be considered part of a cluster. The arbitrary nature of this method prompted us to seek alternative means of identifying clusters. With this objective, we applied a prospective space-time scan statistic to Chicago's 2002 shigellosis surveillance data, and evaluated its utility in objectively describing clusters and assisting in the prioritization of investigations.
METHODS
Chicago is a city of approximately 2.9 million residents and an area of 229 square miles. CDPH investigates cases of reportable diseases, including shigellosis, as defined by the state of Illinois. A staff of field investigators and their supervisors are responsible for collecting illness, risk factor, and exposure data from patients or their close contacts, providing education, and implementing necessary control measures. Coincident with these activities, patients' demographic information, including addresses of residence, are verified or updated.
In 2002, shigellosis data were collected on paper case reports and entered into the CDSS, a Microsoft Access database.17 SAS programs were written to extract shigellosis case data from the CDSS and reformat them for use in other applications.18 The reported residences of shigellosis patients were geocoded to census tracts using Intelligent Dispatcher, a web-based application available through the Chicago Department of Business and Information Services.19 Intelligent Dispatcher searches each given address against a tabular version of all possible geographies within Chicago, and outputs requested identifiers, such as census tract and mapping coordinates. Addresses that initially failed in this process were manually reviewed for typographical errors, or alternatively, paper case reports were reviewed to ensure that the correct address had been entered into the CDSS.
Analyses were conducted using SaTScan, a free software available online that was originally developed to identify cancer clusters.20 The prospective space-time module in SaTScan identifies “live” clusters of disease—cases that are current as of the date of the analysis and strongly associated in place and time.21 Fifty-two separate space-time analyses were run, one for each week of 2002. SaTScan required three data files for each run: one file with the geographic coordinates of the center of each Chicago census tract; one with the population of each tract; and one with case data. The same coordinate and population files were used for each run, but the case files were unique to the week analyzed.
The case file for each week consisted of the total number of cases in a census tract for each date that a case occurred there, beginning with earliest case in January 1999 and ending with the most recent case in the CDSS as of the week of the analysis. Data for cases that could not be geocoded (e.g., due to an unreported or invalid address) were excluded. An analysis was run for each Saturday of 2002, and only cases that had been registered in the CDSS prior to that Saturday were included. For example, in the analysis for Week 1 (pertaining to Saturday, January 5, 2002), the case with the most recent onset that had been entered into the CDSS by this date was December 21, 2001; SaTScan therefore searched for live clustering that included the illness onset date of this most recently occurring case.
The time between exposure and symptom onset for cases of shigellosis may be as long as seven days, so SaTScan was set to delineate live clusters that spanned ≤ 28 days (or four incubation periods) prior to the onset date of the most current case in the case file. The temporal scanning window was maintained at one year for each analysis; the Week 1 case file, for example, whose most recent case had onset on December 21, 2001, adjusted for analyses performed since December 22, 2000. The maximum spatial cluster size was set as a circle with radius ≤ 26,136 cartesian units (4.95 miles); at this setting, SaTScan would not delineate clusters larger than one-third the area of the city.
Clusters identified by SaTScan were described in terms of the range of illness onset dates of the cases in the cluster, the size of the cluster area in terms of the number of census tracts, the number of observed and expected cases in the cluster, the relative risk of being a case in the cluster, and the probability that a cluster of that magnitude or greater would be identified if the null hypothesis of no clusters were true (p-value). Additionally, the investigative findings of each case in the cluster were reviewed and clusters were described in terms of the reported frequency of the following risk factors for shigellosis transmission: a family or household member with diarrhea or confirmed Shigella sonnei infection around the time of the case-patient's illness; direct or indirect contact with a day care center, a summer camp, or international travel; swimming; or association with a known outbreak (defined as >2 laboratory-confirmed Shigella sonnei infections within 28 days in persons residing in separate households for whom a common exposure setting is identified). Graphical representations of the results were generated using ArcMap software.22
RESULTS
From January 1, 1999, to December 31, 2002, 1,261 cases of confirmed Shigella sonnei infection and 46 cases of confirmed Shigella infection with no reported serotype were registered in the CDSS. (In practice, cases with no serotype reported to CDPH are presumed to be Shigella sonnei unless determined otherwise.) Of these 1,307 cases, 1,259 (96%) had address information sufficient for geocoding and were included in the analysis. The 48 excluded cases were significantly more likely than included cases to have been registered in the CDSS in 2001, have a reported age of 20–49 years, and have no reported race.
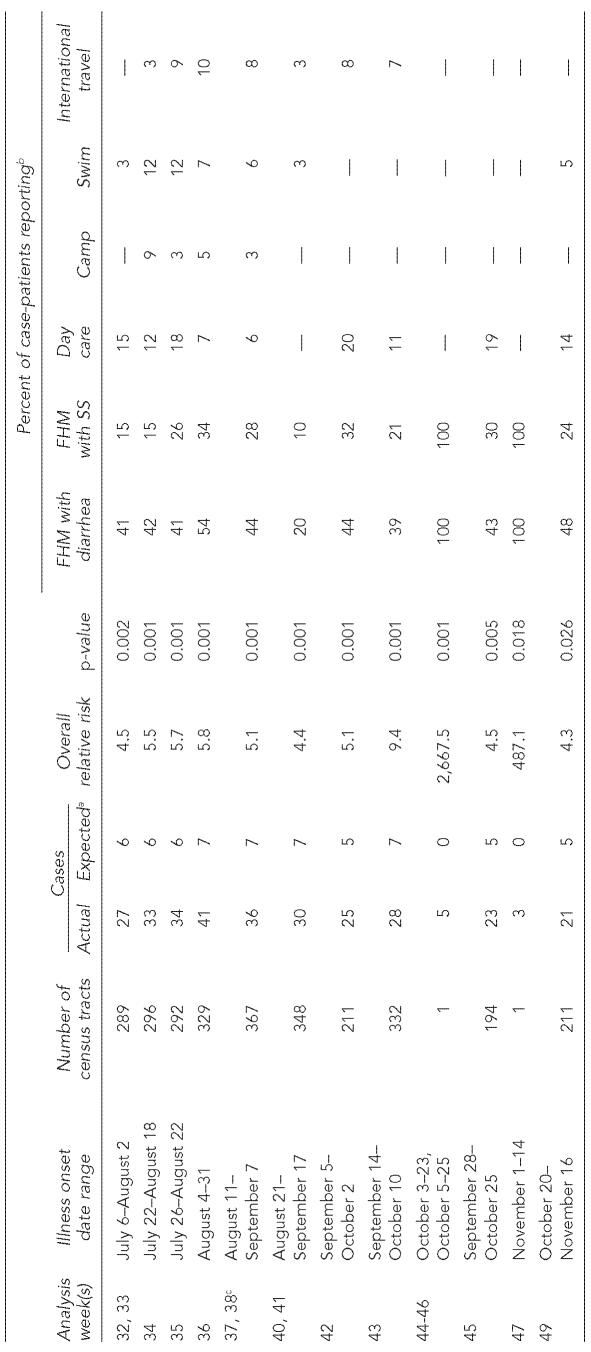
Twelve live clusters of shigellosis were detected at the p<0.05 significance level. These consisted of two single household clusters and 10 community clusters (Table). Four of the clusters were found in consecutive weeks. Many individual cases were located within multiple clusters. The community clusters ranged in size from 194 to 367 census tracts (median5294), and in disease burden from 21 to 41 cases (median=29). The most common investigative finding reported among cluster cases was family or household members with diarrhea around the same time as the case-patient. In the community clusters, the frequency of this report ranged from 20%–54% of all cases in the cluster (median=43%). A median of 25% of cases per cluster were found to have a family or household member with confirmed Shigella sonnei infection, and attendance at a day care center by a case-patient or household or family member was found for a median of 13% of cluster cases.
Table.
Descriptions of live clusters of confirmed shigellosis cases detected using the SaTScan prospective space-time scan statistic, Chicago, 2002
Rounded to nearest whole number
Risk factors are not mutually exclusive. Reported day care attendance, summer camp attendance, and international travel among family or household members (as well as case-patients themselves) are included in these percentages.
Values shown are for Week 38. The expected number of cases, number of census tracts in the cluster, and p-value did not change during the weeks that the cluster was identified, but the observed number of cases and overall relative risk in Week 38 changed from 35 to 36, and from 4.9 to 5.1, respectively.
FHM = family or household member
SS = Shigella sonnei infection
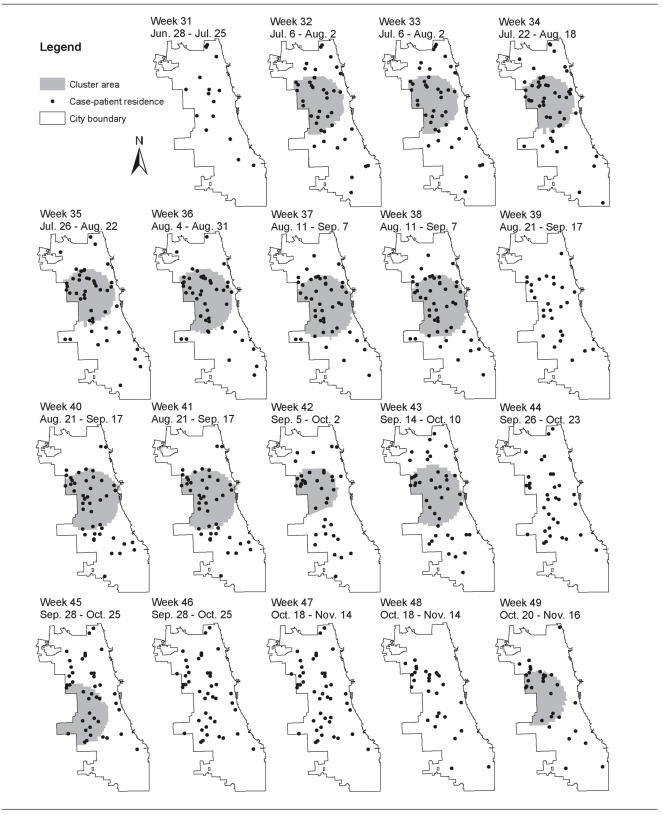
All of the community clusters were found in data pertaining to Weeks 32 to 49 and had a temporal span of 28 days. The Figure displays the areas detected as statistically significant community clusters at the p<0.05 level during Weeks 31 to 49, with the reported patient residences of live cases for that week also plotted. While 17 cases were registered in the CDSS with onset during June 28–July 25 by Week 31, the Week 32 analysis had 43 cases with onset during July 6–August 2, a 2.5-fold increase. The clustering first detected for Week 32 in the west-central part of the city was sustained over several subsequent weeks.
Figure.
Map sequence of Chicago shigellosis case-patient residences and areas delineated as community clusters using the the SaTScan prospective space-time scan statistic for weeks 31–49 of 2002. Dates shown are the illness onset date ranges of the shigellosis cases displayed. Single household clusters are not displayed.
Within the community clusters, 15 different day care centers were identified as potential exposure settings for case-patients or their close contacts. Two of these day care centers had documented outbreaks in 2002, and the earliest culture-confirmed cases in these outbreaks were included in the clusters delineated in the analyses for Weeks 32 and 42, respectively. Two other venue-associated outbreaks were investigated and reported in 2002, but none of the cases involved were identified within the clusters that were significant at the p<0.05 level. The earliest culture-confirmed case associated with one of these outbreaks (linked to a day care center) was included in a cluster identified in the Week 47 analysis that had an overall relative risk of 4.5 and p=0.150. Two cases associated with the fourth venue-associated outbreak of 2002 (linked to a summer camp) were included in a cluster found by the Week 31 analysis; it had an overall relative risk of 46.4, but p=0.99.
DISCUSSION
The prospective space-time scan statistic offers local health departments an objective way of describing clusters of shigellosis cases, and provides information to supplement what is seen when case-patients' residences are merely plotted on a map. The method we implemented could help prioritize the assignment and investigation of cases, particularly when overall shigellosis incidence exceeds expected numbers or when an agency's resources are stressed by other events, such as outbreaks. The West Nile virus disease outbreak that occurred nationally and in Chicago during the summer and fall of 2002 illustrates this situation, in that the epidemiologic, environmental, and public information resources of many local and state health departments in the United States were strained at that time.23
The simulated application of this method to Chicago's 2002 shigellosis surveillance data yielded 10 statistically significant community clusters and the identification of 15 day care centers that may have served as shigellosis transmission settings, including two at which confirmed outbreaks occurred. These findings suggest that prioritization of cases within a space-time cluster might decrease the delay between illnesses and public health interventions that halt spread within the facility and, by extension, in the surrounding community. In addition, in describing the characteristics of case-patients within the cluster, a profile of the kinds of individuals at risk may be developed for the purpose of communicating prevention methods appropriately.
Depending on the characteristics of the jurisdiction and the disease under study, there are several parameters in SaTScan that can be adjusted. These include shortening or lengthening the time period for which a cluster would be considered live, the maximum geographic size of a cluster, and the frequency with which analyses are run. For example, the temporal limit of 28 days used in our evaluation reflects the belief that sources of shigellosis transmission should be detected as soon as possible, and outbreaks that have persisted for longer than a month are likely to have already been recognized through traditional means, but this setting is easily changed in SaTScan. We decided to limit the results presented in this report to clusters that were significant at the p<0.05 level. However, based on the finding of two non-significant clusters that included index cases linked to venue-associated outbreaks, it is recommended that all clusters, regardless of their statistical significance, be considered for prioritization.
The prospective space-time scan statistic first delineated clustering in the 2002 Chicago data in cases registered by Week 32; this cluster in the west-central area of the city continued to be detected over several subsequent weeks. The objective and descriptive nature of these results would have been especially valuable in supporting decisions about prioritizing investigations and interventions at the time that the increase was occurring. The prospective space-time scan statistic may also be useful in detecting and tracking clusters of other reportable endemic diseases, particularly those that—like shigellosis—are easily transmitted via close personal contact. These diseases include, but are not limited to, hepatitis A virus infection, influenza, and pertussis.
In addition to the various software programs used, implementation of this method at CDPH required a wide array of personnel and skills. The resources included people to enter case reports into the CDSS; verify and collect patient data and communicate findings; update data and correct errors in the CDSS; write, maintain, and execute computerized routines to extract, reformat, and merge data for procedures in the different software programs; and generate and interpret the analysis results and maps. At the time of preparation of this manuscript, CDPH had completed the requirements gathering phase of the data collection module of a new Department-wide surveillance system. This effort is part of the National Electronic Disease Surveillance System initiative.24 The new system will geocode Chicago patient residence addresses in real-time, simultaneous with electronic receipt of case reports that are entered into the system by public health reporters, CDPH staff, or via an automated laboratory data transfer. In addition, CDPH recently implemented a West Nile virus surveillance system that incorporates various data sources and analysis tools, and has an automated mapping and reporting process.25 The methods and technologies used to query data, format and deliver them into analysis programs, create maps and reports, and evoke e-mail notifications may be applied in the future to the goal of shigellosis cluster detection as well.
While the prospective space-time scan statistic is a useful epidemiologic tool, it does not replace the human element of case investigations. Tracking down patients or their caretakers and communicating with administrators of at-risk settings such as day care centers cannot be done without investigators. Similarly, a program such as SaTScan is dependent on the accuracy, timeliness, and completeness of case report data. As improvements in electronic communicable disease reporting are implemented, there are likely to be more opportunities at the local level to measure the effect of the prospective space-time scan statistic in detecting clusters and facilitating timely disease control interventions.
Acknowledgments
The authors thank the Geographic Information Systems Division at the Chicago Department of Business and Information Services for its geocoding assistance, Kevin Gibbs of the CDPH Epidemiology Program for his assistance in creating maps, and the following individuals in the CDPH Communicable Disease Program for their work during 2002 in investigating cases and implementing measures to prevent further transmission: Mahasin Al-Amin, Vilma Alicea-Roman, Edgar Gutierrez, Charmaine Latimore, Loretta Miller, Charlayne Guy-Moore, Wilete Ishow, Joyce Jackson, Lula Johnson-White, Diana Laporte, Janetta Prokopowicz, Daisy Ross, and Laura Sparrow-Doyle.
REFERENCES
- 1.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta A, Polyak CS, Bishop RD, Sobel J, Mintz ED. Laboratory-confirmed shigellosis in the United States, 1989–2002: epidemiologic trends and patterns. Clin Infect Dis. 2004;38:1372–7. doi: 10.1086/386326. [DOI] [PubMed] [Google Scholar]
- 3.Mohle-Boetani JC, Stapleton M, Finger R, Bean NH, Poundstone J, Blake PA, Griffin PM. Communitywide shigellosis: control of an outbreak and risk factors in child day-care centers. Am J Public Health. 1995;85:812–6. doi: 10.2105/ajph.85.6.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day-care related outbreaks of rhamnose-negative Shigella sonnei—six states June 2001–March 2003. MMWR Morb Mortal Wkly Rep. 2004;53(3):60–3. [PubMed] [Google Scholar]
- 5.Shane AL, Tucker NA, Crump JA, Mintz ED, Painter JA. Sharing shigella: risk factors for a multicommunity outbreak of shigellosis. Arch Pediatr Adolesc Med. 2003;157:601–3. doi: 10.1001/archpedi.157.6.601-b. [DOI] [PubMed] [Google Scholar]
- 6.Shigellosis outbreak associated with an unchlorinated fill-and-drain wading pool—Iowa, 2001. MMWR Morb Mortal Wkly Rep. 2001;50(37):797–800. [PubMed] [Google Scholar]
- 7.Coles FB, Kondracki SF, Gallo RJ, Chalker D, Morse DL. Shigellosis outbreaks at summer camps for the mentally retarded in New York State. Am J Epidemiol. 1989;130:966–75. doi: 10.1093/oxfordjournals.aje.a115429. [DOI] [PubMed] [Google Scholar]
- 8.Sobel J, Cameron DN, Ismail J, Strockbine N, Williams M, Diaz PS, et al. A prolonged outbreak of Shigella sonnei infections in traditionally observant Jewish communities in North America caused by a molecularly distinct bacterial subtype. J Infect Dis. 1998;177:1405–9. doi: 10.1086/517825. [DOI] [PubMed] [Google Scholar]
- 9.Shigella sonnei outbreak among men who have sex with men—San Francisco, California, 2000–2001. MMWR Morb Mortal Wkly Rep. 2001;50(42):922–6. [PubMed] [Google Scholar]
- 10.Kimura AC, Johnson K, Palumbo MS, Hopkins J, Boase JC, Reporter R, et al. Multistate shigellosis outbreak and commercially prepared food, United States. Emerg Infect Dis. 2004;10:1147–9. doi: 10.3201/eid1006.030599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cromley EK, McLafferty SL. Analyzing the risk and spread of infectious diseases. In: Cromley EK, McLafferty SL, editors. GIS and Public Health. New York: Guilford Press; 2002. [Google Scholar]
- 12.Gatrell AC, Bailey TC. Interactive spatial data analysis in medical geography. Soc Sci Med. 1996;42:843–55. doi: 10.1016/0277-9536(95)00183-2. [DOI] [PubMed] [Google Scholar]
- 13.Yang DH, Bilaver LM, Hayes O, Goerge R. Improving geocoding practices: evaluation of geocoding tools. J Med Syst. 2004;28:361–70. doi: 10.1023/b:joms.0000032851.76239.e3. [DOI] [PubMed] [Google Scholar]
- 14.McKee KT, Shields TM, Jenkins PR, Zenilman JM, Glass GE. Application of a geographic information system to the tracking and control of an outbreak of shigellosis. Clin Infect Dis. 2000;31:728–33. doi: 10.1086/314050. [DOI] [PubMed] [Google Scholar]
- 15.O'Leary DR, Marfin AA, Montgomery SP, Kipp AM, Lehman JA, Biggerstaff BJ, et al. The epidemic of West Nile virus in the United States, 2002. Vector Borne Zoonotic Dis. 2004;4:61–70. doi: 10.1089/153036604773083004. [DOI] [PubMed] [Google Scholar]
- 16. ESRI, Inc., ArcView: Version 8.2. Redlands, CA: ESRI, Inc.; 2001.
- 17. Microsoft Corporation, Microsoft Access 2000. Redmond, WA: Microsoft Corporation; 1999.
- 18. SAS Institute Inc., SAS: Version 8.2 for Windows. Cary, North Carolina: SAS Institute, Inc., 2001.
- 19. Chicago Department of Business and Information Services, Intelligent Dispatcher: Version 1.6. Chicago: City of Chicago, 2002.
- 20.Kulldorf M. Information Management Services. SaTScan: Version 4.0.2, 2003. Available from: URL: http://www.satscan.org.
- 21.Kulldorf M. Prospective time periodic geographical disease surveillance using a scan statistic. J R Statist Soc A. 2001;164:61–72. [Google Scholar]
- 22. ESRI, Inc., ArcMap: Version 9.0. Redlands, CA: ESRI, Inc.; 2004.
- 23.Zohrabian A, Meltzer MI, Ratard R, Billah K, Molinari NA, Roy K, et al. Emerg Infect Dis. 2004. West Nile virus economic impact, Louisiana, 2002; pp. 1736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Electronic Disease Surveillance System Working Group. National Electronic Disease Surveillance System (NEDSS): a standards-based approach to connect public health and clinical medicine. J Public Health Manag Pract. 2001;7(6):43–50. [PubMed] [Google Scholar]
- 25.Gibbs K, Emmanuel A. Comprehensive GIS application for West Nile virus surveillance. Abstract. 2004 Health GIS Conference Proceedings, ESRI, Inc; Washington, DC. 2004. Available from URL: http://gis.esri.com/library/userconf/health04/abstracts/3018.html. [Google Scholar]