SYNOPSIS
Objectives
This study was designed to determine the role of flushing dental water lines for the removal of heterotrophic plate count bacteria, Legionella spp., and free-living protozoa.
Methods
Forty dental offices were surveyed in the study. An initial sample and a sample taken after three minutes of flushing were obtained from the air/water syringe at each location. All samples were quantitatively analyzed for heterotrophic bacteria using three bacteriological procedures. The samples were analyzed for the presence of Legionella spp. using cultural, immunological, and molecular procedures and for the occurrence of free-living protozoa using a killed bacteria plate procedure.
Results
The flushing process reduced the level of heterotrophic plate count bacteria by 1.1 to 1.5 log10 CFU/ml. Compliance with recommendations for bacterial levels varied depending on the methodology employed in the analysis. The flushing process did not reduce the occurrence of Legionella spp. or free-living protozoa.
Conclusion
The results support recent U.S. Centers for Disease Control and Prevention recommendations that the process of flushing dental water lines cannot be relied upon as a sole means of reliably improving the quality of water used in dental treatment.
Dental unit water quality has become an issue of concern in both infection control practices and occupational exposures in dental health care settings. While flushing of dental unit water lines has been recognized as an important strategy for controlling microbial levels in dental water systems, recommendations regarding the efficacy of flushing have changed in recent years. In 1993 the U.S. Centers for Disease Control and Prevention (CDC) recommended that dental water lines be flushed to reduce the microbial load in dental unit water.1 The most recently published CDC guidelines, however, suggest that other strategies beyond flushing are needed to improve water quality.2 The CDC recommended that the number of bacteria in water used as a coolant/irrigant for nonsurgical dental procedures should have an aerobic heterotrophic plate count (HPC) of ≤500 CFU/ml. 2 This number was selected based on recommendations for levels of HPC in potable water. In addressing this issue, the American Dental Association (ADA) has proposed that water used in dental treatment contain a bacterial level of ≤200 CFU/ml.2 The flushing of dental water lines has been shown to decrease the levels of planktonic bacteria in the water, but this practice has not been shown to affect the biofilm that accumulates in the water lines.3,4 To date, the effectiveness of flushing has been measured by comparing HPC levels present in initial and post-flushing samples. The role of flushing in the removal of specific pathogens has not been examined.
Exposure to dental water has not been shown to present a demonstrable health risk to either patients or dental health care providers. However, the presence of pathogens in dental unit water lines is not consistent with accepted infection-control principles.2 Legionella spp. are important pathogenic agents that have been shown to be transmitted in infectious aerosols from potable water. Occurrence studies have demonstrated the presence of Legionella spp. in dental water.4,5 Further, it has been reported that dental health care workers may have substantially higher titers of Legionella antibodies compared to control populations.6,7 In potable water, Legionella spp. are often associated with biofilms and free-living protozoa. The small diameter of dental water lines, accompanied by periods of prolonged water stagnation, provides a suitable ecological niche for biofilm development and the proliferation of both Legionella spp. and protozoa.
This study was designed to specifically address the efficacy of flushing for the removal of Legionella spp. and free-living protozoa and to evaluate the overall effectiveness of this procedure in reducing the total microbial load as measured by three bacteriological procedures.
METHODS
Forty dental offices using municipal drinking water for their water supply were surveyed in this study. Water samples were collected from the air/water syringe from each dental operatory. The two samples consisted of an initial sample and a second sample obtained after the water from the syringe had been allowed to run to waste for three minutes. The time of use prior to sampling varied among the dental offices. Approximately one-half of the samples were collected from syringes that had not been used within the past 24 hours. The remainder were taken from syringes that had been used during the course of the clinic day. Each sample was analyzed for HPC bacteria, Legionella spp., and free-living protozoa.
Three separate procedures were used to determine HPC bacteria. The first procedure was the aerobic pour plate using plate count agar (PCA) incubated at 35°C for 48 hours.8 The second procedure utilized the spread plate technique using R2A agar incubated at 25°C for seven days.8 The third procedure also utilized the spread plate technique, but using 5% sheep blood agar (SBA) incubated at 35°C for 48 hours.9
The presence of Legionella spp. was determined by cultural, immunological, and molecular procedures. Water samples (minimum volume of 10 ml) were filtered through a 0.2 μm porosity polyethersulfone-based membrane.4 The membranes were placed in 2 ml of the sample water and bacteria were dislodged by vortexing for one minute. The procedures for analysis for Legionella spp. were the same as those previously described by Williams et al.5 Briefly, the cultural analysis utilized a BCYEα agar. The immunological assay, a microscopic direct flourescent antibody (DFA) procedure, uses a Legionella polyclonal conjugate system.5 Molecular analysis was completed using the polymerase chain reaction (PCR) procedure. The Legionella PCR kit (Perkin-Elmer) was used to detect the presence of the genus Legionella and to specifically detect the species pneumophila.
Protozoa were detected using a killed bacterial plate procedure. Agar (1.5%) plates were seeded with an overnight culture of Escherichia coli grown at 35°C in tryptic soy broth. The bacteria were swabbed onto the surface of the agar and exposed to ultraviolet irradiation (254 nm) for 20 minutes to inactivate the E. coli culture. Plates were inoculated with 20 ml of the test water and incubated at 20°C for 5–7 days. Plates were observed daily using an inverted microscope (200X). The presence of any protist form (i.e. flagellate, amoeba, etc.) was scored as a positive sample. The protozoa species were not taxonomically identified.
The significance of the flushing procedure on the various parameters was determined by statistical analysis. Bacterial counts were log transformed prior to conducting the analyses. A constant (0.5) was added to any values that equaled zero. The signed rank test was used to compare levels of HPC in the initial and flushed samples. Paired comparisons for occurrence data for Legionella in initial and flushed samples were tested using the McNemar's test for proportions. Cochran's test for related observations was used to determine if there were significant differences between the methods used for the analysis of Legionella spp.
RESULTS
The effect of flushing on the presence of HPC bacteria as determined by the three assay procedures is shown in the Figure. The mean level of bacteria present in the initial samples using PCA was 3.36 log10 CFU/ml and 1.84 log10CFU/ml in flushed samples. The mean level was 4.83 log10CFU/ml in the initial samples and 3.56 log10 CFU/ml in the flushed samples using R2A medium. The SBA medium yielded a mean of 2.20 log10CFU/ml in the initial samples and 1.08 log10CFU/ml in the flushed samples. The differences between the levels of bacteria present in the initial and flushed samples were significant (p=0.0001) for all three assay procedures. The levels of bacteria in the samples showed considerable variation between the analytical methods. While the initial levels of bacteria varied between the three methods, the flushing procedure produced similar results in the decrease in bacterial numbers. Levels of bacteria were reduced by 1.5 log10CFU/ml using PCA, 1.2 log10 CFU/ml for R2A, and 1.1 log10 CFU/ml for SBA.
Legionella spp. were detected in the dental unit water samples (Table 1). Significant differences (p=0.0004) were observed between the three assay procedures. The PCR assay produced the highest number of positive samples, followed by the DFA procedure and the cultural method. Thirteen (32.5%) of the initial samples were positive for the presence of Legionella by the PCR procedure. Three of the samples that gave a positive response for the genus Legionella were also positive for L. pneumophila. Twelve (30%) of the flushed samples gave positive results for the genus Legionella, and the same three samples that were initially positive for L. pneumophila were also positive in the flushed samples. Based on the PCR procedure, only one sample became negative for the presence of Legionella spp. after the flushing process. The six (15%) initial samples that were positive by the DFA procedure remained positive after flushing. The one positive response detected by the culture method was positive in both the initial and flushed sample. These two culture isolates were further analyzed by the PCR assay and were identified as L. pneumophila. There was no significant difference between the presence of Legionella spp. before or after flushing regardless of the assay procedure.
Table 1.
Prevalence of Legionella spp. in dental unit water samples
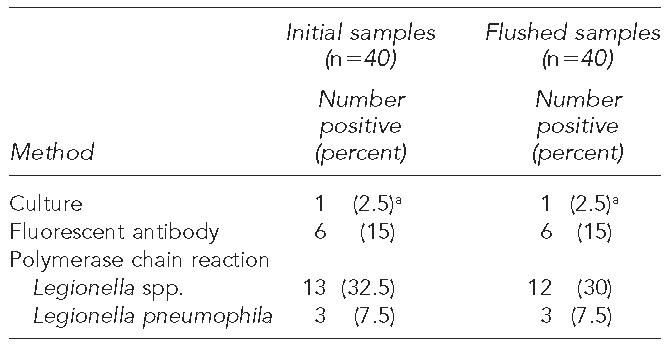
Confirmed by PCR analysis as Legionella pneumophila
Twenty-five (62.5%) of the initial samples were positive for the presence of free-living protozoa. The same 25 samples remained positive after the three-minute flushing procedure.
Both of the Legionella culture positive samples (initial and flushed) contained protozoa. Five (83%) of the six initial samples and five (83%) of the six flushed samples that were positive for Legionella spp. as determined by the DFA procedure contained protozoa. Eight (61.5%) of the 13 samples positive for Legionella spp. by PCR and seven (58.3%) of the 12 positive flushed samples contained protozoa. Two of the three initial samples and two of the three flushed samples that were PCR positive for L. pneumophila were also positive for the presence of protozoa.
DISCUSSION
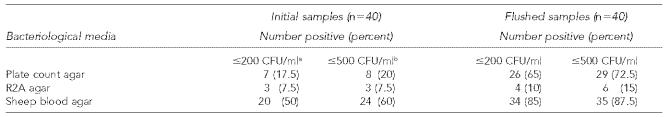
The results of this study indicate that flushing can substantially reduce the level of HPC bacteria present in water used for dental treatment. The 1.1 to 1.5 log10 CFU/ml reduction seen in total counts is similar to the 1.42 log10 CFU/ml reduction reported in a similar study.10 The three different procedures recovered substantially different levels or populations of bacteria. This is of particular importance when decisions about water quality are being based on a given microbial limit. As seen in Table 2, bacterial levels below the two recommended maximum levels (≤200 or ≤500 CFU/ml) varied among the assay procedures. Using the two most common procedures (PCA and R2A) for HPC analysis in drinking water, only 20% or less of the initially drawn samples had bacterial counts below the recommended levels. The ≤500 CFU/ml limit for drinking water was derived from studies that used PCA in the aerobic pour plate procedure. Using this procedure, greater than 70% of the samples were below this limit after flushing. The spread plate procedure using R2A agar has been recommended for the recovery of indigenous oligotrophic aquatic bacteria. This procedure yielded the highest levels of bacteria and the flushing process resulted in a much lower number of samples within the recommended levels. The use of SBA in the spread plate procedure has been proposed for the routine monitoring of potable water for the detection of bacteria with pathogenic potential.9 It is noteworthy that the lowest numbers of bacteria were recovered with this medium.
Table 2.
Levels of heterotrophic bacteria in dental unit water samples
American Dental Association recommended level
Centers for Disease Control and Prevention (CDC)-recommended level
The HPC procedures used in this study are limited to the recovery of cultural planktonic organisms in the water. Bacteria in biofilms in the dental water units would only be detected by these procedures if they were being sloughed off into the bulk water. Further, these procedures are not capable of detecting many of the important waterborne pathogens of particular interest in dental water, viz. Legionella spp. and non-tuberculosis mycobacteria. Regardless of the bacteriological method used, flushing was only able to reduce the microbial numbers by approximately one to one and half orders of magnitude.
As previously reported, Legionella spp.4,5 and free-living protozoa11 were present in the waters examined in the present survey. There were significant differences seen between the analytical methods used to analyze for Legionella spp. The current findings of higher recoveries using molecular and immunological procedures are in agreement with earlier reports.4,5
An important finding of this study was the relative ineffectiveness of the flushing procedure in reducing the occurrence of either Legionella spp. or protozoa. A previous study reported that a two-minute flush reduced the levels of amoebae by 66%.12 In our study, the three-minute flushing procedure may have reduced the numbers present, but did not change the overall occurrence of protozoa in the initial and flushed samples. The close association between these organisms and biofilms in dental unit water lines13 suggests that other strategies beyond flushing would be required to effectively address the issue of biofilm removal. The use of chemical germicides has been recommended for the removal or inactivation of biofilms in dental water lines.2 Commercial devices and procedures that include chemical treatment and microfiltration are currently available for use in dental water treatment.14 The results of this study confirm the latest CDC recommendations that flushing alone is not a reliable procedure for improving water quality used in dental treatment.2
Figure.
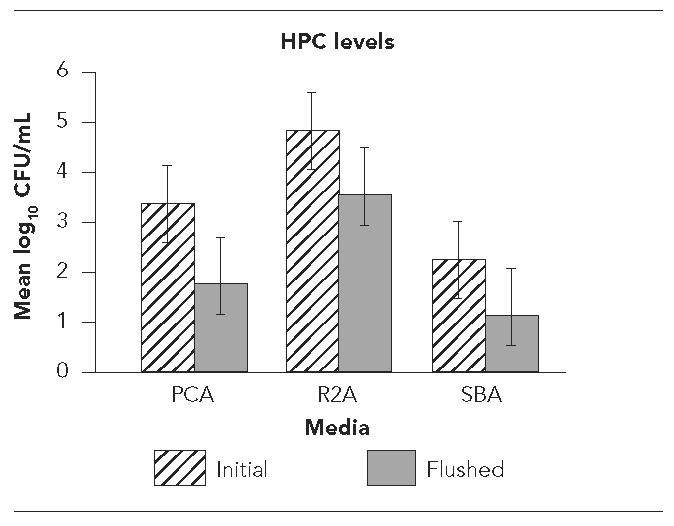
Levels of heterotrophic plate count (HPC) bacteria in initial and flushed dental water samples as determined by plating on plate count agar (PCA), R2A agar, and sheep blood agar (SBA)
NOTE: Error bars denote standard deviations.
Acknowledgments
The U.S. Environmental Protection Agency, through its office of Research and Development, funded the research described here. It has been subjected to agency review and approved for publication.
The authors gratefully acknowledge D. K. Wild for assistance in the statistical analysis for this project.
REFERENCES
- 1.Centers for Disease Control and Prevention (US) Recommended infection control practices for dentistry, 1993. [cited 2006 Jan 18];MMWR Morb Mortal Wkly Rep. 1993 42(RR-8) Also available from: URL: http://www.cdc.gov/mmwr/preview/mmwrhtml/00021095.htm. [Google Scholar]
- 2.Centers for Disease Control and Prevention (US) Guidelines for infection control in dental health-care settings—2003. [cited 2006 Jan 18];MMWR Morb Mortal Wkly Rep. 2003 52(RR-17):1–61. Also available from: URL: http://www.cdc.gov/mmwr/preview/mmwrhtml/rr5217a1.htm. [Google Scholar]
- 3.Williams JF, Johnston AM, Johnson B, Huntington MK, MacKenzie CD. Microbial contamination of dental unit water lines: prevalence, intensity and microbiological characteristics. J Am Dent Assoc. 1993;124:59–65. doi: 10.14219/jada.archive.1993.0205. [DOI] [PubMed] [Google Scholar]
- 4.Atlas RM, Williams JF, Huntington MK. Legionella contamination of dental-unit waters. Appl Environ Microbiol. 1995;61:1208–13. doi: 10.1128/aem.61.4.1208-1213.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams HN, Paszko-Kolva C, Shahamat M, Palmer C, Pettis C, Kelley J. Molecular techniques reveal high prevalence of Legionella in dental units. J Am Dent Assoc. 1996;127:1188–93. doi: 10.14219/jada.archive.1996.0410. [DOI] [PubMed] [Google Scholar]
- 6.Fotos PG, Westfall HN, Snyder IS, Miller RW, Mutchler BM. Prevalence of Legionella-specific IgG, and IgM antibody in a dental clinic population. J Dent Res. 1985;64:1382–5. doi: 10.1177/00220345850640121101. [DOI] [PubMed] [Google Scholar]
- 7.Reinthaler FF, Mascher F, Stunzner D. Serological examinations for antibodies against Legionella species in dental personnel. J Dent Res. 1988;67:942–3. doi: 10.1177/00220345880670061001. [DOI] [PubMed] [Google Scholar]
- 8.Clesceri LS, Greenberg AE, Eaton AD, editors. Standard methods for the examination of water and wastewater. 20th ed. Washington: American Public Health Association, American Water Works Association, and Water Environment Federation; 1998. [Google Scholar]
- 9.Payment P, Coffin E, Paquette G. Blood agar to detect virulence factors in tap water heterotrophic bacteria. Appl Environ Microbiol. 1994;60:1179–83. doi: 10.1128/aem.60.4.1179-1183.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbeau J, Tanguay R, Faucher E, Avezard C, Trudel L, Cote L, et al. Multiparametric analysis of water line contamination in dental units. Appl Environ Microbiol. 1996;62:3954–9. doi: 10.1128/aem.62.11.3954-3959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel R, Borneff M. The significance of amoebae and other protozoa in water conduit systems in dental units. Zentralbl Bakteriol Mikrobiol Hyg [B] 1989;187:312–23. [PubMed] [Google Scholar]
- 12.Barbeau J, Buhler T. Biofilms augment the number of free-living amoebae in dental unit water lines. Res Microbiol. 2001;152:753–60. doi: 10.1016/s0923-2508(01)01256-6. [DOI] [PubMed] [Google Scholar]
- 13.Pederson ED, Stone ME, Ragain JC, Jr, Simecek JW. Water line biofilm and the dental treatment facility: a review. Gen Dent. 2002;50:190–5. [PubMed] [Google Scholar]
- 14.Williams JF, Andrews N, Santiago JI. Microbial contamination of dental unit water lines: current preventive measures and emerging options. Compend Contin Educ Dent. 1996;17:691–4. [PubMed] [Google Scholar]