Abstract
To evaluate the cross-resistance profile of the human immunodeficiency virus type 1 protease inhibitor (PI) atazanavir (BMS-232632), a panel of 551 clinical isolates exhibiting a wide array of PI resistance profiles and a variety of genotypic patterns were assayed for susceptibility to atazanavir and six other PIs: amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir. In general, reductions in atazanavir susceptibility in vitro required several amino acid changes and were relatively modest in degree, and susceptibility was retained among isolates resistant to one or two of the currently approved PIs. There was a clear trend toward loss of susceptibility to atazanavir, as isolates exhibited increasing levels of cross-resistance to multiple PIs. Atazanavir appeared to have a distinct resistance profile relative to each of the other six PIs tested based on susceptibility comparisons against this panel of resistant isolates. Analysis of the genotypic profiles of 943 PI-susceptible and -resistant clinical isolates identified a strong correlation between the presence of amino acid changes at specific residues (10I/V/F, 20R/M/I, 24I, 33I/F/V, 36I/L/V, 46I/L, 48V, 54V/L, 63P, 71V/T/I, 73C/S/T/A, 82A/F/S/T, 84V, and 90M) and decreased susceptibility to atazanavir. While no single substitution or combination of substitutions was predictive of atazanavir resistance (change, >3.0-fold), the presence of at least five of these substitutions correlated strongly with loss of atazanavir susceptibility. Mutations associated with reduced susceptibility to each of the other six PIs were also determined.
Protease inhibitors (PIs) are potent and effective antiretrovirals. However, the extensive use of PIs has led to the emergence of resistant human immunodeficiency virus type 1 (HIV-1) variants that possess various degrees of cross-resistance to other members in the class (13, 33). PI-resistant HIV-1 can be isolated from patients treated with current PIs (35) as well as from patients who have not received any treatment with current PIs (5, 10, 20, 21), indicating that transmission of PI-resistant HIV-1 can occur during primary infection. Drug-resistant HIV-1 variants have been detected in as many as 26% of newly infected, treatment-naïve patients. HIV-1 isolates resistant to nonnucleoside reverse transcriptase inhibitors were the most common (15% to 26%), while those resistant to nucleoside reverse transcriptase inhibitors and PIs were found less often (4% to 8% and 1% to 10%, respectively) (34).
The correlation between HIV-1 genotypic mutations and phenotypic resistance to PIs remains poorly understood (23, 25, 32). Patients on combination regimens that include PIs can fail in the absence of resistance-producing signature mutations (2, 9, 12), and the extent to which a variety of mutational combinations can lead to PI resistance is not well defined (3, 18). While many of the available PIs have signature mutations predictive of decreased susceptibility, secondary substitutions appear to play a major role in determining the level of resistance and extent of cross-resistance. Both phenotype- and genotype-guided treatment for HIV-1 infection in patients failing active antiretroviral therapy was of benefit compared to no resistance testing (4, 6, 7). However, no consensus interpretation algorithm for genotypes is available, and all algorithms are prone to misclassifying individual viruses due to incomplete understanding of the relationship between genotype, phenotype, and clinical response. The use of phenotypic monitoring appears to be a more reliable approach, given the complex nature of amino acid substitutions involved in PI resistance.
Atazanavir (BMS-232632) is an azapeptide inhibitor of the HIV-1 protease currently in phase III clinical development (27, 29). Atazanavir is one of the most potent PIs, having a 50% effective concentration (EC50) of 3 to 5 nM against a variety of HIV-1 isolates in different cell types and is a highly selective and effective inhibitor of the HIV-1 protease (Ki of <1 nM) (29). Comparative anti-HIV-1 studies in vitro suggest that atazanavir is more potent than currently approved HIV-1 PIs, even in the presence of 40% human serum (29). Furthermore, clinical studies have demonstrated that atazanavir possesses the pharmacokinetic properties that enable once-daily dosing in the absence of added ritonavir (E. M. O'Mara, J. Smith, S. J. Olsen, T. Tanner, A. E. Schuster, and S. Kaul, 38th Intersci. Conf. Antimicrob. Agents Chemother., 1998, abstr. I-242).
In vitro passage of HIV-1 in the presence of atazanavir results in the selection of resistant variants (11). Genotypic analysis of three different HIV strains resistant to atazanavir indicated that an N88S substitution in the viral protease appeared first during the selection process in two of the three strains, along with an I50L substitution in one of the strains. An I84V change appeared to be an important substitution in the third strain used, and all three variants required multiple changes to achieve significant resistance levels. Mutations were also observed at the protease cleavage sites following drug selection. The evolution to resistance seemed somewhat distinct for each of the three strains utilized, suggesting that multiple pathways to resistance are possible and confirming the importance of viral genetic background in resistance generation (30).
The objective of this study was to obtain a greater understanding of the overall resistance profile of atazanavir relative to other available PIs in comparative studies with standardized phenotypic assays against a large panel of clinical isolates resistant to other PIs. In addition, genotypic patterns were evaluated in an attempt to define amino acid substitutions and patterns predictive of decreased atazanavir susceptibility.
MATERIALS AND METHODS
Compounds.
Atazanavir was synthesized by Bristol-Myers Squibb (27, 29). The other six PIs were provided by the testing sites.
Viral isolates.
A panel of 950 HIV-1 recombinant clinical isolates was profiled at either ViroLogic, Inc. (South San Francisco, Calif.), LabCorp (Raleigh, N.C.) or Virco NV. (Mechelen, Belgium). The vast majority of clinical isolates, 890, were randomly acquired during the prescreening enrollment process for clinical study AI424-009 (atazanavir/saquinavir combination treatment of PI-experienced subjects, N = 138), AI424-043 (comparison of atazanavir against lopinavir/ritonavir in PI-experienced subjects, N = 302), and AI424-045 (atazanavir in combination with ritonavir or saquinavir against lopinavir/ritonavir in PI-experienced subjects, N = 450). The remaining 60 isolates were initially selected so as to generate a panel with diverse patterns of resistance and cross-resistance to the approved PIs. The ViroLogic isolates (N = 32) were cloned by retransforming bacteria with patient-derived resistance test vector pools, picking colonies, and isolating resistance test vector DNA, which was then screened for functionality, PI susceptibility, and genotype. For the Virco isolates (N = 28), protease and reverse transcriptase coding sequences were amplified from patient-derived viral RNA with HIV-1-specific primers. After homologous recombination of amplicons into a protease-reverse transcriptase-deleted proviral clone, the resulting recombinant viruses were harvested, counted, and used for in vitro susceptibility testing.
Phenotypic assays.
All isolates were assayed for susceptibility to atazanavir, amprenavir, nelfinavir, ritonavir, and saquinavir in addition to either indinavir (Virco, Virologic, and AI424-009 samples) or lopinavir (AI424-043 and AI424-045 samples) with ViroLogic's PhenoSense (24) or Virco's Antivirogram (14, 15) phenotypic assays. Reference atazanavir EC50s were 0.9 to 2.0 nM (ViroLogic) and 6.5 to 8.1 nM (Virco), with the observed differences likely accounted for by the single (ViroLogic) or multicycle (Virco) infection assay used by each company. Both assay systems were previously shown to give comparable fold-change results for other drugs (26). Reference strains were NL4-3 (ViroLogic) and HXB2 (Virco).
Sequence analysis.
Genotypic profiles were determined for all clinical isolates at either ViroLogic, Virco, or LabCorp (Raleigh, N.C.) with the GenoSure assay (J. Sebastian and T. M. Alcorn, 2002, Advance for Medical Laboratory Professionals, http://www.advanceforal.com/pastarticles/nov11_02feature4.html). Genotypes of isolates were determined by dye terminator sequencing and analysis on the ABI 3700 automatic sequencer and are reported as amino acid differences compared to NL4-3 (Virologic) or HXB2 (Virco and LabCorp). The sequencing methodology allows the detection of the simultaneous presence of different amino acids at any position of the region sequenced. The level of detecting such mixtures is on the order of 10 to 20% of the less abundant sequence. Whenever such a mixture of sequences was observed in a sample, the clinical isolate was catalogued as being mutant. The only positions in the protease gene that differ between the reference sequences are 3 and 37. Subsets of susceptible and resistant isolates containing individual substitutions (Table 2) were compared with P values based on Fisher's exact test.
TABLE 2.
Genotypic resistance profile: correlation of amino acid changes with loss of PI susceptibilitya
| PIa | Isolates | No. of strains | Primary substitutions
|
Secondary substitutions
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 N | 48 V | 50 V | 82 A/F/S/T | 84 V | 90 M | 10 I/V/F | 20 R/M/I | 24 I | 33 I/F/V | 36 I/L/V | 46 I/L | 54 V/L | 63 P | 71 V/T/I | 73 C/S/T/A | 88 D/S/T | 93 L | |||
| ATV | Total no. of isolates | 943 | 117 | 33 | 12 | 220 | 97 | 307 | 410 | 170 | 44 | 81 | 369 | 245 | 175 | 634 | 349 | 86 | 122 | 417 |
| No. (%) susceptible | 634 | 89 (14) | 7 (1) | 11 (2) | 69 (11) | 9 (1) | 115 (18) | 174 (27) | 80 (13) | 10 (2) | 32 (5) | 223 (35) | 93 (15) | 45 (7) | 372 (59) | 144 (23) | 11 (2) | 72 (11) | 262 (41) | |
| No. (%) resistant | 309 | 28 (9) | 26 (8) | 1 (<1) | 151 (49) | 88 (28) | 192 (62) | 236 (76) | 90 (29) | 34 (11) | 49 (16) | 146 (47) | 152 (49) | 130 (42) | 262 (85) | 205 (66) | 75 (24) | 50 (16) | 155 (50) | |
| P | 0.0349 | <0.0001 | 0.117 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0005 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0491 | 0.0119 | ||
| APV | No. (%) susceptible | 719 | 107 (15) | 20 (3) | 1 (<1) | 114 (16) | 17 (2) | 165 (23) | 234 (33) | 111 (15) | 23 (3) | 33 (5) | 275 (38) | 114 (16) | 84 (12) | 444 (62) | 208 (29) | 31 (4) | 106 (15) | 314 (44) |
| No. (%) resistant | 224 | 10 (4) | 13 (6) | 11 (5) | 106 (47) | 80 (36) | 142 (63) | 176 (79) | 59 (26) | 21 (9) | 48 (21) | 94 (42) | 131 (58) | 91 (41) | 190 (85) | 141 (63) | 55 (25) | 16 (7) | 103 (46) | |
| P | <0.0001 | 0.038 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0003 | 0.0004 | <0.0001 | 0.3469 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0029 | 0.5898 | ||
| NFV | No. (%) susceptible | 465 | 21 (5) | 5 (1) | 8 (2) | 26 (6) | 5 (1) | 73 (16) | 104 (22) | 52 (11) | 7 (2) | 22 (5) | 165 (35) | 34 (7) | 15 (3) | 246 (53) | 78 (17) | 9 (2) | 20 (4) | 168 (36) |
| No. (%) resistant | 478 | 96 (20) | 28 (6) | 4 (1) | 194 (41) | 92 (19) | 234 (49) | 306 (64) | 118 (25) | 37 (8) | 59 (12) | 204 (43) | 211 (44) | 160 (33) | 388 (81) | 271 (57) | 77 (16) | 102 (21) | 249 (52) | |
| P | <0.0001 | <0.0001 | 0.2578 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0276 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| RTV | No. (%) susceptible | 539 | 89 (17) | 4 (1) | 3 (1) | 14 (3) | 2 (<1) | 66 (12) | 124 (23) | 55 (10) | 5 (1) | 24 (4) | 189 (35) | 46 (9) | 12 (2) | 314 (58) | 101 (19) | 7 (1) | 82 (15) | 225 (42) |
| No. (%) resistant | 404 | 28 (7) | 29 (7) | 9 (2) | 206 (51) | 95 (24) | 241 (60) | 286 (71) | 115 (28) | 39 (10) | 57 (14) | 180 (45) | 199 (49) | 163 (40) | 320 (79) | 248 (61) | 79 (20) | 40 (10) | 192 (48) | |
| P | <0.0001 | <0.0001 | 0.0361 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0037 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0184 | 0.085 | ||
| SQV | No. (%) susceptible | 685 | 99 (14) | 1 (<1) | 9 (1) | 115 (17) | 7 (1) | 117 (17) | 205 (30) | 95 (14) | 25 (4) | 38 (6) | 249 (36) | 127 (19) | 67 (10) | 414 (60) | 168 (24) | 17 (2) | 91 (13) | 300 (44) |
| No. (%) resistant | 257 | 18 (7) | 32 (12) | 3 (1) | 105 (41) | 90 (35) | 190 (74) | 205 (80) | 75 (29) | 19 (7) | 43 (17) | 120 (47) | 118 (46) | 108 (42) | 220 (86) | 181 (70) | 69 (27) | 31 (12) | 117 (46) | |
| P | 0.0018 | <0.0001 | 1 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0228 | <0.0001 | 0.0044 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.6642 | 0.6588 | ||
| IDV | Total no. of isolates | 198 | 32 | 5 | 4 | 64 | 18 | 59 | 90 | 33 | 17 | 16 | 67 | 74 | 50 | 134 | 75 | 15 | 32 | 82 |
| No. (%) susceptible | 111 | 27 (24) | 4 (4) | 4 (4) | 5 (5) | 2 (2) | 19 (17) | 26 (23) | 10 (9) | 0 | 10 (9) | 39 (35) | 16 (14) | 6 (5) | 62 (56) | 29 (26) | 1 (1) | 22 (20) | 41 (37) | |
| No. (%) resistant | 87 | 5 (6) | 1 (1) | 0 | 59 (68) | 16 (18) | 40 (46) | 64 (74) | 23 (26) | 17 (20) | 6 (7) | 28 (32) | 58 (67) | 44 (51) | 72 (83) | 46 (53) | 14 (16) | 10 (11) | 41 (47) | |
| P | 0.0004 | 0.3872 | 0.1322 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0018 | <0.0001 | 0.7937 | 0.7624 | <0.0001 | <0.0001 | <0.0001 | 0.0002 | <0.0001 | 0.1244 | 0.1907 | ||
| LPV | Total no. of isolates | 745 | 85 | 28 | 8 | 156 | 79 | 248 | 320 | 137 | 27 | 65 | 302 | 171 | 125 | 500 | 274 | 71 | 90 | 335 |
| No. (%) susceptible | 553 | 78 (14) | 11 (2) | 0 | 43 (8) | 12 (2) | 127 (23) | 166 (30) | 72 (13) | 9 (2) | 25 (5) | 214 (39) | 65 (12) | 33 (6) | 338 (61) | 147 (27) | 25 (5) | 80 (14) | 242 (44) | |
| No. (%) resistant | 192 | 7 (4) | 17 (9) | 8 (4) | 113 (59) | 67 (35) | 121 (63) | 154 (80) | 65 (34) | 18 (9) | 40 (21) | 88 (46) | 106 (55) | 92 (48) | 162 (84) | 127 (66) | 46 (24) | 10 (5) | 93 (48) | |
| P | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0883 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0005 | 0.2744 | ||
Numbers in parentheses indicate the percentage of isolate subsets of susceptible or resistant isolates containing the indicated substitution.
RESULTS
The susceptibility profile for atazanavir and five of the six approved PIs (amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir) was evaluated against a diverse set of 950 HIV-1 clinical isolates. Sixty of the HIV-1 clinical isolates were initially selected from the ViroLogic and Virco collections as being representative of viruses displaying reduced PI susceptibility to one or more inhibitors. Data on an additional 890 isolates were generated as a result of the preenrollment screening process for entry into clinical studies AI424-009, AI424-043, and AI424-045, which examined the effectiveness of atazanavir in various combinations in patients experiencing virologic failure on PI-containing regimens. Clade analysis of 624 of these isolates showed that 163 (26.2%) were non-B clade isolates, with representative viruses from clades A, C, AE, AG, D, F, G, and J.
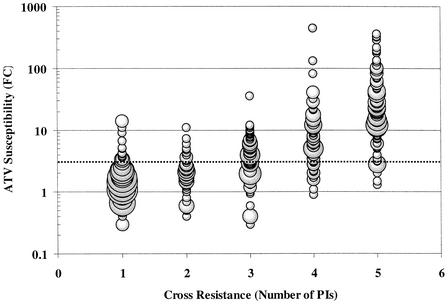
Preliminary analysis of the variability of atazanavir susceptibility in genotypically wild-type samples suggests that the vast majority (>99%) of such viruses have an atazanavir fold change (FC) of 2.3-fold or less. For comparative purposes, resistance was arbitrarily defined as a change in susceptibility of >3.0-fold, as precise clinically relevant resistance levels have yet to be established for these PIs. Of the 950 isolates analyzed, 551 displayed susceptibility levels >3.0-fold higher than the reference strain to one or more of the approved PIs and composed the panel of PI-resistant isolates that were evaluated. Figure 1 displays the spectrum of observed atazanavir FCs across this panel of 551 resistant isolates. Atazanavir susceptibility was retained against isolates resistant to one to two PIs, with a clear trend toward loss of atazanavir susceptibility as isolates become resistant to three, four, and five PIs. The vast majority of the isolates resistant to upwards of four PIs showed intermediate FCs of between 3- and 10-fold, while most of the isolates resistant to at least five PIs showed atazanavir FCs of >10-fold. The median atazanavir FC for isolates resistant to one to five PIs was 1.6-, 2.1-, 4.0-, 6.2-, and 22.0-fold, respectively.
FIG. 1.
Relationship of atazanavir susceptibility and cross-resistance to other PIs. The atazanavir (ATV) FC for all 551 isolates listed in Table 1 are plotted based on their cross-resistance profile for five of the six marketed PIs (amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir) and susceptibility to atazanavir. Diameter of plotted points corresponds directly to the number of isolates displaying the indicated value. The dotted line identifies the position of a 3.0-fold change in susceptibility.
Table 1 lists the cross-resistance groupings of the 551 clinical isolates included in the panel. Of the PI-resistant clinical isolates evaluated, 157 exhibited >3.0-fold changes in susceptibility levels to a single marketed PI (predominantly nelfinavir). The high percentage of nelfinavir-resistant isolates is most likely reflective of extensive prior nelfinavir usage. The 121 isolates specifically resistant to nelfinavir and included here are a representative subset of an even larger number of nelfinavir-resistant isolates identified and were chosen based on diversity of genotypic patterns. While modest decreases in atazanavir susceptibility (median FC = 4.0-fold) were observed with 18 of the nelfinavir-resistant isolates, there was no obvious correlation with the magnitude of nelfinavir resistance or mutational patterns (data not shown), and atazanavir susceptibility was retained against 85% of these nelfinavir-resistant isolates. Atazanavir susceptibility (82%) was also retained among a subset of 44 isolates exhibiting nelfinavir-specific resistance which did not contain the nelfinavir signature substitution D30N, whose presence has been previously reported to confer susceptibility to other PIs (22).
TABLE 1.
Susceptibility to ATV of a panel of PI-resistant clinical isolates
| Resistance group and PI | No. of isolates | No. (%) of ATV- susceptible isolates | ATV-resistant isolates
|
||
|---|---|---|---|---|---|
| No. | FC range | Median FC | |||
| 1 of 5 PIs | |||||
| APV | 2 | 2 | 0 | NAa | NA |
| NFV | 121 | 103 | 18 | 3-14 | 4.0 |
| RTV | 27 | 26 | 1 | 3.3 | NA |
| SQV | 7 | 7 | 0 | NA | NA |
| Total | 157 | 138 (88%) | 19 | ||
| 2 of 5 PIs | |||||
| APV/LPV | 1 | 1 | 0 | NA | NA |
| NFV/SQV | 5 | 2 | 3 | 4-7 | 4.8 |
| IDV/NFV | 3 | 1 | 2 | 5-6 | 5.5 |
| APV/RTV | 5 | 5 | 0 | NA | NA |
| IDV/RTV | 3 | 2 | 1 | 4.0 | NA |
| NFV/RTV | 30 | 25 | 5 | 3-11 | 3.6 |
| LPV/RTV | 2 | 2 | 0 | NA | NA |
| RTV/SQV | 8 | 8 | 0 | NA | NA |
| Total | 57 | 46 (81%) | 11 | ||
| 3 of 5 PIs | |||||
| NFV/RTV/SQV | 28 | 7 | 21 | 3-36 | 5.5 |
| NFV/RTV/LPV | 32 | 13 | 19 | 3-9 | 4.5 |
| APV/NFV/RTV | 2 | 1 | 1 | 4.3 | NA |
| APV/LPV/RTV | 5 | 5 | 0 | NA | NA |
| IDV/NFV/SQV | 1 | 0 | 1 | 5.4 | NA |
| IDV/NFV/RTV | 21 | 6 | 15 | 4-12 | 6.5 |
| APV/RTV/SQV | 9 | 1 | 8 | 3-12 | 7.4 |
| LPV/NFV/SQV | 1 | 1 | 0 | NA | NA |
| Total | 99 | 34 (34%) | 65 | ||
| 4 of 5 PIs | |||||
| NFV/LPV/RTV/SQV | 25 | 5 | 20 | 3-449 | 7.0 |
| APV/SQV/RTV/LPV | 1 | 0 | 1 | 11 | NA |
| APV/IDV/NFV/RTV | 18 | 6 | 12 | 3-12 | 5.6 |
| IDV/NFV/RTV/SQV | 10 | 0 | 10 | 3-82 | 10.2 |
| APV/NFV/LPV/RTV | 17 | 4 | 13 | 3-18 | 5.9 |
| LPV/NFV/RTV/SQV | 25 | 0 | 25 | 5-41 | 12.0 |
| Total | 96 | 15 (16%) | 81 | ||
| 5 of 5 PIs | |||||
| APV/NFV/LFV/RTV/SQV | 111 | 5 | 106 | 3-362 | 23.0 |
| APV/IDV/NFV/RTV/SQV | 31 | 2 | 29 | 4-41 | 16.0 |
| Total | 142 | 7 (5%) | 135 | ||
NA, not applicable; APV, amprenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; RTV, ritonavir; SQV, saquinavir.
Apart from the observation of some cross-resistance between atazanavir and 18 of the nelfinavir-resistant isolates, atazanavir susceptibility was retained against this subset of 157 isolates resistant to a single PI (Table 1), and there was no obvious pattern of cross-resistance between atazanavir and any of the other PIs. This was further illustrated by the finding that 46 of 57 (81%) isolates in the panel that were cross-resistant to two marketed PIs remained susceptible to atazanavir (Table 1). Of the 11 isolates displaying modest reductions in atazanavir susceptibility, 10 were resistant to nelfinavir and either indinavir, ritonavir, or saquinavir and 1 was resistant to ritonavir and indinavir. Only two of these isolates showed an FC in atazanavir susceptibility of >6-fold. As HIV-1 isolates exhibited increased cross-resistance to multiple marketed PIs, atazanavir susceptibility also decreased. Of the 99 isolates resistant to three marketed PIs (predominantly including nelfinavir plus ritonavir and either indinavir, lopinavir, or saquinavir), 34 (34%) retained atazanavir susceptibility and 65 isolates showed some cross-resistance (FC of 4.1- to 15-fold) to atazanavir, although it is important to note that the median FC was only 4.0-fold for this panel of isolates (Table 1). While susceptibility to atazanavir was readily lost when isolates were cross-resistant to five of the other approved PIs tested, seven isolates retained atazanavir susceptibility despite being resistant to all of the other PIs tested. Fifteen (16%) of the 96 isolates that were resistant to four marketed PIs also retained susceptibility to atazanavir.
Overall, atazanavir susceptibility changes of 3.0-fold or less were exhibited by 184 (86%) of the 214 isolates that were resistant to one to two PIs and by 49 (25%) of the 195 isolates that were resistant to three or four PIs. For comparative purposes, amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir susceptibility was retained against 96%, 89% (46 of 52 isolates), 98% (159 of 162 isolates), 26%, 65%, and 91%, respectively, for the same group of isolates resistant to one to two PIs and 61%, 11% (6 of 56), 42% (58 of 139), 8%, 1%, and 49%, respectively, for the 195 isolates cross-resistant to three to four PIs. In general, resistance levels to atazanavir were relatively modest in degree compared to the other PIs tested, with only 23% of the 176 atazanavir-resistant isolates (from the panel of isolates resistant to one to four PIs) displaying changes in susceptibility of greater than 10-fold. A similar analysis of isolates resistant to amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir yielded values of 24%, 46%, 39%, 57%, 60%, and 42%, respectively. The atazanavir median FC for all 551 resistant isolates was 3.9-fold, with a median FC of 9.5-fold for the subset of 311 isolates showing an atazanavir FC of >3.0-fold (Table 1).
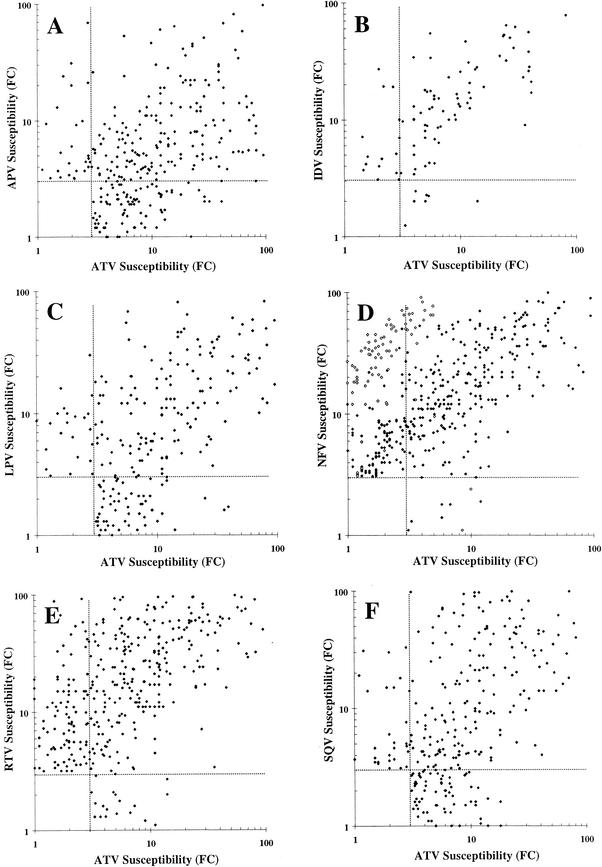
A review of the resistance patterns for isolates resistant to one to three PIs suggests that atazanavir has a distinct resistance profile, in that there is no obvious pattern of atazanavir cross-resistance with any particular PI. This distinctiveness readily evaporated when isolates became cross-resistant to four or more PIs. To examine this further, a series of scatter plots were generated comparing the susceptibility patterns of atazanavir against each of the six other PIs against the panel of PI-resistant isolates. In each panel of Fig. 2, subsets of the 551 PI-resistant isolates were identified which showed >3.0-fold decreased susceptibility to atazanavir and/or the comparator PI analyzed, with isolates susceptible to both PIs excluded. Each graph is divided into four regions: top left, isolates susceptible to atazanavir and resistant to the second PI; bottom right, isolates susceptible to the second PI and resistant to atazanavir; and top right, isolates showing some degree of cross-resistance to both PIs.
FIG. 2.
Comparative scatter plots of resistant isolates. Each panel plots individual isolates displaying a ≤3.0-fold change in susceptibility to atazanavir (ATV) and/or amprenavir (APV; panel A, 348 isolates), indinavir (IDV; panel B, 94 isolates), lopinavir (LPV; panel C, 265 isolates), nelfinavir (NFV; panel D, 492 isolates), ritonavir (RTV; panel E, 434 isolates), and saquinavir (SQV; panel F, 349 isolates). Open squares in panel D represent isolates containing D30N substitutions. Dotted lines identify the position of a 3.0-fold change in susceptibility to each of the PIs.
The percentage of isolates in the top left region of panels A through F (amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir, respectively) were 10.6%, 18.1%, 11.7%, 36.8%, 28.3%, and 10.9%, respectively. Similarly, the percentage of isolates in the lower right region of panels A through F was 34.8%, 7.4%, 26.4%, 2.2%, 5.5%, and 24.9%, respectively. In no case was a strong alignment of isolates along a diagonal line evident, suggesting a distinct resistance profile for atazanavir. One possible exception is a subpopulation of nelfinavir-resistant, and perhaps indinavir-resistant, viruses that are also resistant to several other PIs. Seventy of the 181 nelfinavir-resistant isolates susceptible to atazanavir contained a D30N substitution, leaving well over 100 isolates without a D30N change which were resistant to nelfinavir and still susceptible to atazanavir. Exclusion of the D30N-containing isolates reduces the percentage of isolates in this quadrant of the graph from 36.8% to 22.6%.
Overall, two distinctive patterns were observed. The first, exemplified by amprenavir (Fig. 2, panel A), lopinavir (panel C), and saquinavir (panel F), showed a high proportion of atazanavir-resistant isolates remaining susceptible (lower right quadrant) to the second PI examined. The second and opposite pattern was exhibited by nelfinavir (panel D), ritonavir (panel E), and, to a lesser extent, indinavir (panel B) and suggests that a high percentage of isolates resistant to the second PI retained susceptibility to atazanavir. All six panels are suggestive of a relatively distinct resistant profile for atazanavir based on the distribution of isolates plotted. The scarcity of isolates showing a >10-fold change in atazanavir susceptibility in the lower right quadrants again illustrates that atazanavir resistance levels were modest in degree for those isolates that are highly cross-resistant to multiple PIs.
Previous studies (11) involving in vitro passage of HIV-1 in the presence of atazanavir indicated that multiple pathways to atazanavir resistance were possible, depending on the HIV-1 strain used. Several amino acid substitutions, including N88S, I84V, and I50L were identified as key changes. To better understand the relationship between genotypes and atazanavir phenotypic cross-resistance in a clinical setting, the mutational patterns for 943 clinical isolates were profiled and characterized. This panel consisted of 544 of the 551 resistant isolates (genotypes were not available for seven resistant isolates from study AI424-045) shown in Table 1 and Fig. 1 and 2 and an additional set of 399 isolates that displayed susceptibility changes of ≤3.0-fold against all of the PIs employed in this study. The 399 susceptible isolates contained 1 to 17 substitutions and an average of 7.7 amino acid changes, while the number of substitutions present in the 544 resistant isolates averaged 12.4 per isolate and ranged from 4 to 23 changes. All 793 isolates had at least one amino acid change present compared to the reference strains used.
Each of the isolates was evaluated for the presence of “primary” (D30N, G48V, I50V, V82A/F/S/T, I84V, and L90M) and “secondary” (L10I/F/V, K20R/M/I, L24I, V32I, M36I/L/V, M46I/L, I47V, F53L, I54V/L, L63P, A71V/T/I, G73C/S/T/A, and N88D/S/T) substitutions as designated by the International AIDS Society-USA resistance panel (16, 17). For this analysis, mixtures of wild-type and resistance substitutions were scored as resistance mutations. Several additional residue changes were also evaluated, including L33I/F/V, V77I, and I93L. Of these, only changes at residue 33 appeared to play a differentiating role in resistance and were added to the list of secondary substitutions. While changes at additional amino acid positions were also noted in nearly all isolates, they were not evaluated further because they represented either polymorphisms or changes specific to the reference strains used or had no obvious correlation with loss of susceptibility. In addition, three of the designated secondary substitutions, V32I, I47V, and F53L, were also not considered further because they appeared in less than 5% of the isolates. As a result, subsequent analysis was performed by evaluating the frequency of the specific changes noted above for six primary residues (30, 48, 50, 82, 84, and 90) and 12 secondary residues (10, 20, 24, 33, 36, 46, 54, 63, 71, 73, 88, and 93).
Table 2 summarizes the incidence of the primary and secondary substitutions listed above for each of the 943 isolates, with the data sorted by the presence of the indicated amino acid substitutions and further divided into susceptible and resistant groups for each of the seven PIs. Because we were looking for relationships highly predictive of resistance, a highly conservative approach was taken in identifying those changes displaying statistical relevance to PI resistance, with P values of <0.0005 as a cutoff for significance. The association of reduced susceptibility to each of the marketed PIs with primary and secondary amino acid changes previously reported further validated this approach (31).
Changes at primary residues 82, 84, and 90 and secondary residues 10, 46, 54, 63, 71, and 73 were predictive of resistance to all seven PIs evaluated in this evaluation. In addition, atazanavir resistance appears to correlate with additional amino acid substitutions at primary residue 48 and secondary residues 20, 24, 33, and 36. As indicated in Table 2, the pattern of critical substitutions was somewhat common for each of the PIs: an M36I/L/V change was important only for atazanavir, I50V was important only for amprenavir and lopinavir, and N88D/S/T and I93L were important for nelfinavir. The presence of D30N correlates with susceptibility to amprenavir, indinavir, lopinavir, and ritonavir, while susceptibility to lopinavir also correlated with the presence of an N88D/S/T substitution. Interestingly, the presence of substitutions at residue 88 has no significant relationship with atazanavir resistance even though this is a key residue identified during in vitro selection (11). However, of the 10 isolates containing N88S (no mixtures) in this overall panel of isolates, 8 showed an atazanavir FC of >3.0-fold (range, 0.3- to 36-fold; median, 6.4-fold). These results are in full alignment with the in vitro selection data (11), indicating a key role for the I84V and N88S substitutions, along with changes at residues 10, 33, 46, 63, 71, and 84, in atazanavir resistance.
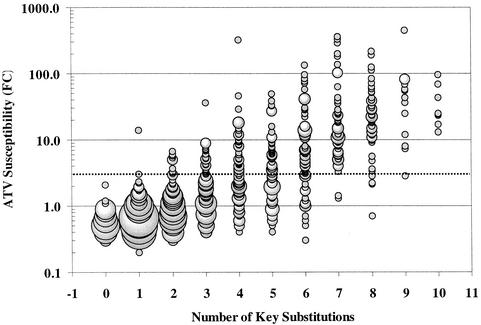
The data presented in Table 2 clearly show that signature mutations such as D30N for nelfinavir and I50V for amprenavir are not absolutely required for resistance to these PIs and that multiple pathways to resistance are possible for all of the PIs. As a result, genotypic predictions of resistance based on the presence or absence of specific amino acid changes or combinations of changes are particularly difficult. Since predicting the susceptibility of atazanavir based solely on the presence of specific amino acids was equally challenging, an attempt was made to correlate the overall number of key substitutions with decreased susceptibility to atazanavir. For this analysis, substitutions at amino acid residues 10, 20, 24, 33, 36, 46, 48, 54, 63, 71, 73, 82, 84, and 90 were considered “key” atazanavir resistance changes because of their strong correlation with loss of susceptibility to atazanavir.
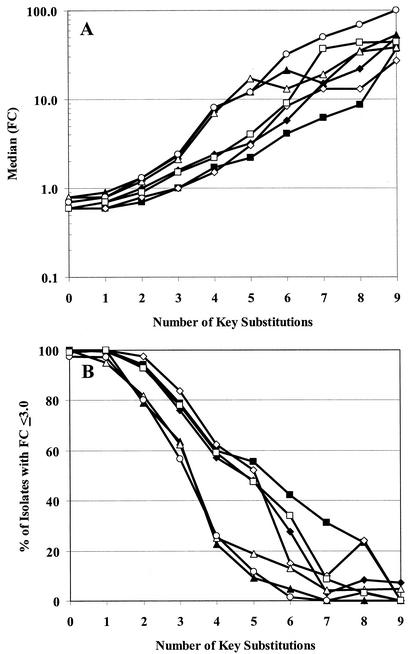
Figure 3 is a plot of FC in atazanavir susceptibility for each of the 943 isolates evaluated against the number of the 14 key atazanavir substitutions present. Results suggest that an accumulation of five of these changes is required for the majority of isolates to display an FC of >3.0-fold. For atazanavir, the median FC for isolates with five key substitutions was 3.2-fold, and 57% of isolates with four changes remained susceptible with the resistance cutoff criteria chosen. Overall, 563 (88.2%) of 638 isolates with more than four key substitutions remained susceptible to atazanavir, versus only 71 (18.7%) of 380 isolates with more than five substitutions. A similar analysis of the data for the other PIs suggested that saquinavir also requires an accumulation of at least five key substitutions to achieve similar resistance levels, while amprenavir and lopinavir require six changes and indinavir, nelfinavir, and ritonavir require only four key substitutions to achieve these levels. These results are illustrated in Fig. 4A (median FC) and 4B (percent of isolates with ≤3.0-fold change) and support the observation that several amino acid changes are required to achieve significant resistance levels for all PIs.
FIG. 3.
Relationship of atazanavir susceptibility and number of key substitutions. The atazanavir (ATV) FCs for 943 resistant and susceptible isolates are plotted based on the number of identified atazanavir key substitutions (Table 2) present. The dotted line identifies the position of a 3.0-fold change in susceptibility.
FIG. 4.
Relationship between number of key substitutions and susceptibility to PIs. Panel A plots the range of FCs for all seven PIs relative to a number of key substitutions present. The dotted line identifies the position of a 3.0-fold change in susceptibility. Panel B plots the percentage of isolates displaying a ≤3.0-fold change in susceptibility to each of the PIs for isolates containing one to eight key substitutions: atazanavir (⧫), amprenavir (▪), indinavir (▴), lopinavir (◊), nelfinavir (▵), ritonavir (○), and saquinavir (□).
DISCUSSION
Atazanavir is a potent inhibitor of HIV protease with demonstrated effectiveness in clinical trials (I. Sanne, P. Cahn, L. Percival, P. Phanuphak, T. Kelleher, M. Giordano, and G. Pantaleo, 41st Intersci. Conf. Antimicrob. Agents Chemother., 2001, abstr. I-667; K. E. Squires, A. Thiry, and M. Giordano, 42nd Intersci. Conf. Antimicrob. Agents Chemother., 2002, abstr. H-1076). Its excellent oral bioavailability and pharmacokinetic profile enable once-daily dosing and low pill burden in the absence of added ritonavir. The results of this study suggest that atazanavir has a resistance profile that is distinct from that of six other marketed PIs with regard to isolates displaying resistance to more than three PIs. For isolates resistant to four or more PIs, significant cross-resistance to atazanavir was observed. Overall atazanavir resistance levels were generally modest in degree, and significant susceptibility was retained by those isolates showing resistance to one or two PIs (Table 1).
Phenotypic analysis of drug susceptibility in vitro has been hampered by the lack of a standard definition of resistance and by the lack of standardized methodology (1, 28). In this study of 551 PI-resistant HIV-1 clinical isolates, we have arbitrarily defined resistance as a reduction in susceptibility of greater than 3.0-fold to ensure that the data were above any assay variability. Correlating fold changes with actual loss of effectiveness in the clinic remains a significant challenge, complicated by the use of combination therapy, limited measurements of trough exposure levels, and different methods for determining EC50s in the presence and absence of serum proteins. Atazanavir is ≈86% protein bound, and the addition of 40% human serum to the conventional cell protection assay decreased potency fivefold (29). Trough levels (Cmin) observed with the once daily 400-mg dosing regimen of atazanavir were determined to be >200 ng/ml (>10-fold higher than the protein-adjusted EC50s of 8 to 20 ng/ml and 5 to 10 ng/ml for atazanavir against reference strains with a conventional multicycle cell infection assay and the ViroLogic PhenoSense single-cycle assay, respectively).
The results reported in this study suggest that atazanavir is likely to remain effective against isolates that are resistant to one or two of the currently marketed PIs. However, the significance of the relationships described here to efficacy in a clinical setting will need to await the results of ongoing clinical trials involving treatment of PI-experienced patients with atazanavir-containing regimens. The very high proportion of nelfinavir-resistant isolates among those selected during prescreening for clinical studies AI424-009, AI424-043, and AI424-045 was not unexpected in light of the wide use of this particular PI in recent years. Despite the observation that atazanavir susceptibility decreased modestly (median FC = 4.0-fold) in 18 of the 121 isolates resistant to nelfinavir alone, there was some suggestion of relatedness in Fig. 2D for nelfinavir-resistant viruses without the D30N substitution. Interestingly, calculated linear correlation values from the isolates plotted in each of the panels of Fig. 2 yielded values of 0.53, 0.68, 0.14, 0.31, 0.39, and 0.46 for atazanavir versus amprenavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir, respectively, suggesting that the greatest relatedness was between atazanavir and indinavir and the least was between atazanavir and lopinavir. Eliminating isolates containing a D30N substitution from Fig. 2D increased the linear correlation slightly, to 0.34.
Atazanavir susceptibility decreased as isolates became cross-resistant to three or more PIs, although only modest changes were noted in many of these isolates. Significant loss of atazanavir susceptibility was observed in all but seven of the 142 isolates that were cross-resistant to five marketed PIs. Amprenavir and saquinavir both displayed excellent coverage of isolates resistant to one to three PIs but, like atazanavir, exhibited decreased susceptibility as isolates showed higher levels of resistance and cross-resistance to multiple PIs.
The profiles presented in Fig. 2 appear to sort into distinct groups based on whether a significant number of atazanavir-resistant isolates retained susceptibility to another PI (amprenavir, lopinavir, and saquinavir) or whether atazanavir susceptibility was retained against a significant proportion of PI-resistant (indinavir, nelfinavir, and ritonavir) isolates. It is important to note that this type of two-PI analysis has limited utility with regard to providing useful information on sequencing of PIs, since comparisons are between two PIs irrespective of whether a given isolate is cross-resistant to one or five PIs.
Genotypic analysis of 943 susceptible and resistant isolates identified amino acid changes that correlate strongly with decreased susceptibility to atazanavir and the other six PIs. While individual amino acid substitutions had limited predictive value, the accumulation of several key substitutions identified for each of the PIs was more predictive of resistance. For atazanavir, an accumulation of five or more substitutions at any of the 14 key amino acid changes located at residues 10, 20, 24, 33, 46, 48, 54, 71, 73, 82, 84, and 90 was predictive of an atazanavir FC of >3-fold. A similar analysis for the other PIs identified 10 to 16 key residues (Table 2) for each PI and a correlation with an accumulation of as few as four substitutions predictive of decreased susceptibility (Fig. 4) to indinavir, nelfinavir, and ritonavir. Many of the key substitutions identified were also noted in an earlier study examining genotypic changes in 112 resistant isolates (19).
Multiple combinations of mutations can give rise to atazanavir resistance, an observation that is consistent with earlier observations from cell culture selection experiments (11). Five of the amino acid residues identified in the current analysis, 10, 33, 46, 63, 71, and 84, also emerged during in vitro selection of resistant viruses. Interestingly, initial data from ongoing atazanavir clinical trials suggest that the novel amino acid change I50L, which appeared in one HIV-1 strain in vitro, emerged in several atazanavir-treated patients who experienced virological failure and reduced susceptibility to atazanavir (8). Furthermore, all of these isolates exhibited increased susceptibility to other PIs. The I50L substitution is very rare and was not present in any of the 943 isolates in the current panel.
A better understanding of the overall resistance profile and its relationship to other PIs will need to await reciprocal studies in which a sufficient number of isolates resistant to atazanavir can be identified and tested for susceptibility to the other PIs.
Acknowledgments
We thank Kurt Hertogs of Virco bvba (a division of Tibotec-Virco), Mechelen, Belgium, for his efforts in selecting Virco isolates and discussions on resistance patterns; Gabriele Heilek-Snyder and Heather King for assistance in generating phenotypic and genotypic data; and Dona Morse for preparation of data tables.
REFERENCES
- 1.Angarano, G., and L. Monno. 2000. Genotype and phenotype resistance: an overview. J. Biol. Regul. Homeost. Agents 14:11-14. [PubMed] [Google Scholar]
- 2.Atkinson, B., J. Isaacson, M. Knowles, E. Mazabel, and A. K. Patick. 2000. Correlation between human immunodeficiency virus genotypic resistance and virologic response in patients receiving nelfinavir monotherapy or nelfinavir with lamivudine and zidovudine. J. Infect. Dis. 182:420-427. [DOI] [PubMed] [Google Scholar]
- 3.Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retroviruses 16:1209-1213. [DOI] [PubMed] [Google Scholar]
- 4.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, and T. C. Merigan. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS 14:F83-93. [DOI] [PubMed] [Google Scholar]
- 5.Boden, D., A. Hurley, L. Zhang, Y. Cao, Y. Guo, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 6.Clevenbergh, P., J. Durant, P. Halfon, P. del Giudice, V. Mondain, N. Montagne, J. M. Schapiro, C. A. B. Boucher, and P. Dellamonica. 2000. Persisting long-term benefit of genotype-guided treatment for HIV-infected patients failing HAART. The Viradapt study: week 48 follow-up. Antivir. Ther. 5:65-70. [PubMed] [Google Scholar]
- 7.Cohen, C. J., S. Hunt, M. Sension, C. Farthing, M. Conant, S. Jacobson, J. Nadler, W. Verbiest, K. Hertogs, M. Ames, A. R. Rinehart, and N. M. Graham. 2002. A randomized trial assessing the impact of phenotypic resistance testing on antiretroviral therapy. AIDS 16:579-588. [DOI] [PubMed] [Google Scholar]
- 8.Colonno, R. J., J. Friborg, R. E. Rose, E. Lam, and N. Parkin. 2002. Identification of amino acid substitutions correlated with reduced atazanavir susceptibility in patients treated with atazanavir-containing regimens. Antiviral Ther. 7:S6. [Google Scholar]
- 9.Descamps, D., P. Flandre, V. Calvez, G. Peytavin, V. Meiffredy, G. Collin, C. Delaugerre, S. Robert-Delmas, B. Bazin, J. P. Aboulker, G. Pialoux, F. Raffi, and F. Brun-Vezinet. 2000. Mechanisms of virologic failure in previously untreated HIV-infected patients from a trial of induction-maintenance therapy. Trilege (Agence Nationale de Recherches sur le SIDA 072 Study Team). JAMA 283:205-211. [DOI] [PubMed] [Google Scholar]
- 10.Gehringer, H., J. R. Bogner, F. D. Goebel, H. Nitschko, and K. von der Helm. 2000. Sequence analysis of the HIV-1 protease coding region of 18 HIV-1-infected patients prior to HAART and possible implications on HAART. J. Clin. Virol 17:137-141. [DOI] [PubMed] [Google Scholar]
- 11.Gong, Y. F., B. S. Robinson, R. E. Rose, Y. F. Gong, B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P. F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havlir, D. V., N. S. Hellmann, C. J. Petropoulos, J. M. Whitcomb, A. C. Collier, M. S. Hirsch, P. Tebas, J. P. Sommadossi, and D. D. Richman. 2000. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. JAMA 283:229-234. [DOI] [PubMed] [Google Scholar]
- 13.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. Van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 14.Hertogs, K., M. P. de Béthune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertogs, K., S. Kemp, S. Bloor, K. Van Acker, J. Hansen, W. De Beukeleer, C. Roelant, B. Larder, and R. Pauwels. 1998. Comprehensive HIV drug resistance monitoring with rapid, high-throughput phenotypic and genotypic assays with correlative data analysis. AIDS (London) 12:S11. [Google Scholar]
- 16.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 17.Kantor, R., W. Fessel, J. Andrew, R. Zolopa, d. Israelski, N. Shulman, J. G. Montoya, M. Harbour, J. M. Schapiro, and R. W. Shafer. 2002. Evolution of primary protease inhibitor resistance mutations during protease inhibitor salvage therapy. Antimicrob. Agents Chemother. 46:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karmochkine, M., A. Si Mohamed, C. Piketty, C. Ginsburg, G. Raguin, V. Schneider Fauveau, L. Gutmann, M. D. Kazatchkine, and L. Belec. 2000. The cumulative occurrence of resistance mutations in the HIV-1 protease gene is associated with failure of salvage therapy with ritonavir and saquinavir in protease inhibitor-experienced patients. Antiviral Res. 47:179-188. [DOI] [PubMed] [Google Scholar]
- 19.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little, S. J. 2000. Transmission and prevalence of HIV resistance among treatment-naive subjects. Antivir. Ther. 5:33-40. [DOI] [PubMed] [Google Scholar]
- 21.Little, S. J., E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. Pitt, E. S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 22.Markowitz, M., M. Conant, A. Hurley, R. Schluger, M. Duran, J. Peterkin, S. Chapman, A. Patick, A. Hendricks, G. J. Yuen, W. Hoskins, N. Clendeninn, and D. Ho. 1998. A preliminary evaluation of nelfinavir mesylate, an inhibitor of human immunodeficiency virus (HIV)-1 protease, to treat HIV infection. J. Infect. Dis. 177:1533-1540. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Picado, J., A. V. Savara, L. Shi, L. Sutton, and R. T. D'Aquila. 2000. Fitness of human immunodeficiency virus type 1 protease inhibitor-selected single mutants. Virology 275:318-322. [DOI] [PubMed] [Google Scholar]
- 24.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pieniazek, D., M. Rayfield, D. J. Hu, J. Nkengasong, S. Z. Wiktor, R. Downing, B. Biryahwaho, T. Mastrob, A. Tanuri, V. Soriano, R. Lal, and T. Dondero. 2000. Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. HIV Variant Working Group. AIDS 14:1489-1495. [DOI] [PubMed] [Google Scholar]
- 26.Qari, S. H., R. Respess, H. Weinstock, E. M. Beltrami, K. Hertogs, B. A. Larder, C. J. Petropoulos, N. Hellmann, and W. Heneine. 2002. Comparative analysis of two commercial phenotypic assays for drug susceptibility testing of human immunodeficiency virus type 1. J. Clin. Microbiol. 40:31-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabasseda, X., J. Silvestre, and J. Castañer. 1999. BMS-232632. Drugs Future 24:375-380. [Google Scholar]
- 28.Richman, D. D. 2000. Principles of HIV resistance testing and overview of assay performance characteristics. Antivir. Ther. 5:27-31. [PubMed] [Google Scholar]
- 29.Robinson, B. S., K. A. Riccardi, Y. F. Gong, Q. Guo, D. A. Stock, W. S. Blair, B. J. Terry, C. A. Deminie, F. Djang, R. J. Colonno, and P. F. Lin. 2000. BMS-232632, a highly potent human immunodeficiency virus protease inhibitor that can be used in combination with other available antiretroviral agents. Antimicrob. Agents Chemother. 44:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose, R. E., Y. F. Gong, J. A. Greytok, C. M. Bechtold, B. J. Terry, B. S. Robinson, M. Alam, R. J. Colonno, and P. F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antiviral News 8:65-91. [Google Scholar]
- 32.Schmidt, B., H. Walter, B. Moschik, C. Paatz, K. van Vaerenbergh, A.-M. Vandamme, M. Schmitt, T. Harrer, K. Uberla, and K. Korn. 2000. Simple algorithm derived from a geno-/phenotypic database to predict HIV-1 protease inhibitor resistance. AIDS 14:1731-1738. [DOI] [PubMed] [Google Scholar]
- 33.Voelker, R. 2000. HIV drug resistance. JAMA 284:169.. [PubMed] [Google Scholar]
- 34.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to antiretroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 35.Winters, M. A., J. D. Baxter, D. L. Mayers, D. N. Wentworth, M. L. Hoover, J. D. Neaton, and T. C. Merigan. 2000. Frequency of antiretroviral drug resistance mutations in HIV-1 strains from patients failing triple drug regimens. The Terry Beirn Community Programs for Clinical Research on AIDS. Antivir. Ther. 5:57-63. [PubMed] [Google Scholar]