Abstract
Objective: This paper focuses on the first two years of operation of Genetics Home Reference (GHR), a Web-based resource <http://www.ghr.nlm.nih.gov> for the general public that helps to explain the health implications of findings from the Human Genome Project.
Methods and Findings: Key challenges of Web-based consumer health communication encountered in the growth and maintenance of GHR are discussed: prioritizing topics for GHR, streamlining the development process while keeping genetic information accurate, and designing a system that helps consumers navigate complex genetic relationships. Various strategies are used to address these challenges. Tying content development to topics of national priority and addressing topics requested by users makes the site increasingly important for both consumers and health professionals. Informatics methods are essential for quality control, particularly for genetic information that changes frequently. Indexing and hierarchical browsing features help to facilitate navigation.
Conclusions: GHR is a credible, dynamic Website that uses lay language to explain the effects of genetic variation on human health. Informatics strategies are key to effective management of a large and expanding body of genetics information. Feedback from formal and informal sources indicates increasing usage and favorable acceptance of GHR.
Highlights
Genetics Home Reference (GHR) is a consumer-friendly Website that explains the effects of genetic variations on human health.
The GHR team is challenged to add content at a steady pace, keep its information accurate, and simplify access to a wide spectrum of relevant information.
The GHR team uses several strategies to streamline content development, enhance quality control, and facilitate navigation.
Informatics techniques are used to implement these strategies.
Implications for practice
GHR is a tool the health care community can use to meet the challenge of communicating the complexities of human genetics.
Informatics methods are essential for streamlining content development, enhancing quality control, and facilitating navigation.
INTRODUCTION
Increasingly, the public is seeking health information online [1–3], including information about inherited disorders [4, 5]. Consumers report that genetics Websites are often confusing, difficult to understand, and hard to navigate [6, 7]. The Human Genome Project has also amplified interest in genetics and is propelling medicine into an era in which genetic knowledge will contribute to optimal health care [8, 9]. The surge of genetic information generated by the Human Genome Project can be overwhelming and often leaves the public struggling to understand the role of genetics in health care [8, 10]. The health care community is challenged to communicate complex developments in human genetics in a way that the public can freely access, easily understand, and appropriately apply [7].
To address this challenge, the Lister Hill National Center for Biomedical Communications (LHNCBC), part of the National Library of Medicine (NLM), initiated the Genetics Home Reference (GHR) project [11], a Web-based resource for the general public. Launched in 2003, GHR <http://www.ghr.nlm.nih.gov> fills a unique niche by using lay language to interpret the health implications of genome sequencing. In addition, it spans a broad spectrum of information from lay-level questions to the details of gene function and structure. Prior to the launch of GHR, online genetic resources addressed the needs of professionals with clinical and technical content that was often difficult for the general public to understand. GHR uniquely meets the needs of consumers seeking information that explains the effect of genetic variation on health.
This paper focuses on the first two years of GHR operations, the ongoing challenges of delivering genetic information to the public, and the strategies used to address these challenges.
BACKGROUND AND CURRENT STATUS
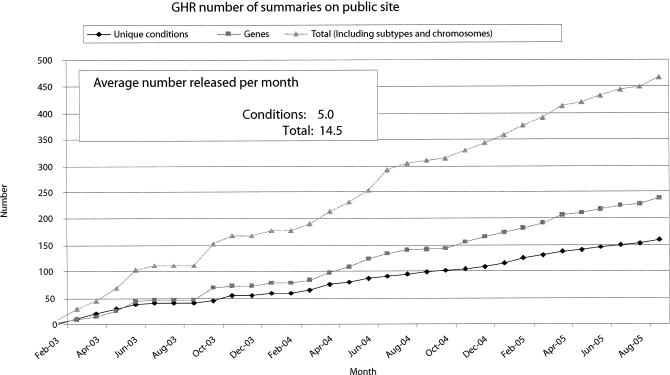
GHR became operational in April 2003 with about a dozen health conditions plus their related genes. Mitchell et al. have previously described the system, design principles, data model, and initial system architecture [11]. After two and a half years of operation, the content has grown steadily (Figure 1) to include all human chromosomes, more than 175 genetic conditions, about 275 gene summaries, and 7 chapters of a tutorial (Help Me Understand Genetics Handbook). Since January 2004, about 15 new health conditions and related gene topics have been added monthly. The content is developed by staff <http://www.ghr.nlm.nih.gov/ghr/ProjectStaff/> with educational backgrounds and experience in human genetics and molecular biology. The staff also regularly monitors the content to ensure that it is accurate and up to date. External experts <http://www.ghr.nlm.nih.gov/ghr/ExpertReviewers/> in genetics review each topic page before it is posted to the Website and annually thereafter.
Figure 1.
Genetics Home Reference content development statistics
Recognizing that a lay audience may have a limited science background, GHR offers tools to help the motivated learner. Each condition, gene, and chromosome summary provides a glossary list of terms used on the page, with direct links to their definitions. In addition, a link to a searchable glossary of genetic and medical terms appears on all pages. The illustrated handbook provides information about ways genes work, types of gene mutations, patterns of inheritance, the role of a genetics professional, genetic testing, gene therapy, pharmacogenomics, and the Human Genome Project. The handbook is a dynamic document, and new topics and illustrations are added as needed to support content in the condition, gene, and chromosome summaries.
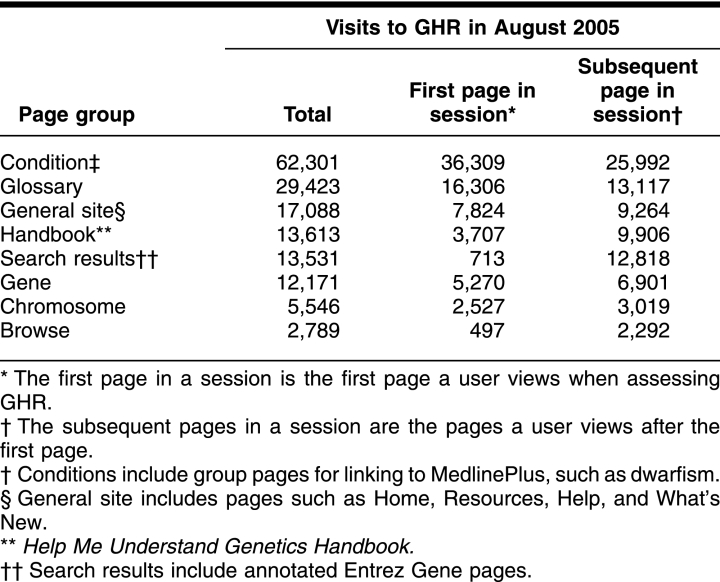
Website traffic, user email messages, and formal user evaluations indicate increasing usage and favorable acceptance of the GHR site. Site traffic has increased rapidly since the site became operational and, by September 2005, was averaging 3.5 million hits monthly. In general, summaries of health conditions are viewed most often (Table 1). These summaries are visited about twice as frequently as the glossary, which is viewed next in frequency. General pages such as the home page, the handbook, search results, gene summaries, chromosome summaries, and browse follow in usage. Email comments from users anecdotally indicate much appreciation of GHR. Further, GHR is becoming known to journalists and has been linked to from online news sources for science and health stories for the public, notably CNN, MSNBC, and Nature.
Table 1 Visits to Genetics Home Reference (GHR) pages
CHALLENGES AND STRATEGIES
To create, maintain, and expand a lay-friendly genetics resource, the GHR staff addresses three ongoing challenges. Due to the large number of known genetic conditions, one challenge is prioritizing topic selection. Another challenge is streamlining content development while ensuring accuracy. Lastly, GHR must help the lay public navigate the complex relationships between health conditions and genetic factors. Additional strategies are required to accelerate the pace of content development and maintenance and to keep abreast of emerging genetic information.
Challenge 1: Prioritize topic development
With thousands of inheritable conditions as candidates for inclusion in GHR, a multifaceted approach is used to prioritize topic development. These strategies allow for both breadth and depth in the collection of topics that GHR ultimately presents to the public.
Strategy 1a: Focus on genetic topics of national import
In 2005, GHR developed materials to support the Department of Health and Human Services newborn screening initiative [12], through which a national committee of genetics and public health experts recommended a panel of conditions to include in statewide newborn screening programs. To support this initiative, GHR developed materials for the twenty-nine genetic conditions in the recommended screening panel (Table 2) and is pursuing methods to bring these topics to the attention of parents and providers.
Table 2 Draft recommended newborn screening core panel (March 2005)
Strategy 1b: Coordinate with other genetics-related US government projects
The GHR staff collaborates with other federal agencies and prioritizes content development to support complementary genetics projects. For example, staff at the Genetics and Rare Diseases Information Center [13], established by National Human Genome Research Institute and the Office of Rare Diseases, provided a list of twenty disorders for which information is most often requested. GHR summaries are currently available for most of these disorders. The remaining conditions with a known genetic cause have been added to the GHR high-priority development list.
Strategy 1c: Address topics requested by Genetics Home Reference (GHR) users
The GHR staff performs a monthly review of users' search requests to find new topics to add to the Website. Topics requested by users of NLM's consumer health Website, MedlinePlus [14], are also considered. GHR and MedlinePlus staff members collaborate regularly to develop complementary content for each Website.
Challenge 2: Streamline content development
Several informatics strategies contribute to efficient processes for content development and quality control. All are based on the principle that automated data extraction and analyses can be used to locate, collate, and integrate relevant information during content development. Even though all content is reviewed by genetic experts before its presentation, automated tools decrease the time needed to find and assess facts. Two strategies used to streamline content development and quality control are presented below. Long-term studies are underway to expand these efforts.
Strategy 2a: Use structured data for the foundation of GHR content
Structured data are stored internally and used to create a document-based presentation for the public. The data elements of the initial version of GHR have been previously described [11], but modifications to accommodate new information are ongoing. Using data fields to capture specific information minimizes interpretation errors. For example, the gene symbol “CAT” is not confused with the word “cat” when the value is stored in a gene symbol field. Also, fields in the GHR data structure are linked with other data sources. For example, the cytogenetic location code in GHR gene summaries is linked with the cytogenetic location code in Entrez Gene [15]. This linkage lays the groundwork for harvesting data from existing data sources.
Strategy 2b: Harvest data from existing databases to create quality-control procedures and system enhancements
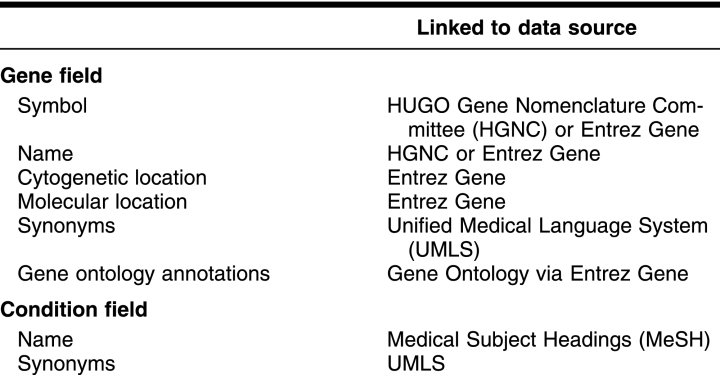
GHR's tools are based on harvesting information from Entrez Gene [15], the HUGO Gene Nomenclature Committee (HGNC) [16], GeneReviews and GeneTests [17], and MedlinePlus [14]. Table 3 shows the data sources for specific GHR data elements. Quality control includes comparing the data harvested from these sources with the data presented on the GHR site. An automated system compares GHR with source data and alerts content developers to any differences to evaluate and address. Weekly data harvesting and analyses keep GHR content synchronized with shifts in genetic information in the primary research databases, such as changes in gene symbols or names.
Table 3 Data sources in specific fields of GHR
The process of creating GHR gene records illustrates how data harvesting streamlines the content development process. To create a new gene record, the content developer enters a gene symbol into the GHR Content Manager (the software used to collect and store GHR's data). The software searches data from Entrez Gene [15] and HGNC [16] to find the gene of interest. Often, many candidates are found. These possibilities are presented to the content developer to choose the correct match. If a match is selected, the software automatically fills the gene record with the official gene symbol and name, gene location codes, synonyms, and functional activities from the Gene Ontology (GO) Consortium [18]. Links to Entrez Gene [15], the Online Mendelian Inheritance in Man (OMIM) catalog [19], GeneCards [20], and HGNC [16] are also automatically inserted. This information is then used as the basis to develop the new gene topic.
Challenge 3: Help the public to navigate complex genetic relationships
GHR is designed to ease navigation within the site and to help locate relevant information on other Websites. Of particular interest is the relationship of genes or chromosomes to health conditions. From each condition topic, GHR provides navigational links to related genetic factors, promoting a better understanding of the genetic etiology of disorders. In addition, GHR links to explanations of relevant concepts in the Help Me Understand Genetics Handbook. The underlying data model is designed to help consumers navigate these gene-disease-chromosome relationships. Several informatics strategies help consumers find relevant information.
Strategy 3a: Use semantic tools to enhance information retrieval
GHR uses an NLM-developed search engine [21, 22] that applies semantic expansion to find relevant topics that would not otherwise be retrieved. For example, controlled vocabulary from the Unified Medical Language System (UMLS) [23] is used to expand searches to find content using different words or phrases for the same concept. As a result, when a user searches GHR for “cancer,” the search engine expands the search to find content containing the phrases “neoplasm,” “malignancy,” “malignant tumoral disease,” and other UMLS cancer synonyms.
Further, GHR content is indexed using NLM's Medical Subject Headings (MeSH) [24]. MeSH provides a bridge linking to relevant information in other resources—such as ClinicalTrials.gov [25], MedlinePlus [14], and PubMed [26]—that also index using MeSH. MeSH indexing provides a fast and accurate mechanism to synchronize information.
Strategy 3b: Encourage browsing of the content
Even though most consumers will use the search feature for extended exploration, many novice users first become intrigued through browsing the content. GHR users can find condition, gene, and chromosome topics by browsing alphabetical lists of names and gene symbols or by browsing chromosome numbers. Alphabetical lists include the primary name chosen for GHR topics as well as synonyms. Hierarchical browsing features are provided for condition and gene summaries. The content-indexing techniques described above serve as aides for browsing the GHR content. The browse hierarchy for a genetic condition is loosely based on MeSH disease concepts. For example, GHR's cancers category roughly maps to MeSH's neoplasms concept. For gene topics, the browsing hierarchy is automatically derived from upper levels of GO [18]. For example, the GO hierarchy shows that both the APOE gene, which is associated with Alzheimer disease, and the TPO gene, which causes one form of congenital hypothyroidism, have a molecular function related to antioxidant activity.
Strategy 3c: Provide a range of additional online resources related to particular conditions, genes, and chromosomes
Links to other Websites are selected for a wide range of audiences including patients, family members, clinicians, educators, students, and genetic researchers. These links are grouped to help cue the reader to the kind of information available from each source. At a glance, a user is able to see that OMIM [19], which is written for health care professionals and researchers, presents more challenging information than MedlinePlus [14], which is designed for the general public. This linking and annotation strategy is certainly not unique to GHR, but the strategy is well worth emulating. The criteria used for selecting Websites as additional resources are available at <http://www.ghr.nlm.nih.gov/ghr/page/Disclaimers/>.
Additionally, GHR offers search results from GHR as well as MedlinePlus [14], GeneTests and GeneReviews [17], and Entrez Gene [15]. Augmenting search results with links to other sites helps guide users to reliable sources of information on genetic topics. Such augmentation is especially useful when topics have not yet been fully developed for GHR. For example, if a user looks for information on juvenile diabetes, the search results offer a link to the MedlinePlus page about this condition. Similarly, when a user searches GHR for a gene that the site does not yet include, links to annotated Entrez Gene data are presented. The annotations add context to help the lay audience interpret the highly technical information in Entrez Gene.
Strategy 3d: Apply consumer feedback to improve effectiveness
In the first year of the GHR project, informal surveys of online users and first-year college students were conducted to help assess early layout designs, as well as content substance, reading level, and understandability. Feedback from these surveys helped shape the glossary content, the question-and-answer layout of summaries, the placement and types of links, the prioritization of search results, and the topics for the handbook. System-level tests to ensure compliance with accessibility regulations and to stress system capacity were also conducted.
In early 2004, a survey was conducted to obtain systematic consumer feedback about GHR and to use the findings to prepare a more comprehensive evaluation. Members of the Genetic Alliance, an international coalition that represents individuals with genetic conditions, were invited to provide a critical assessment of the site via an online survey conducted from late February to mid-April 2004. Participation was voluntary, and 374 respondents completed the survey. The findings were not generalizable to all Genetics Alliance members or the general population because of the nonrandom nature of the survey. The survey demographics, however, were similar to the profile recently identified by the Pew Internet and American Life Project [2, 3], in which health-information seekers were predominately female, were well-educated, and sought information using the Internet. The survey results indicated that GHR's perceived credibility and overall consumer satisfaction were high. Respondents also found GHR to be authoritative, accurate, unbiased, pertinent, up to date, and informative. Full details of this study are reported elsewhere [27].
FUTURE PLANS
As GHR continues to improve and expand, it will be in a position to explore new paradigms for genes, health conditions, drugs, diet, and environmental factors [6]. Continued development of the site will guide the use of informatics techniques and drive innovative research to assist with topic selection, content development, quality control, and information retrieval. Text harvesting techniques [28, 29] can be used to find new information for GHR content. Summarization and question-answering research [30] may help to focus and condense relevant information for content development and annual updates. Biomedical ontologies may play a role in integrating disparate biomedical resources [31]. Consumer health research [32, 33] will continue to focus the project on what needs the intended users have and whether they can find the information of interest. Future evaluation will include surveys of motivated health-seeking Internet consumers to clarify GHR's perceived readability, usability, and image. Evaluations among special populations of potential GHR users may shape and guide GHR's support for targeted initiatives.
CONCLUSIONS
GHR is a credible, dynamic Website that uses lay language to interpret the health implications of the Human Genome Project. This site uniquely meets the needs of consumers seeking genetic information. The strategies developed to address the challenges posed by GHR's focus are key to the effective management of a large and expanding body of genetics information. Feedback from formal and informal evaluations have helped shape GHR's layout, navigational design, and level of content. Continued site development will guide the use of informatics techniques and drive new research to assist with topic selection, content development, and quality control and to help consumers navigate the complex world of genetics.
Acknowledgments
The authors thank others who contribute to the Genetics Home Reference project: Sherri Calvo, May Cheh, Erik Dorfman, John Gillen, Nicholas Ide, Russell Loane, Robert Logan, Stephanie Morrison, Diane Mucci, and Phillips Wolf.
Contributor Information
Joyce A. Mitchell, Email: Joyce.mitchell@hsc.utah.edu.
Cathy Fomous, Email: fomous@nlm.nih.gov.
Jane Fun, Email: fun@nlm.nih.gov.
REFERENCES
- Cline RJ, Haynes KM. Consumer health information seeking on the Internet: the state of the art. Health Educ Res. 2001 Dec; 16(6):671–92. [DOI] [PubMed] [Google Scholar]
- Fox S, Fallows D. Internet health resources. [Web document]. Washington, DC: Pew Internet and American Life Project, 2003. [cited 5 Oct 2005]. <http://www.pewinternet.org/pdfs/PIP_Health_Report_July_2003.pdf>. [Google Scholar]
- Fox S. Health information online. [Web document]. Washington, DC: Pew Internet and American Life Project, 2005. [cited 7 Dec 2005]. <http://www.pewinternet.org/pdfs/PIP_Healthtopics_May05.pdf>. [Google Scholar]
- Guttmacher AE.. Human genetics on the Web. Annu Rev Genomics Hum Genet. 2001;2:213–33. doi: 10.1146/annurev.genom.2.1.213. [DOI] [PubMed] [Google Scholar]
- Taylor MR, Alman A, and Manchester DK. Use of the Internet by patients and their families to obtain genetics-related information. Mayo Clin Proc. 2001 Aug; 76(8):772–6. [DOI] [PubMed] [Google Scholar]
- Mitchell JA, McCray A, Bodenreider O.. From phenotype to genotype: issues in navigating the available information resources. Methods Inf Med. 2003;42(5):557–63. doi: 10.1267/METH03050557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt JM, Lariesy RA, Parrott RL, Silk KJ, and Felter EM. Perceived barriers to Internet-based health communication on human genetics. J Health Commun. 2002 Jul–Sep; 7(4):325–40. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Collins FS. Welcome to the genomic era. N Engl J Med. 2003 Sep 4; 349(10):996–8. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Collins FS. Genomic medicine—a primer. N Engl J Med. 2002 Nov 7; 347(19):1512–20. [DOI] [PubMed] [Google Scholar]
- Khoury MJ. Genetics and genomics in practice: the continuum from genetic disease to genetic information in health and disease. Genet Med. 2003 Jul–Aug; 5(4):261–8. [DOI] [PubMed] [Google Scholar]
- Mitchell JA, Fun J, and McCray AT. Genetics Home Reference: a new NLM consumer health resource. J Med Inform Assoc. 2004 Nov–Dec; 11(6):439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Department of Health and Human Services, Health Resources and Services Administration, Maternal and Child Health Bureau. Newborn screening: toward a uniform screening panel and system. executive summary. [Web document]. The Department. [cited 5 Oct 2005]. <ftp://ftp.hrsa.gov/mchb/genetics/screeningdraftsummary.pdf>. [Google Scholar]
- Office of Rare Diseases, National Institutes of Health. The Genetic and Rare Diseases Information Center. [Web document]. The Institutes. [cited 12 Dec 2005]. <http://rarediseases.info.nih.gov/html/resources/gard_brochure.html>. [Google Scholar]
- Miller N, Lacroix EM, and Backus JE. MEDLINEplus: building and maintaining the National Library of Medicine's consumer health Web service. Bull Med Libr Assoc. 2000 Jan; 88(1):11–7. [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, and Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005 Jan 1; 33(database issue):D54–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povey S, Lovering R, Bruford E, Wright M, Lush M, and Wain H. The HUGO Gene Nomenclature Committee (HGNC). Hum Genet. 2001 Dec; 109(6):678–80. [DOI] [PubMed] [Google Scholar]
- Pagon RA, Tarczy-Hornock P, Baskin PK, Edwards JE, Covington ML, Espeseth M, Beahler C, Bird TD, Popovich B, Nesbitt C, Dolan C, Marymee K, Hanson NB, Neufeld-Kaiser W, Grohs GM, Kicklighter T, Abair C, Malmin A, Barclay M, and Palepu RD. GeneTest-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002 May; 19(5):501–9. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierback B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, and White R. Gene ontology consortium. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004 Jan 1; 32(database issue):D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger J, Bocchini CA, and McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nuclei Acids Res. 2005 Jan 1; 33(database issue):D514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, Ben-Dor U, Esterman N, Rosen N, Peter I, Olender T, Chalifa-Caspi V, and Lancet D. GeneCards 2002: towards a complete, object-oriented human gene compendium. Bioinformatics. 2002 Nov; 18(11):1542–3. [DOI] [PubMed] [Google Scholar]
- McCray AT, Loane RF, Browne AC, and Bangalore AK. Terminology issues in user access to Web-based medical information. Proc AMIA Symp 1999;107–11. [PMC free article] [PubMed] [Google Scholar]
- Browne AC, Divita G, Aronson AR, and McCray AT. UMLS language and vocabulary tools. Proc AMIA Symp 2003 Nov;798. [PMC free article] [PubMed] [Google Scholar]
- Humphreys BL, Lindberg DA, Schoolman HM, and Barnett GO. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998 Jan–Feb; 5(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb CE. Medical Subject Headings (MeSH) [historical notes]. Bull Med Libr Assoc. 2000 Jul; 88(3):265–6. [PMC free article] [PubMed] [Google Scholar]
- McCray AT, Ide NC. Design and implementation of a national clinical trials registry. Am Med Inform Assoc. 2000 May–Jun; 7(3):313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US National Library of Medicine, National Institutes of Health. PubMed®: MEDLINE® retrieval on the World Wide Web. [Web document]. The Library. [cited 12 Dec 2005]. <http://www.nlm.nih.gov/pubs/factsheets/pubmed.html>. [Google Scholar]
- Peng Z, Logan RA. Content quality, usability, affective evaluation, and overall satisfaction of online health information. Presented at: Health Communications Division, International Communication Association Annual Meeting; 2005 May. [Google Scholar]
- Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. Proc AMIA Symp 2001;17–21. [PMC free article] [PubMed] [Google Scholar]
- Rindflesch TC, Tanabe L, Weinstein JN, and Hunter L. EDGAR: extraction of drugs, genes and relations from the biomedical literature. Pac Symp Biocomput 2000;517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszman M, Rindflesch TC, and Kilicoglu H. Abstraction summarization for managing the biomedical research literature. Proc HLT-NAACL Workshop Comp Lex Sem 2004;76– 83. [Google Scholar]
- Bodenreider O, Mitchell JA, and McCray AT. Biomedical ontologies. Pac Symp Biocomput 2005;76–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RA.. Evaluating consumer informatics: learning from health campaign research. Medinfo. 2004;11(pt 2):1147–51. [PubMed] [Google Scholar]
- Soergel D, Tse T, Slaughter L.. Helping healthcare consumers understand: an “interpretive layer” for finding and making sense of medical information. Medinfo. 2004;11(pt 2:):931–5. [PubMed] [Google Scholar]