Abstract
Setting: The University of Washington Health Sciences Libraries and Information Center BioCommons serves the bioinformatics needs of researchers at the university and in the vibrant for-profit and not-for-profit biomedical research sector in the Washington area and region.
Program Components: The BioCommons comprises services addressing internal University of Washington, not-for-profit, for-profit, and regional and global clientele. The BioCommons is maintained and administered by the BioResearcher Liaison Team. The BioCommons architecture provides a highly flexible structure for adapting to rapidly changing resources and needs.
Evaluation Mechanisms: BioCommons uses Web-based pre- and post-course evaluations and periodic user surveys to assess service effectiveness. Recent surveys indicate substantial usage of BioCommons services and a high level of effectiveness and user satisfaction.
Next Steps/Future Directions: BioCommons is developing novel collaborative Web resources to distribute bioinformatics tools and is experimenting with Web-based competency training in bioinformation resource use.
Highlights
Data from every level of biological organization are being acquired and digitized at exponential rates, which, along with massive advances in informatics and synthetic chemistry, is changing the nature of biomedical research in fundamental ways.
The University of Washington (UW) Health Sciences Libraries and Information Center (HSLIC) BioCommons is a robust and flexible umbrella organization that has evolved to provide the biomedical research community with current information and tools to promote speed and efficiency in research.
The BioCommons supports the bioinformation needs of more than 2,500 basic biology researchers at the UW, provides support to the regional business and not-for-profit community, and collaborates with regional and national organizations to provide bioinformation support.
The BioCommons is experimenting with advanced Web-based publishing, bioinformatics tools, and training to further increase the efficiency and speed of the UW, regional, and global biomedical research enterprise.
Implications for practice
Advanced Web-based information systems, bioininformatics tools, and training can enable a small team of specialists with limited funding to meet the growing needs of a large and diverse biomedical research community.
The biomedical community is highly diverse and individualistic in its research interests and needs and is simultaneously local, regional, and global.
Collaboration with for-profit and not-for-profit sectors, in addition to governmental sectors, is critical for success.
INTRODUCTION
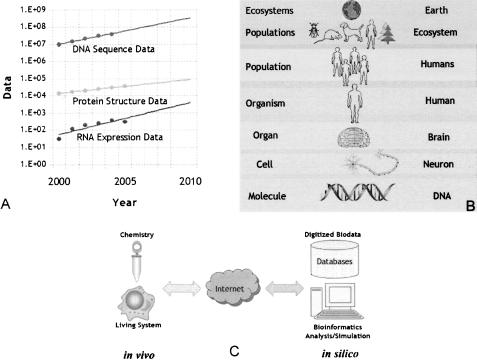
Bioinformatics is now driven by the exponential acquisition and digitization of vast amounts of data at all levels of biological organization (Figure 1A and 1B) [1, 2], leading to the rapid development of new and evermore sophisticated and powerful tools and data resources. Bioinformation needs have moved beyond the “classical bioinformatics” of sequence alignments and protein structures to the “new bioinformatics,” encompassing gene expression, cell and organism anatomy, metabolic pathway analysis, brain function, and even ecosystems [2]. The inherently digital nature of biological information [3–8] is also leading to the development of novel approaches to understanding biological systems, where in silico models are used to reconstitute in vivo living systems through advanced chemistry (Figure 1C) [9–11].
Figure 1.
(A) Exponential growth of DNA sequence data, RNA expression data, and protein structural data; (B) the levels of biological organization and information; and (C) the interplay between in vivo and in silico biology
An early leader in recognizing the importance of bioinformatics for medical libraries, the University of Washington (UW) Health Sciences Libraries and Information Center (HSLIC) began developing a bioinformatics program to enhance access to molecular sequence databases and other biological information resources in 1995 [12]. The UW HSLIC serves a diverse user base of clinicians, researchers, and students in the health and biological sciences across Seattle and beyond. The UW is one of the nation's leading research centers and has ranked among the top five federally funded research institutions for the past twenty-five years. Several major genomic mapping and sequencing programs are based at the UW, and the university serves as an incubator for the vibrant and growing biotechnology industry in the Seattle and Washington area [13]. As detailed in the Bio21 plan, Washington's extensive efforts in biotechnology will broadly involve many biology-based technologies including imaging, diagnostics, and informatics and will require significant bioinformation support. Table 1 provides uniform resource locators (URLs) of resources and companies listed.
Table 1 Web addresses of resources and companies listed
To help meet the needs of the UW in bioinformatics and bioinformation resources, as well the anticipated needs of the state's growing biology-based industries, the UW HSLIC has created a collection of responsive services, tools, and personnel assembled under the BioCommons umbrella organization.
THE BIOCOMMONS STRUCTURE
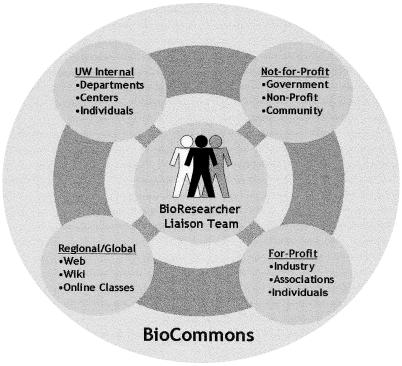
The BioCommons is composed of four service sectors: (1) UW internal, (2) not-for-profit, (3) for-profit, and (4) regional or global, maintained and administered by the BioResearcher Liaison Team (Figure 2). The BioCommons architecture provides a highly flexible structure capable of adapting to constantly changing resources and needs. Currently funded entirely by grants and non-state funds, the BioCommons primarily serves the UW basic research community and secondarily supports the needs of regional and global organizations. As the BioCommons develops, its role in the greater regional biomedical research enterprise should grow and perhaps evolve to supporting both basic biological and translational research.
Figure 2.
Structure of the University of Washington Health Sciences Libraries BioCommons
THE BIORESEARCHER LIAISON TEAM
Central to the BioCommons is the BioResearcher Liaison Team, composed of individuals with a wide range of expertise applicable to the BioCommons mission. The team leader is a biologist with a doctoral degree and “bench” experience in academic, governmental, and industrial research organizations as well as experience in the information technology (IT) industry and extensive contacts in both communities. The team leader acts as a liaison for the HSLIC to the bioresearch community both within and outside of UW, serves in the library's liaison pool, and was recently appointed an affiliate instructor in the UW Department of Biomedical and Health Informatics (BHI) to facilitate collaborations with research faculty on grants and publications. Additionally, the BioResearcher Liaison Team consists of a programmer and a network specialist and draws on the expertise of the library's other liaison teams (Table 1). Together, the BioResearcher Liaison Team provides the core support for the four services and resource components of the BioCommons (Figure 2).
UNIVERSITY OF WASHINGTON INTERNAL COMPONENTS
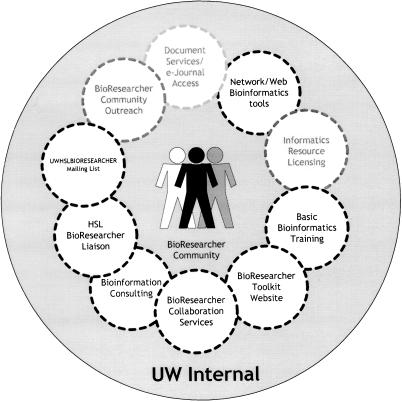
This team provides a number of different services (Figure 3), including regular training through the quarterly BioResearcher Tune-Up, the BioResearcher Toolkit network and Web bioinformatics resources (i.e., Sequencher and GeneSifter), and individual bioinformatics consulting. The BioResearcher Liaison Team provides a mix of standard and customized ad hoc services adapted to meet the needs of the UW research community in a rapidly changing environment.
Figure 3.
Services provided by the BioCommons BioResearch Liaison Team to the University of Washington BioResearcher Community
Bioresearcher Tune-Up course
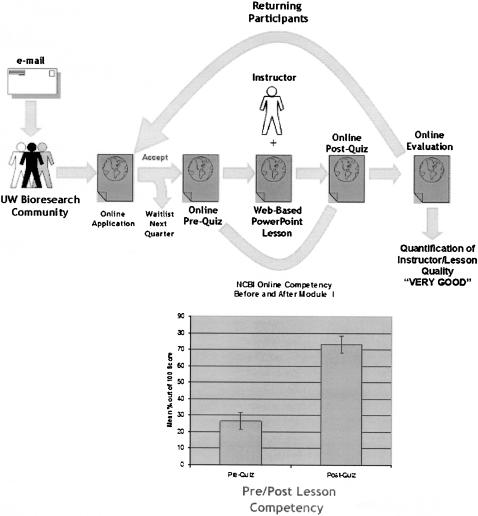
A standard BioCommons service is the HSLIC's quarterly BioResearcher Tune-Up course with participants recruited using the roughly 2,500-subscriber UWHSLBIORESEARCHER opt-in mailing list (Figure 4). The 3 course modules (5 hours each) cover topics ranging from the basics of National Center for Biotechnology Information (NCBI) resources to advanced topics such as metagenomics (Table 1).
Figure 4.
The BioResearcher Tune-Up bioinformation resource immersion course process
Course module I uses Web-based pre- and posttests and a single gene example as the basis for a Web-based, interactive PowerPoint lesson plan, a highly effective approach for providing participants with practical knowledge in using basic NCBI resources as illustrated by pre- and post-quiz data for two separate quarterly modules (Figure 4). Participants, who on average rated their skill at using both the Basic Local Alignment Search Tool (BLAST) and Entrez as “intermediate,” only scored 20% on the basic 5 question pre-quiz, but scored 75% on the similar post-quiz given immediately following the class
Three versions of module I have been produced, using Huntington disease (HD) [14, 15], Holt-Oram syndrome (TBX5) [16, 17], and dyserythropoietic anemia with thrombocytopenia (GATA-1) as examples [18–21]. Each model gene provides a simple path through the use of BLAST and Entrez and furthermore demonstrates the power of NCBI's online resources to discover new knowledge. The HD model, for example, reveals the existence of a new set of neurological disorders caused by polyglutamine mutations in the TATA Binding Protein (TBP) [22–24] and a previously underappreciated relationship between type I diabetes and Huntington disease [25–27].
Module II is a workshop given by one of the providers of commercial bioinformatics tools distributed through the BioResearcher Toolkit Website. Finally, module III focuses on advanced topics such as RNAi, gene annotation software, and gene regulatory mechanisms that are of general interest to researchers. Overall, the BioResearcher Tune-Up is highly regarded and well attended, with typical evaluations of “Very Good” and “Excellent” (Figure 4).
BioResearcher Toolkit and network and Web bioinformatics tools
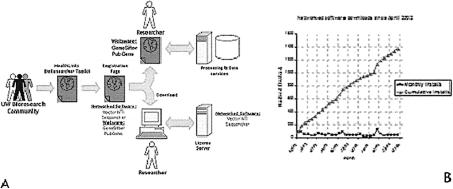
The BioCommons also provides UW bioresearchers with access to network and Web-based bioinformatics tools via the BioResearcher Toolkit Website. Using an automated Web-based registration process (Figure 5) that allows access only to UW users, researchers at the university are able to employ commercial resources such as molecular sequence analysis tools (Vector NTI and Sequencher), expression analysis tools (GeneSifter), and data mining tools (PubGene). These resources are heavily used, with nearly 2,500 unique registered users since 2002.
Figure 5.
(A) Schema for provision of bioinformatics tools via the BioResearcher Toolkit and (B) usage statistics for such tools
Bioinformation consulting and other services
BioCommons consulting through the BioResearcher Liaison Team is also highly utilized by UW researchers and increasingly by the regional business and nonprofit bioresearch community, with over 100 consults provided in the 2004/05 fiscal cycle. The nature of such contacts varies, ranging from simple issues surrounding the installation of software to the complex analysis of data in support of grants. Following each consult, users are requested to complete a short online evaluation form. These evaluations indicate a high degree of satisfaction with BioCommons consulting, with typical ratings of “Very Good” and “Excellent” and satisfactory resolution of issues.
In addition to the training and consultation services available internally to the UW bioresearch community, other services include (1) copyright and intellectual property assistance to clarify such issues and verify UW affiliate compliance, (2) advocacy for the acquisition of resources relevant to bioresearchers, and (3) production and maintenance of the HealthLinks BioResearcher Toolkit Website (Table 1) [12, 28].
SUPPORT FOR NOT-FOR-PROFIT ORGANIZATIONS
The BioCommons has strong alliances with governmental, educational, and nonprofit organizations. The team leader has been involved in the development and implementation of NCBI training materials, including modules on nonhuman genomes and expression resources as part of the “NCBI Advanced Workshop for Bioinfomation Specialists (NAWBIS)” course, and the UW HSLIC has hosted NCBI training courses. Such outside activities are a powerful means of remaining current in the field and creating a network of contacts to draw on for assistance. This work has also led to collaborations with NCBI development groups, such as the Gene Expression Omnibus (GEO) group, and provides a valuable feedback loop for tool users and developers. The BioCommons has also promoted highly specialized training in the use of open source microarray software by the UW Center for Expression Arrays (CEA) and has facilitated advanced training from other organizations such as the European Bioinformatics Institute (EBI).
The BioCommons maintains an active presence in the regional biotech community by direct participation in a nonprofit educational organization, the Pacific Northwest Bio/Technologies Alliance, which acts as a quarterly forum for networking and presentations from the UW, local biotechnology companies (such as AMGEN-Seattle), nonprofit research organizations (such as the Seattle Biomedical Research Institute [SBRI] and the Institute for Systems Biology [ISB]), and other interested groups and individuals (e.g., local law firms and computer gaming companies).
Project LARIAT, a National Institutes of Health (NIH)/National Center for Research Resources (NCRR)–supported virtual community of biomedical researchers and educators in several US Western states (Alaska, Hawaii, Idaho, Montana, Wyoming, New Mexico, and Nevada), has also partnered with the BioCommons to provide access to restricted UW HSLIC bioinformation resources through the BioResearcher Toolkit and has used the Project LARIAT Access Grid Node (AGN) to provide BioCommons BLAST training to remote graduate students [2].
RELATIONSHIPS WITH THE FOR-PROFIT SECTOR
The BioCommons has cultivated relationships with the commercial sector to provide the regional bioresearch community with access to state-of-the-art technologies. For example, the BioCommons has worked with Seattle-based VizX Labs to provide that company's advanced Web-based microarray analysis service (GeneSifter.net) to UW users, resulting in a mutually beneficial collaboration wherein VizX labs and the research community benefit from feedback that leads to improvements in the GeneSifter.net services.
The BioResearcher Liaison Team has consulted on bioinformatics issues for the investment community in the interest of encouraging the development of new commercial research tools. The group has worked with local leaders such as Ignition and Trimergent to develop new ideas and concepts for products and services supporting the bioinformation needs of the community. The Washington Technology Center (WTC) also makes use of BioCommons expertise in areas such as proteomics to evaluate projects under consideration for funding. Additionally, the BioCommons has begun discussions with the UW Center for Nanotechnology on projects involving the intersection between bio- and nanotechnology, in particular in the area of commercialization of diagnostic devices and agriculture. Nearly 10% (N = 100) of BioCommons consult requests now come from the regional business community
Finally, the UW BHI and the BioCommons have cohosted informal forums for the discussion of the history of bioinformatics with science historian George Dyson and panel discussions on the local business of bioinformatics from Seattle area bioinformatics leaders, including the chief technology officers of VizX Labs (a bioinformatics software development company), InSilicos (a proteomics informatics company), and Arkitek (a company specializing in advanced animations of molecular and cellular biology processes) [29]. Such forums hopefully will promote novel collaborations to develop new bioinformatics tools, such as sophisticated three-dimensional animation techniques in simulating biochemical pathways [30].
REGIONAL AND GLOBAL COMPONENTS
The BioResearcher Toolkit component of the UW HSLIC HealthLinks Website receives nearly 60,000 hits annually and is the second most viewed component of the HealthLinks site. The BioResearcher Toolkit [28], and its predecessor site the Molecular Biologist Toolkit [12], primarily consist of searchable lists of annotated links to molecular and other bioinformatics resources organized according to the levels of biological information as depicted in Figure 1B. The links are largely freely available, while the Computer and Laboratory and Training and Education sections of the Toolkit contain resources restricted to UW affiliates. While the site was originally established as a local resource, it has gained prominence as a national resource and is receiving increasing international traffic.
Working with the geographically dispersed UW research community; the Washington, Wyoming, Alaska, Montana, and Idaho states; and Project LARIAT, the BioCommons is also evaluating and testing teleconferencing technologies for use in distance learning and collaboration. For example, with Project LARIAT, the BioCommons conducted an experiment in distance training using AGN technology to teach a “virtual” course on the use of BLAST to biochemistry graduate students in seven Western states. With more than thirty participants in three one-hour presentations, this experiment indicated that the AGN technology could be used effectively for highly interactive distance training for a very complex bioinformatics tool. Plans for similar, more complex training are now underway.
ASSESSMENT OF THE BIOCOMMONS
In a UW HSLIC survey of the UW bioresearch community in 2005, more than 53.2% (N = 250) of those surveyed (1,717 surveys emailed with a 15.1% response rate) indicated that the BioResearcher Toolkit was an important research resource. Access to bioinformatics tools was seen as critical to their research by 66% (N = 250) of respondents, with several documented instances of such access resulting in favorable grant reviews. Eighty percent (N = 237) of respondents considered it important and useful for the UW HSLIC to identify approaches to supporting the biological information needs of the state biotech industry and nonprofit research organizations. Given this finding, the importance of establishing mechanisms that will allow for greater access to UW HSLIC bioinformation resources by regional and global organizations cannot be overemphasized. Finally, 76% (N = 231) of respondents felt that funding for resources such as the BioCommons should come directly from the UW and Washington State.
THE FUTURE
It is important for the BioCommons to make resources easier to use and to promote more collaboration among bioresearchers. To this end, the BioResearcher Liaison Team has begun research to develop new tools to promote collaboration and to increase the ease of use of and access to bioinformation resources. The BioCommons and the Institute for Systems Biology are now experimenting with a wiki to allow users with appropriate permissions to post and modify bioinformatics-related information. For example, users could create a page on the BioCommons wiki and post a description of a new bioinformatics algorithm and a link to their servers for downloading the appropriate code. Other users could then try the algorithm and add comments to the wiki page, potentially leading to direct collaborations to improve the new bioinformatics tool. Similarly, reviews of topics such as transcription mechanisms could be posted, reviewed, and updated by all members, creating a constantly current collection of data and commentary about a constantly changing subject. Because all pages are archived and contributor identities are logged, the wiki site is under a form of constant peer review.
The BioCommons is also developing a toolbar to allow secure tools to be added directly to a Web browser and provide users with a simple means of manipulating, viewing, and analyzing Web-based information and data directly through their Web browsers [31–33]. Access rights to restricted UW HSLIC resources could be built directly into the toolbar, potentially allowing it to serve as a means for controlled distribution of proprietary resources. The BioCommons Toolbar prototype (Figure 6A) is now being tested in limited release with select users.
Figure 6.
The future: toolbar showing built-in access to (A) eJournals, (B) bioinformatics tools and microarray analysis components (such as GeneSifter), and (C) “hybrid” training process
The continuously changing field of bioinformatics also requires more effective and efficient means of education and training. The current BioResearcher Tune-Up does not allow for self-paced learning, and plans are underway to develop a hybrid system that would allow users to learn from an online lesson with built-in testing, while still having the option of direct training with an instructor (Figure 6C).
CONCLUSIONS
The BioCommons is a flexible way to provide for the bioinformation needs of both the internal UW biomedical research enterprise and those of the larger regional, national, and global bioresearch communities. While universities will continue to be an important component of bioresearch, for-profit and not-for-profit organizations will increasingly play critical roles in moving biomedical research forward. It is important that resources such as the BioCommons be made available beyond the internal UW research community as they will increase the efficiency and speed of biomedical research. The speed at which both data and knowledge about biological systems are acquired, shared, and analyzed has already impacted a number of modern health crises [34–36].
Medical libraries can play an important role in bioresearch by managing access to online resources, making such resources easier to use, and providing training in their use as they evolve. Librarians, via liaison and consulting activities, can bring reliable, objective assessment of such resources to all users, an aspect of bioinformation librarianship that will likely be important as commercial interests gain more control over bioinformatics tools and data sources in the coming era of personalized medicine [37, 38].
Acknowledgments
The authors thank James S. Brockenbrough, Debra Revere, Stuart Yarfitz, Eric Olson, Bob Cottingham, Elon Gasper, and everyone at VizX Labs; Seamus Reagan, Rob Arnold, and the other directors of the Pacific Northwest Bio/Technologies Alliance; Beth Etschied and Kim Emmonds of the Washington Research Foundation; our colleagues at the UW HSLIC; and many others for their help in establishing the UW HSLIC BioCommons and producing this paper. We especially thank our colleagues in the NCBI EDUCOLLAB Group and our associates at the NCBI and the NLM for their support.
Contributor Information
Mark Minie, Email: meminie@u.washington.edu.
Stuart Bowers, Email: stuartmb@u.washington.edu.
Peter Tarczy-Hornoch, Email: pth@u.washington.edu.
Edward Roberts, Email: ejnr@u.washington.edu.
Rose A. James, Email: rajames@u.washington.edu.
Neil Rambo, Email: nrambo@u.washington.edu.
Sherrilynne Fuller, Email: sfuller@u.washington.edu.
REFERENCES
- Bergeron B. Bioinformatics computing. 1st ed. Upper Saddle River, NJ: Prentice Hall PTR, 2002. [Google Scholar]
- Tarczy-Hornochm P, Minie M. Bioinformatics challenges and opportunities. In: Chen H, Fuller S, Friedman C, Hersh W, eds. Medical informatics: knowledge management and data mining in biomedicine. New York, NY: Springer, 2005:647. [Google Scholar]
- Auffray C, Imbeaud S, Roux-Rouquie M, and Hood L. From functional genomics to systems biology: concepts and practices. C R Biol. 2003 Oct–Nov; 326(10–11):879–92. [DOI] [PubMed] [Google Scholar]
- Hood L. Leroy Hood expounds the principles, practice and future of systems biology. Drug Discov Today. 2003 May 15; 8(10):436–8. [DOI] [PubMed] [Google Scholar]
- Hood L. Systems biology: integrating technology, biology, and computation. Mech Ageing Dev. 2003 Jan; 124(1):9–16. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L.. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–72. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- Morris RW, Bean CA, Farber GK, Gallahan D, Jakobsson E, Liu Y, Lyster PM, Peng GC, Roberts FS, Twery M, Whitmarsh J, and Skinner K. Digital biology: an emerging and promising discipline. Trends Biotechnol. 2005 Mar; 23(3:):113–7. [DOI] [PubMed] [Google Scholar]
- Nurse P. Systems biology: understanding cells. Nature. 2003 Aug 21; 424(6951):883. [DOI] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA, Pfannkoch C, and Venter JC. Generating a synthetic genome by whole genome assembly: PhiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci U S A. 2003 Dec 23; 100(26):15440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, and Venter JC. Global transposon mutagenesis and a minimal mycoplasma genome. Science. 1999 Dec 10; 286(5447):2165–9. [DOI] [PubMed] [Google Scholar]
- Check E. Venter aims for maximum impact with minimal genome. Nature. 2002 Nov 28; 420(6914):350. [DOI] [PubMed] [Google Scholar]
- Yarfitz S, Ketchell DS. A library-based bioinformatics services program. Bull Med Libr Assoc. 2000 Jan; 88(1):36–48. [PMC free article] [PubMed] [Google Scholar]
- Kim A. New life-sciences fund lifts hopes of state's researchers. Seattle Times 2005 Jul 11. [Google Scholar]
- Landles C, Bates GP. Huntington and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep. 2004 Oct; 5(10):958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA. Huntington's disease: new paths to pathogenesis. Cell. 2004 Jul 9; 118(1):4–7. [DOI] [PubMed] [Google Scholar]
- Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet. 2003 Apr 1; 12(spec. no. 1):R37–44. [DOI] [PubMed] [Google Scholar]
- Packham EA, David Brook J. Interaction makes the heart grow stronger. Trends Mol Med. 2003 Oct; 9(10):407–9. [DOI] [PubMed] [Google Scholar]
- Cantor AB. GATA transcription factors in hematologic disease. Int J Hematol. 2005 Jun; 81(5):378–84. [DOI] [PubMed] [Google Scholar]
- Del Vecchio GC, Giordani L, De Santis A, De Mattia D.. Dyserythropoietic anemia and thrombocytopenia due to a novel mutation in GATA-1. Acta Haematol. 2005;114(2):113–6. doi: 10.1159/000086586. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, and Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000 Mar; 24(3):266–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Crispino JD. Differential requirements for the activation domain and FOG-interaction surface of GATA-1 in megakaryocyte gene expression and development. Blood. 2005 Aug 15; 106(4):1223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, and Hartl FU. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004 Jul 2; 15(1):95–105. [DOI] [PubMed] [Google Scholar]
- van Roon-Mom WM, Reid SJ, Faull RL, Snell RG.. TATA-binding protein in neurodegenerative disease. Neuroscience. 2005;133(4):863–72. doi: 10.1016/j.neuroscience.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Stevanin G, Fujigasaki H, Lebre AS, Camuzat A, Jeannequin C, Dode C, Takahashi J, San C, Bellance R, Brice A, and Durr A. Huntington's disease-like phenotype due to trinucleotide repeat expansions in the TBP and JPH3 genes. Brain. 2003 Jul; 126(pt. 7):1599–603. [DOI] [PubMed] [Google Scholar]
- Farrer LA. Diabetes mellitus in Huntington disease. Clin Genet. 1985 Jan; 27(1):62–7. [DOI] [PubMed] [Google Scholar]
- Podolsky S, Leopold NA, and Sax DS. Increased frequency of diabetes mellitus in patients with Huntington's chorea. Lancet. 1972 Jun 24; 1(7765):1356–8. [DOI] [PubMed] [Google Scholar]
- Ristow M. Neurodegenerative disorders associated with diabetes mellitus. J Mol Med. 2004 Aug; 82(8):510–29. [DOI] [PubMed] [Google Scholar]
- Helms AJ, Bradford KD, Warren NJ, and Schwartz DG. Bioinformatics opportunities for health sciences librarians and information professionals. J Med Libr Assoc. 2004 Oct; 92(4):489–93. [PMC free article] [PubMed] [Google Scholar]
- Anderson B.. Lights …camera …action! 3-D animations allow researchers, and everyone else, to visualize biology. The Scientist 26 Sept. 2005;19(18):38–42. [Google Scholar]
- Grimm D. 2004 visualization challenge. multimedia—interactive. Science. 2004 Sep 24; 305(5692):1906. [DOI] [PubMed] [Google Scholar]
- Schirra F, Suzuki T, Richards SM, Jensen RV, Liu M, Lombardi MJ, Rowley P, Treister NS, and Sullivan DA. Androgen control of gene expression in the mouse meibomian gland. Invest Ophthalmol Vis Sci. 2005 Oct; 46(10):3666–75. [DOI] [PubMed] [Google Scholar]
- Richards SM, Liu M, Jensen RV, Schirra F, Yamagami H, Lombardi MJ, Rowley P, Treister NS, Suzuki T, Sullivan BD, and Sullivan DA. Androgen regulation of gene expression in the mouse lacrimal gland. J Steroid Biochem Mol Biol. 2005 Sep; 96(5):401–13. [DOI] [PubMed] [Google Scholar]
- Richards SM, Jensen RV, Liu M, Sullivan BD, Lombardi MJ, Rowley P, Schirra F, Treister NS, Suzuki T, Steagall RJ, Yamagami H, and Sullivan DA. Influence of sex on gene expression in the mouse lacrimal gland. Exp Eye Res 2005 Jun 23. [DOI] [PubMed] [Google Scholar]
- Gallaher S. SARS: what we have learned so far. Dimens Crit Care Nurs. 2005 Mar–Apr; 24(2):51. 4;quiz 55–6. [DOI] [PubMed] [Google Scholar]
- Kliger Y, Levanon EY, and Gerber D. From genome to antivirals: SARS as a test tube. Drug Discov Today. 2005 Mar 1; 10(5):345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski DM, Astell C, Brunham RC, Low DE, Petric M, Roper RL, Talbot PJ, Tam T, Babiuk L.. Severe acute respiratory syndrome (SARS): a year in review. Annu Rev Med. 2005;56:357–81. doi: 10.1146/annurev.med.56.091103.134135. [DOI] [PubMed] [Google Scholar]
- Molidor R, Sturn A, Maurer M, and Trajanoski Z. New trends in bioinformatics: from genome sequence to personalized medicine. Exp Gerontol. 2003 Oct; 38(10):1031–6. [DOI] [PubMed] [Google Scholar]
- Weston AD, Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004 Mar–Apr; 3(2):179–96. [DOI] [PubMed] [Google Scholar]