Abstract
A multiplex PCR procedure for detecting the aminoglycoside resistance genes aac(6′)-Ie-aph(2")-Ia, aph(2")-Ib, aph(2")-Ic, aph(2")-Id, aph(3′)-IIIa, and ant(4′)-Ia was evaluated and found to determine accurately the presence of these genes in enterococci.
High-level aminoglycoside resistance (MIC, ≥500 μg/ml) in enterococci is usually mediated by aminoglycoside-modifying enzymes, which eliminate the synergistic bactericidal effect of combined exposure to a cell wall-active agent and an aminoglycoside. Previously, among enterococci there had been only one gene, aac(6′)-Ie-aph(2")-Ia, known to mediate high-level resistance to gentamicin (6). In recent years, three new aminoglycoside resistance genes that also mediate resistance to gentamicin have been detected in enterococci: aph(2")-Ib, aph(2")-Ic, and aph(2")-Id (3, 10, 18). The aph(3′)-IIIa and ant(4′)-Ia genes also encode resistance in enterococci to various aminoglycosides but not to gentamicin (1, 7, 13, 17). Epidemiologic studies to detect the presence of the aac(6′)-Ie-aph(2")-Ia, aph(2")-Ib, aph(2")-Ic, and aph(2")-Id gentamicin resistance genes in clinical and animal enterococcal isolates have either utilized single PCR primer pairs for each gene in separate reactions or, at most, used PCR primers for two of these gentamicin resistance genes in one reaction (11). We describe a multiplex PCR procedure that enables detection of these four gentamicin resistance genes, plus the aph(3′)-IIIa and ant(4′)-Ia genes, in enterococcal isolates in a single reaction.
(This work was presented in part at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 16 to 19 December 2001.)
Bacterial strains.
One hundred thirteen enterococci from human (n = 57), farm animal (n = 35), and retail food (n = 21) sources were evaluated for six aminoglycoside resistance genes by multiplex PCR. Ninety-three of 113 had intermediate to high-level resistance to gentamicin (MIC range, 128 to ≥2,000 μg/ml) and 20 had low-level resistance to gentamicin (MICs, ≤16 μg/ml). Isolates were collected from 1986 to 2000 and included 65 of Enterococcus faecium, 45 of E. faecalis, 2 of E. gallinarum, and 1 of E. casseliflavus. Human isolates were from separate patients from 11 hospitals in three states. Separate animal and retail food isolates were obtained from 17 locations in seven states. In preliminary experiments, the aminoglycoside resistance gene content of all 113 isolates had been determined by conventional PCR methods with one pair of primers per reaction for each of six aminoglycoside resistance genes [aac(6′)-Ie-aph(2")-Ia, aph(2")-Ib, aph(2")-Ic, aph(2")-Id, aph(3′)-IIIa, and ant(4′)-Ia]. Of the 93 gentamicin-resistant isolates, all contained either the aac(6′)-Ie-aph(2")-Ia, aph(2")-Ib, aph(2")-Ic, or aph(2")-Id gene [one isolate contained both aac(6′)-Ie-aph(2")-Ia and aph(2")-Ic] (Table 1). The aph(3′)-IIIa gene was present in 80 of 113 isolates, and the ant(4′)-Ia gene was present in 26 of 113 isolates (Table 1). Five of the 20 isolates with low-level resistance to gentamicin contained none of the six genes studied (Table 1).
TABLE 1.
Aminoglycoside resistance gene content of 93 enterococci with intermediate to high-level gentamicin resistance and 20 enterococci with low-level gentamicin resistance (LLGR)
| No. of strains with gene(s) | Presence of gene:
|
|||||
|---|---|---|---|---|---|---|
| aac(6′)-Ie- aph(2")-Ia | aph(2")- Ib | aph(2")- Ic | aph(2")- Id | aph(3′)- IIIa | ant(4′)- Ia | |
| 17 | + | − | − | − | − | − |
| 8 | + | − | − | − | + | − |
| 1 | + | − | + | − | + | − |
| 7 | − | + | − | − | − | − |
| 12 | − | + | − | − | + | + |
| 2 | − | − | + | − | − | − |
| 20 | − | − | + | − | + | − |
| 1 | − | − | − | + | − | − |
| 12 | − | − | − | + | + | − |
| 13 | − | − | − | + | + | + |
| 14 (LLGR) | − | − | − | − | + | − |
| 1 (LLGR) | − | − | − | − | − | + |
| 5 (LLGR) | − | − | − | − | − | − |
Multiplex PCR.
Bacterial cells were lysed to obtain genomic DNA for PCR as follows: three to five bacterial colonies were emulsified in 25 μl of an 0.25% sodium dodecyl sulfate-0.05 N NaOH solution and boiled for 15 min. Then 200 μl of H2O was added to the mixture, and 5 μl of the diluted mixture was used in the PCR. PCR experiments were performed in a volume of 50 μl with the following in a reaction tube: 5 μl of DNA template (obtained as described above), 1.5 mM MgCl2, 0.1 mM (each) deoxynucleoside triphosphate, 1× PCR buffer, 2.5 U of Taq DNA polymerase (Invitrogen, Carlsbad, Calif.), and primer sets for the six aminoglycoside resistance genes (total of 12 primers; see Table 2). PCR primers were synthesized by Invitrogen. The amount of each primer in the PCR was as follows: 25 pmol for aac(6′)-Ie-aph(2")-Ia, 25 pmol for aph(2")-Ib, 3.5 pmol for aph(2")-Ic, 5 pmol for aph(2")-Id, 3 pmol for aph(3′)-IIIa, and 2 pmol for ant(4′)-Ia. PCR was performed in a Perkin-Elmer GeneAmp 2400 thermal cycler with an initial lysing step of 3 min at 94°C; 35 cycles of 40 s at 94°C, 40 s at 55°C, and 40 s at 72°C; and a final extension step of 2 min at 72°C. The nucleotide sequences of primer sets for each gene evaluated are shown in Table 2. PCR primers were designed with the aid of OMIGA (version 2.0) software (Genetics Computer Group, Madison, Wis.). PCR products were analyzed by electrophoresis at 100 V for 1 to 1[1/2] hours in a 2% agarose gel stained with ethidium bromide. DNAs from three strains (E. faecium SF11770, E. faecalis WBH18190, and E. faecium WBH17002) that together contained the six genes of interest were placed in a single tube for each set of multiplex PCR experiments, and the resulting six PCR products were used as positive controls and size markers in gel electrophoresis.
TABLE 2.
Nucleotide sequences of primer sets used to amplify aminoglycoside resistance genes in multiplex PCR experiments
| Aminoglycoside resistance gene | Product size (bp) | Primer sequence (5′ → 3′) |
|---|---|---|
| aac(6′)-Ie-aph(2")-Ia | 369 | CAGGAATTTATCGAAAATGGTAGAAAAG |
| CACAATCGACTAAAGAGTACCAATC | ||
| aac(6′)-Ie-aph(2")-Ia | 348 | CAGAGCCTTGGGAAGATGAAG |
| CCTCGTGTAATTCATGTTCTGGC | ||
| aph(2")-Ib | 867 | CTTGGACGCTGAGATATATGAGCAC |
| GTTTGTAGCAATTCAGAAACACCCTT | ||
| aph(2")-Ic | 444 | CCACAATGATAATGACTCAGTTCCC |
| CCACAGCTTCCGATAGCAAGAG | ||
| aph(2")-Id | 641 | GTGGTTTTTACAGGAATGCCATC |
| CCCTCTTCATACCAATCCATATAACC | ||
| aph(3′)-IIIa | 523 | GGCTAAAATGAGAATATCACCGG |
| CTTTAAAAAATCATACAGCTCGCG | ||
| ant(4′)-Ia | 294 | CAAACTGCTAAATCGGTAGAAGCC |
| GGAAAGTTGACCAGACATTACGAACT |
In preliminary experiments, the primer concentration used for each gene was the same (5 pmol of each primer), and two enterococcal colonies were inoculated directly into the thermal cycler for each multiplex reaction. This resulted in some ambiguities for a few isolates tested in the multiplex PCRs. Expected PCR products were visualized much more consistently when we changed the primers for each gene tested to the various concentrations provided in the above paragraph, and when we included a short sodium dodecyl sulfate-NaOH lysis step at the beginning of the procedure as stated above. The cell walls of a minority of enterococci are more difficult to lyse than those of other enterococci. The inclusion of a short lysis step improved the accuracy of the method without sacrificing an inordinate amount of time.
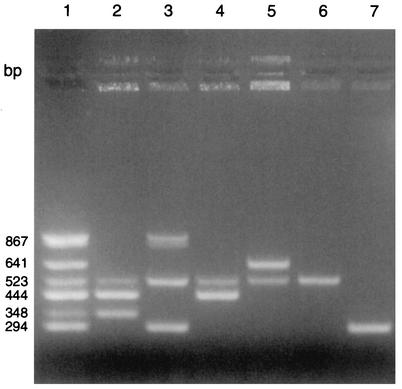
Since the aac(6′)-Ie-aph(2")-Ia gene has been associated with a transposon in E. faecalis (Tn5281), Staphylococcus aureus (Tn4001), and Staphylococcus epidermidis (Tn4031) (8, 15, 16), primer pairs 5′-CAGGAATTTATCGAAAATGGTAGAAAAG-3′ and 5′-CACAATCGACTAAAGAGTACCAATC-3′ with an expected PCR product of 369 bp were used in the multiplex PCR (Table 2). One primer is located near the end of the structural gene, and the other is located downstream of the structural gene and includes a portion of the transposon common to these three gram-positive bacteria. This would enable us to attempt to confirm whether the transposon was always associated with aac(6′)-Ie-aph(2")-Ia in our isolates. For the 26 isolates that were known to contain the aac(6′)-Ie-aph(2")-Ia gene, multiplex PCR consistently detected this gene in 23 isolates. The PCR product of one isolate was slightly smaller than the expected size (∼320 bp, rather than 369 bp). Subsequent nucleotide sequencing of this product showed that 54 bases had been deleted in a portion of the transposon. It is possible that, for the three isolates in which multiplex PCR did not detect the aac(6′)-Ie-aph(2")-Ia gene, there may have been a mutation in the location where one of the primers was designed to anneal. Multiplex PCR correctly detected the aph(2")-Ib, aph(2")-Ic, aph(2")-Id, aph(3′)-IIIa, and ant(4′)-Ia genes in all isolates compared to the conventional PCR method with one pair of primers per reaction for each of the aminoglycoside resistance genes. A new primer pair for the aac(6′)-Ie-aph(2")-Ia gene with an expected PCR product of 348 bp was then designed (Table 2) and used in the multiplex reaction for the six genes. Both new primers are located within the structural gene. The new primer pair enabled the aac(6′)-Ie-aph(2")-Ia gene to be visualized consistently, including from the three isolates in which the aac(6′)-Ie-aph(2")-Ia gene had not been detected with the previous primer pair. Figure 1 shows PCR products in a representative gel electrophoresis photo that includes isolates that together contain the six genes in various combinations, as well as the three isolates that made up our positive-control reaction (six genes in a single tube).
FIG. 1.
Agarose gel electrophoresis of multiplex PCR products from enterococcal isolates tested for six aminoglycoside resistance genes: aac(6′)-Ie-aph(2")-Ia (348 bp), aph(2")-Ib (867 bp), aph(2")-Ic (444 bp), aph(2")-Id (641 bp), aph(3′)-IIIa (523 bp), and ant(4′)-Ia (294 bp). Lane 1, positive control from a single reaction with PCR products from three isolates that together contain the six genes studied (see text). Lanes 2 to 7, six representative isolates, respectively, that possess one or more of the six genes studied.
The aac(6′)-Ie-aph(2")-Ia gene, which encodes the AAC(6′)-APH(2") bifunctional aminoglycoside-modifying enzyme, had been the only gene known to mediate high-level gentamicin resistance among enterococci. The AAC(6′)-APH(2") enzyme modifies essentially all the clinically available aminoglycosides, except streptomycin, and so the presence of high-level gentamicin resistance (MIC, ≥500 μg/ml) had been thought to eliminate the therapeutic use of all aminoglycosides other than streptomycin. However, the aph(2")-Ib, aph(2")-Ic, and aph(2")-Id genes, which encode other aminoglycoside-modifying enzymes, are now also known to mediate resistance to gentamicin in enterococci, but not resistance to some other aminoglycosides, such as amikacin (3, 10, 18). Thus, high-level gentamicin resistance in enterococci may not in all instances preclude the therapeutic use of other aminoglycosides (2). Previously, we had utilized primer pairs for each gentamicin resistance gene separately in molecular epidemiologic studies of gentamicin-resistant enterococci, which proved quite time-consuming. The present multiplex PCR assay can screen all four gentamicin resistance genes in one reaction.
Besides the six genes that we studied, three other aminoglycoside resistance genes that encode aminoglycoside-modifying enzymes have been found in enterococci: aac(6′)-Ii, ant(3")-Ia, and ant(6′)-Ia (4, 5, 9, 12, 14). We did not include aac(6′)-Ii in the multiplex PCR assay, since this gene is known to be intrinsic to all E. faecium isolates and has not been found to date in other enterococcal species (5, 11). The ant(3")-Ia and ant(6′)-Ia genes also were not included in the multiplex PCR assay. Unlike the other aminoglycoside resistance genes, each of which mediates resistance to multiple aminoglycosides, ant(3")-Ia and ant(6′)-Ia mediate only high-level streptomycin resistance (MIC, ≥1,000 μg/ml) in enterococci and not resistance to any of the other aminoglycosides used clinically. Thus, the presence of either of these two genes in enterococci could easily be detected by conventional MIC testing in clinical microbiology laboratories.
A previous study of the epidemiology of aminoglycoside resistance genes in enterococci used either three or four pairs of primers in each PCR and did not include primers for the aph(2")-Ib or aph(2")-Id genes (11). Also, the presence of the aminoglycoside resistance genes had not been previously verified by individual reactions prior to the multiplex PCR assay. The multiplex PCR method described herein is a convenient method for rapid detection of the presence of six aminoglycoside resistance genes in enterococci. However, it should be noted that, due to the intrinsic limitations of any PCR assay, a negative result may not always signify the absence of a gene and a positive result, in the absence of correlative MIC data, may not always signify the presence of an intact gene. For example, a single mutation in the region of the gene responsible for primer annealing may significantly decrease efficiency of annealing and produce less or no PCR product; furthermore, nucleotide insertions or deletions in regions outside the PCR product that may inactivate the gene would not be detected. Nonetheless, multiplex PCR remains a useful molecular epidemiologic tool to aid detection of aminoglycoside resistance genes and, in a few selected cases, may also prove helpful in determining what aminoglycoside might be efficacious therapeutically in a clinical setting (2).
Acknowledgments
This study was supported in part by a Merit Review grant from the Medical Research Service of the Department of Veterans' Affairs.
REFERENCES
- 1.Carlier, C., and P. Courvalin. 1990. Emergence of 4′,4"-aminoglycoside nucleotidyltransferase in enterococci. Antimicrob. Agents Chemother. 34:1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow, J. W. 2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31:586-589. [DOI] [PubMed] [Google Scholar]
- 3.Chow, J. W., M. J. Zervos, S. A. Lerner, L. A. Thal, S. M. Donabedian, D. D. Jaworski, S. Tsai, K. J. Shaw, and D. B. Clewell. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, N. C., Ø. Olsvik, J. M. Swenson, C. A. Spiegel, and F. C. Tenover. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 43:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa, Y., M. Galimand, R. Leclercq, J. Duval, and P. Courvalin. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37:1896-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray, G. S., and W. M. Fitch. 1983. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol. Biol. Evol. 1:57-66. [DOI] [PubMed] [Google Scholar]
- 8.Hodel-Christian, S. L., and B. E. Murray. 1991. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob. Agents Chemother. 35:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingshead, S., and D. Vapnek. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13:17-30. [DOI] [PubMed] [Google Scholar]
- 10.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2")-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 44:2876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi, N., M. Alam, Y. Nishimoto, S. Urasawa, N. Uehara, and N. Watanabe. 2001. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 126:197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krogstad, D. J., R. R. Korfhagen, R. C. Moellering, Jr., C. Wennersten, and M. N. Swartz. 1978. Aminoglycoside-inactivating enzymes in clinical isolates of Streptococcus faecalis: an explanation for antibiotic synergism. J. Clin. Investig. 62:480-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura, M., Y. Katakura, T. Imanaka, and S. Aiba. 1984. Enzymatic and nucleotide sequence studies of a kanamycin-inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J. Bacteriol. 160:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ounissi, H., and P. Courvalin. 1987. Nucleotide sequences of streptococcal genes, p. 275, appendix B. In J. J. Ferretti and R. Curtiss III (ed.), Streptococcal genetics. American Society for Microbiology, Washington, D.C.
- 15.Rouch, D. A., M. E. Byrne, Y. C. Kong, and R. A. Skurray. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J. Gen. Microbiol. 133:3039-3052. [DOI] [PubMed] [Google Scholar]
- 16.Thomas, W. D., and G. L. Archer. 1989. Mobility of gentamicin resistance genes from staphylococci isolated in the United States: identification of Tn4031, a gentamicin resistance transposon from Staphylococcus epidermidis. Antimicrob. Agents Chemother. 33:1335-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 18.Tsai, S. F., M. J. Zervos, D. B. Clewell, S. M. Donabedian, D. F. Sahm, and J. W. Chow. 1998. A new high-level gentamicin resistance gene, aph(2")-Id, in Enterococcus spp. Antimicrob Agents Chemother. 42:1229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]