Abstract
Background: The information landscape in biological and medical research has grown far beyond literature to include a wide variety of databases generated by research fields such as molecular biology and genomics. The traditional role of libraries to collect, organize, and provide access to information can expand naturally to encompass these new data domains.
Methods: This paper discusses the current and potential role of libraries in bioinformatics using empirical evidence and experience from eleven years of work in user services at the National Center for Biotechnology Information.
Findings: Medical and science libraries over the last decade have begun to establish educational and support programs to address the challenges users face in the effective and efficient use of a plethora of molecular biology databases and retrieval and analysis tools. As more libraries begin to establish a role in this area, the issues they face include assessment of user needs and skills, identification of existing services, development of plans for new services, recruitment and training of specialized staff, and establishment of collaborations with bioinformatics centers at their institutions.
Conclusions: Increasing library involvement in bioinformatics can help address information needs of a broad range of students, researchers, and clinicians and ultimately help realize the power of bioinformatics resources in making new biological discoveries.
Highlights
Molecular biology databases and retrieval and analysis tools are a fundamental part of the information landscape in biology and medicine.
The role of libraries can expand naturally to encompass these data domains.
It is a challenge for users to keep up with the rapidly growing array of bioinformatics resources and to use them effectively.
Libraries have opportunities to provide end-user education and support in the use of bioinformatics resources.
Issues relevant to the establishment of bioinformatics end-user services are discussed in this paper.
Implications for practice
Bioinformatics and genetics end-user education and support represent new service roles for libraries that can benefit the research, clinical, and consumer health communities.
Library involvement in this area can contribute to the process of scientific discovery through the facilitation of more effective use of bioinformatics resources.
INTRODUCTION
Molecular biology databases and analysis software programs, broadly referred to in this paper as bioinformatics resources, are now essential tools in biological and medical research, based on their demonstrated power, rapid growth, and widespread use. Less well established are: (1) an understanding of the range of user groups and their informatics needs, (2) a knowledge of how well these users employ these resources, (3) an identification of the organizational units in an institution that provide centralized bioinformatics educational and end-user support programs, and (4) a specification of the roles medical and science libraries can play in facilitating access to and effective use of this vast array of bioinformatics resources.
This paper addresses the first two issues based on empirical evidence and the author's eleven years' experience in user services at the National Center for Biotechnology Information (NCBI). A separate study by Messersmith et al. [1] investigates the third question. With respect to the fourth issue, this paper provides an overview, from a national perspective, of the role libraries have begun to play in bioinformatics and explores a broad range of issues to consider for library involvement in the field.
BIOINFORMATICS RESOURCES AS FUNDAMENTAL TOOLS IN BIOLOGY AND MEDICINE
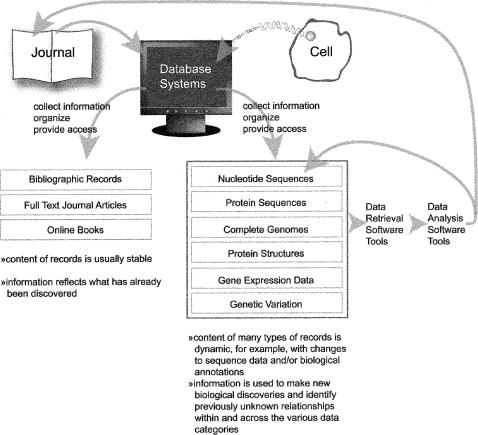
A variety of measures demonstrate the increasing influence of molecular approaches to biological research, the rapidly growing number and variety of corresponding databases and software tools, and the widespread use of software programs to manage and analyze primary research data. For example, in the MEDLINE database, the percentage of articles tagged with the Medical Subject Headings (MeSH) term “Genetic Processes [G05]” grew from 1% of articles with publication dates of 1960 to 1969 to 10% of articles with publication dates of 2000 to 2005. Beyond the increase in genetics-focused literature, many new types of data are generated by fields of study such as molecular biology, genomics, and proteomics. Figure 1 provides examples of these new information domains and suggests the library's role in collecting, organizing, and facilitating access to them is a natural extension of its traditional roles with literature.
Figure 1.
The expanding landscape of biological information sources and data types; traditional factual content from the scientific literature is now complemented by an explosive growth in sequence-related databases containing both primary as well as highly annotated data
Growth in databases and retrieval and analysis tools
The size and number of databases continues to grow exponentially. As a single example of a primary research data repository, GenBank [2] has grown by a factor of approximately 105 in number of sequence records and base pairs (bp) from December 1982 to August 2005 [3]. During a similar period of time, the number of sequence collections described in the Nucleic Acids Research (NAR) journal has grown from 4 [4–7] to 719 [8]. From July 2003 until July 2005, the number of publicly available, peer-reviewed Web servers described by NAR for data retrieval and analysis grew from 131 to 166 [9, 10].
Frequency of usage
An indicator of the high frequency use of these tools is the number of times that papers describing one of the field's major analysis tools have been cited. For example, the papers describing the original [11] and gapped [12] versions of the Basic Local Alignment Search Tool (BLAST) are currently the most-cited scientific papers published since 1988 and 1995, respectively [13, 14]. As an example of usage, the algorithm described in the papers has been implemented at hundreds of sites worldwide, and the BLAST server at NCBI handles over 100,000 searches per day.
Application and impact of bioinformatics resources
In contrast to literature, which is generally used as a source of information about what has been discovered in the past, the new data domains and associated software tools are used as sources for making new biological discoveries (Figure 1). For example, BLAST has been used in combination with repositories of sequence data to identify previously unknown relationships between organisms and genes. An early example was the discovery of the relationship between the human NF1 gene, associated with neurofibromatosis, and well-studied yeast genes involved in the regulation of cell proliferation [15]. Establishing these relationships through a computational approach can shave years and dollars off research time, enabling researchers to identify putative gene functions and therefore more efficiently design experiments to characterize function. Other examples of the application of bioinformatics resources include the identification of potential drug targets to control pathogens [16] and the elucidation of molecular signatures of cancer [17].
In the clinical arena, Guttmacher and Collins [18] and Gibbs [19] are among the authors who have discussed the impact that genomics and bioinformatics will have on medicine. For example, genetic tests can increasingly be used as diagnostic tools [20, 21], and the study of pharmacogenetics aims to identify relationships between genetic variations and drug response [22, 23].
Such evidence indicates that bioinformatics resources are now fundamental tools in basic research and are increasingly being used in the training of biologists and in clinical medicine. However, given the burgeoning array of molecular biology databases as well as data retrieval and analysis tools, users are challenged daily to identify the resources that best fit their needs and to use them effectively. This raises questions about the demographics of bioinformatics users, their needs, and libraries' roles in meeting those needs.
IDENTIFICATION OF USERS AND THEIR NEEDS
Relatively few studies have characterized bioinformatics user groups and their information needs [24]. Several studies have: (a) examined information retrieval processes used by researchers facing a specific scientific problem, such as functional analysis of a gene [25]; (b) presented classifications of bioinformatics tasks that can be considered in the design of a general bioinformatics query system [26]; or (c) developed ontologies of tasks and resources as tools to facilitate queries for multiple databases [27].
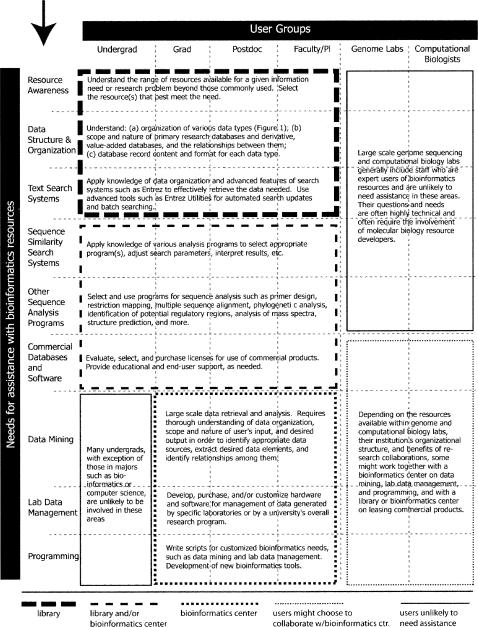
For the purpose of examining the potential role of libraries in bioinformatics, however, it is useful to take a broad look at the wide range of user groups who currently access bioinformatics resources and their corresponding wide range of informatics needs and tasks. Figures 2 and 3 provide summaries of user groups and needs in the research community and in consumer health and clinical communities, respectively. The figures represent a model categorization based on 11 years of work in NCBI User Services, which handles questions from over 300 users per week. Of course, both figures can have overlap between users and needs, but they are separated into 2 figures for simplicity and clarity.
Figure 2.
Model distribution of bioinformatics educational and end-user support services for the research community
Figure 3.
Model distribution of genetics educational and end-user support services for the consumer health and clinical communities
In the research community, Figure 2 illustrates a range of user groups whose needs can be as basic as identifying which resources best meet their specific needs or as complex as programming techniques for large-scale data retrieval and analysis. Among the clinical and consumer health communities, Figure 3 shows a range of user groups with needs varying from gaining an awareness of relevant genetics resources, such as those described by Ohles [28], and obtaining concise information about genetic conditions to diagnosing and managing those conditions and identifying or using relevant resources for clinical genetics research.
Both figures are discussed in further detail in the subsequent section on “Goal Setting in the Establishment of New Services.”
Complexity of user questions
User questions range from basic, factual questions (e.g., what is a CDS?) to complex questions about data organization and retrieval (e.g., how to obtain detailed information about the genes represented by sequence fragments on a specific microarray chip?). Programming questions have become more common (e.g., how to write scripts to extract desired data elements from a number of different data files?), and, finally, users often want assistance in interpreting results from analysis tools.
Similarly, the types of services needed by users range from search assistance (e.g., obtain sequence, mapping, and phenotype information about a given gene) to participation in the research process (e.g., based on an understanding of a research problem, identify and extract relevant data from one or more sources and identify relationships among the data).
VARIATION IN USER KNOWLEDGE AND SKILLS
The ubiquitous presence and widespread use of these resources does not imply that all users are equally knowledgeable of and skilled in the effective use of these tools. Many molecular biology databases and software tools are in the public domain and freely available to all users [29]. However, wide variation in user knowledge and skills in effect creates great inequalities in the ability to discover key information resources, understand them, and use them properly. Three sample indicators of user expertise level include awareness of resources, skill in searching, and critical analysis of data.
Awareness of bioinformatics resources
The NCBI has periodically conducted informal requests for information (RFIs) from users to gain information about the visibility and usefulness of NCBI information resources and to obtain feedback for purposes of future development of services and resources. The RFIs reported here, though older, shed light on the lag time between the release of a bioinformatics resource and the general awareness of users about a small portion of the resources available.
An April 1994 RFI obtained feedback about NCBI's early molecular biology data retrieval systems from forty researchers at the National Institutes of Health, Cold Spring Harbor Laboratory, and Washington University. A January 2000 RFI obtained feedback on human genome resources from forty-three researchers at the University of California, Los Angeles, and the University of Tennessee, Memphis, prior to their attendance at the “NCBI Field Guide” [30]. Respondents included graduate students, postdoctoral fellows, research associates, principal investigators, and department heads. Both RFIs included some general questions to assess users' awareness and usage frequency of example resources, using a Likert scale to record the responses. Appendix A (available only online) contains a brief survey instrument, adapted from these RFIs.
In the 1994 RFI, 42% (N = 17) of respondents were not yet aware of Entrez, which had been available on CD-ROM since 1991 and as a client-server software program since 1993 [31]. As the Web came into common use and as the application of bioinformatics resources in research became more prevalent, awareness of some of these early resources increased. By the time of the 2000 RFI, only 9% (N = 4) of respondents were not yet aware of the Entrez Nucleotide database. A higher percentage of users, though, were unaware of more recently released resources. Thirty percent (N = 13) were not aware of UniGene or LocusLink/RefSeq [32], available since 1996 and 1999, respectively, and 44% (N = 19) were not aware of more specialized resources such as Clusters of Orthologous Groups [33], known as “COGs” and available since 1997.
User skill and sophistication in searching
Few users need to access the hundreds of databases cataloged by NAR, and many users' needs can be satisfied by the use of a small number of resources closely targeted to their research problem. Even within a single resource, however, users are often not aware of the scope and types of records available, the sometimes nuanced relationships among them, and the search system features that can help to effectively mine the data.
In Entrez [34], an NCBI integrated search system that handles more than 3 million searches daily, 70% of the searches are done as simple queries [35], in which terms are entered without field specifiers, Boolean queries, or other advanced search techniques that can be applied to achieve more meaningful search results. Just 21% of the searches use Boolean operators, 13% use field specifiers, and 1% each use wildcard, range searching, and the History function. The statistics for some specific data domains in the Entrez system, such as Nucleotide and Protein, show an even higher frequency (75% and 89%, respectively) of simple searches.
Though a small fraction of users, such as researchers in large scale genome sequencing labs and computational biologists, are experts in the use of bioinformatics resources and in some cases help develop sophisticated analytical software tools and databases, these kinds of statistics bring into question how well most users understand the scope of a particular data domain and the software features that can help them to hone their search precisely to their needs and how effectively or efficiently they are currently mining the data.
A single data domain, such as Entrez Nucleotide or Protein, can contain records from a wide range of primary (archival) and derivative (curated) source databases. Simple queries can therefore result in very large and redundant retrieval that includes a wide variety of record types (e.g., expressed sequence tags, sequence data from patents, high throughput genomic sequence data, complete chromosomes, characterized genes) and qualities (archival and curated). The lack of a controlled vocabulary in most of the databases can further complicate retrieval.
To help make such expansive data sets more manageable, Entrez includes software features designed to help users hone retrieval to specific data subsets or record types. It also includes features to facilitate the biological discovery process, such as the ability to retrieve related records in a database and traverse across databases to retrieve associated records [36]. While it is challenging to present a simple user interface for a complicated and powerful system, many of these options exist as check boxes and pop-up menus on easily accessible pages such as such as Limits, Preview/Index, and History. Yet, these aids are seldom used.
Critical analysis of data
Once data are retrieved from a system, it is important that users understand the limitations of the data in order to use them appropriately. As noted in Figure 1, the content of records in molecular biology databases is dynamic. Sequence data and biological annotations in both archival and curated records can change over time, and it is helpful to view their content critically as a work in progress. As also shown in the figure, the results of data analyses can in turn influence the content of database records, so it is important to use the data judiciously.
While sequence records in both primary (archival) and derivative (curated) databases undergo varying levels of quality control and review [2, 37], records can sometimes contain errors. For example, some of the biological annotations in primary sequence records might have been computationally predicted but not yet experimentally proven. Therefore, annotations that are incorrect can have a domino effect if they are used as the basis for annotations on similar, newly sequenced data. Galperin and Koonin [38] provide examples of dubious functional assignments being used as the basis for subsequent functional predictions.
As a final example of cautions to be exercised in the use of bioinformatics resources, the quality of data in some domains, such as gene expression, in great part depends on the experimental methodology used to generate the data. Minie [39], for example, cites a review by Churchill [40] that discusses the proper design and analysis of microarray experiments and points out pitfalls of some of the published microarray work to date.
GROWTH OF LIBRARY INVOLVEMENT IN BIOINFORMATICS
In recognition of the fundamental importance of these new data domains, medical and science libraries have become increasingly involved in this area over the last two decades. In 1988, NCBI was established in the National Library of Medicine (NLM) to collect, organize, and provide access to the molecular biology data [41, 42]. While few academic libraries are currently involved in collecting or organizing data, they have gradually become involved in facilitating access.
In August 1994, an RFI questionnaire (Appendix B, available only online) posted to MEDLIB-L sought information about the presence and nature of molecular biology services in medical libraries. From among the 1,412 subscribers at that time, 18 responded, each representing a different institution. Twelve (67%) of the institutions provided assistance with text-base searches of sequence databases, 6 (33%) provided assistance with sequence similarity searches, and 11 (61%) provided instruction for searching sequence databases. Librarians providing these services were generally self-taught through reading documentation and experimenting with the systems and provided these services in their “spare time.”
In May 2001, a similar RFI questionnaire (Appendix C, available only online) developed at NCBI was posted to the Association of Academic Health Sciences Libraries (AAHSL) mailing list. Thirty-six of the 134 member institutions on the list responded: 14 (39%) stated they provided user support for nonbibliographic databases and tools, 7 (19%) provided training to users in molecular biology databases and tools, and 6 (17%) of the institutions had library staff whose time was officially dedicated to these services. The percentage of time dedicated ranged from “on demand” and “4%” to “80%.”
While the number of universities reporting bioinformatics services in both surveys did not change dramatically and was relatively small in proportion to the number of universities with medical schools or graduate programs in biology, the surveys suggested some libraries were taking on a new service role.
Another indicator of library involvement, or preparation for involvement, in bioinformatics is the number of staff who have attended relevant training programs. As an example, 567 individuals attended the 1-day MLA continuing education course, “Molecular Biology Information Resources” [43], that was offered from 1997 through 2001, and 235 attended the expanded 3-day version of that course [44] from November 2002 through November 2005. Approximately 95% of participants were library staff. Forty-seven people attended the 5-day “NCBI Advanced Workshop for Bioinformatics Information Specialists (NAWBIS)” [45] since it was first offered in August 2002; 38 (81%) of the participants were library staff. Participation in these courses represents a significant investment by libraries and their staff in preparing for a role in bioinformatics services.
Based on the author's anecdotal experience, the actual application of the courses by library staff has varied depending on factors such as the range of an individual's other responsibilities, percent of time dedicated to bioinformatics, institutional support from library administrators, individual confidence in bioinformatics knowledge and skills, and risk taking.
Many of the participants attended the one-day introductory course primarily to increase their general awareness of molecular biology resources, in anticipation of future questions they might receive. Some participants in that course and the subsequent, longer courses began to provide selected services, such as developing and presenting introductory workshops on resources such as Entrez and BLAST [46], collaborating with university faculty on curriculum integrated instruction [47, 48], participating in curriculum development programs [49], and training other librarians [50–52]. Participants in the “NAWBIS” advanced course generally have a significant percentage of their time formally allocated to bioinformatics support and have established broader ranges of services. Subsequent papers in this symposium provide details on some of these programs.
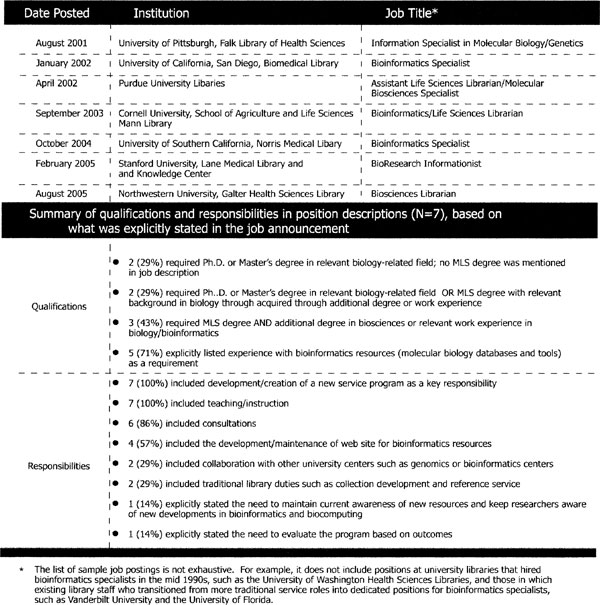
A final indicator of growing library involvement is the establishment of full-time, dedicated positions for bioinformatics. While specific job titles vary, they are broadly referred to here as “bioinformatics specialists.” An early leader in the establishment of a dedicated position was the University of Washington, Seattle, which hired a full time molecular biologist in 1995 [53]. In some libraries, a staff member who initially provided traditional library services was later transitioned into a dedicated position [54]. Figure 4 shows some additional examples of positions that have been posted between 2001 and 2005 and indicates the gradual, although continued, efforts of libraries to establish dedicated positions.†
Figure 4.
Examples of dedicated positions in libraries for bioinformatics specialists
RECRUITMENT, TRAINING, AND MANAGEMENT OF STAFF
Recruitment
To become involved in bioinformatics, a library must have staff with the appropriate expertise, skills, and desire to build new service programs. Libraries considering such programs face a number of choices. Should existing staff take on new roles or should new staff be hired? If new staff can be hired, whom to hire: librarian or scientist? What knowledge and skills are necessary? Should there be an individual specialist whose position is dedicated solely to bioinformatics services or teams whose members allocate a portion of their time to bioinformatics while also continuing to handle more traditional library roles? The choices that are possible in a given institution of course depend on available resources and management philosophies, and various models have been implemented.
Early leaders in providing library-based bioinformatics support, who formed the Molecular Biology and Genomics Special Interest Group of the Medical Library Association (MLA) [55] as a result of the 1994 RFI to MEDLIB-L, came from backgrounds including the humanities as well as science and often developed new services in their personal time, typically with little or no time officially dedicated to bioinformatics. The scope of their work was limited primarily by the amount of time they could dedicate to bioinformatics in relation to their other responsibilities.
Some of these leaders eventually transitioned into specialized positions for bioinformatics, expanding their services accordingly. Some specialists trained additional library staff who now work together to provide bioinformatics services through a tiered approach [50]. Other libraries have chosen a team approach that combines the various skills of staff, such as those with backgrounds in science, administration, and computer systems development [56].
Given the breadth of bioinformatics resources and the range and complexity of user questions, more libraries are recognizing the need to establish dedicated positions. Individuals who incorporate bioinformatics into their many job responsibilities still can and do provide valuable services. However, libraries that have implemented a robust program of in-depth services have generally done so with dedicated staff [50, 53, 54, 57–59]. This is also consistent with the philosophy behind the growing development of informationist positions in the library community [60, 61].
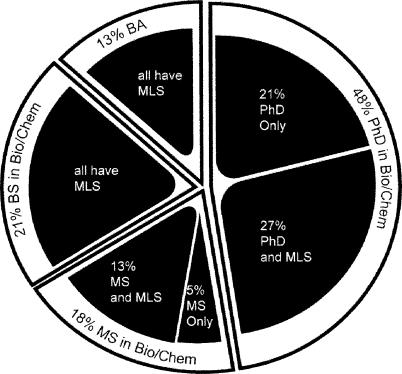
As the educational backgrounds of “NAWBIS” participants demonstrate (Figure 5), bioinformatics support can be provided by individuals with varied backgrounds. Regardless of how acquired, a knowledge of molecular biology, relevant lab techniques, bioinformatics resources, and the research culture are essential to understanding user questions and resources and to establishing credibility with clientele. But a strong background in science is not enough. Because of the need for end-user education, even a scientist with a doctoral degree must demonstrate excellent teaching and communication skills and a desire to teach and provide research support. Depending on the responsibilities of the position, it is generally essential for the specialist to also have strong skills in program planning and it might be desirable for the specialist to have Web development skills. Finally, as implied in the subsequent section on outreach, it is essential that the specialist have marketing skills to communicate the library services to clientele.
Figure 5.
Educational backgrounds of library staff who have attended the “NCBI Advanced Workshop for Bioinformatics Information Specialists”
While it can be challenging to find individuals with such combinations, a number of avenues can be followed to recruit potential candidates. Libraries interested primarily in candidates with graduate degrees in biosciences can post job announcements to journals such as Science, electronic mailing lists such as “Employment” in the BioSci newsgroups <http://www.bio.net>, and postdoctoral fellow or departmental mailing lists. Schools of library and information science or medical informatics can also consider some of these avenues to recruit biology students who would then become the rare and sought after dual-trained information specialists. A number of bench scientists sometimes seek different career avenues, including bioinformatics and education. Libraries interested in hiring individuals with a library degree and science background acquired through formal education or work experience may already have talented staff able to develop services, if placed in a dedicated position.
Training
Once library staff have been assigned or hired to establish bioinformatics support services, they must become familiar with a wide range of databases and retrieval and analytical tools. Staff who are expected to offer a limited, introductory set of services can begin by learning about a small set of comprehensive, commonly used databases, such as those in Entrez, and the corresponding search system features. They can also prepare for general questions about other resources such as sequence similarity search programs, genome browsers, and three-dimensional structures. An example of this type of introductory training is the NCBI course, “Introduction to Molecular Biology Information Resources” [44]. Staff in need of additional science background may need to consider an undergraduate biology course prior to learning about the bioinformatics resources.
Staff who fill dedicated positions will need to prepare for more intensive and technical questions that span a wider range of topics. They, too, may benefit from information covered in an introductory course to enhance their understanding of the relationships and nuances among the various databases and to learn about some of the advanced features in the search systems. In addition, they can benefit from an extended overview of bioinformatics topics and resources. Even individuals with doctorates in biological science are generally familiar with a small segment of the available bioinformatics resources, usually those directly relevant to their research. They will need to prepare for questions from clientele with varied research interests. The “NAWBIS” course mentioned above is one example of an extended resource overview [45].
Additional depth and breadth in knowledge and skills with these and other resources comes gradually over time from interactions with users. Those interested in also pursuing more in-depth training can attend additional technical workshops such as NCBI's “PowerTools” [62] and academic programs in bioinformatics to acquire skills such as programming and large-scale data mining. Finally, because new resources are constantly being introduced, considerable self-study and continuing education courses are necessary to keep pace with this rapidly growing and changing field.
Management
After bioinformatics support specialists are hired and trained, long-term success of the program depends in part on supportive management of the program and staff. Some management considerations include: acquiring resources to establish a new position; using hard versus soft money to fund the position; encouraging career growth, particularly if a scientist is hired; addressing cultural differences between library staff and scientists in the library; establishing a reporting structure that will facilitate the work of the bioinformatics specialist and grant them the degree of autonomy necessary to establish and implement programs based on their insight into their institution's unique needs; and adding more staff as needed. Many of these issues are addressed in Epstein's editorial [63].
ASSESSMENT OF USER NEEDS AND SKILLS IN A GIVEN COMMUNITY
Before implementing a new service program for bioinformatics, a formal assessment of user needs in a given institution is recommended. While Figures 2 and 3 provide a broad overview of user groups and needs, various tools have been developed to identify the needs of the particular clientele that a library will serve. Two examples are the survey by Yarfitz and Ketchell [53] and Appendix A of this paper.
While useful, such surveys have limitations. Users do not know what they do not know, so their responses can often represent perceived needs bounded by their current knowledge of what is available or possible. They cannot pose a question about a resource they do not know exists or an informatics analysis they do not know is possible.
Another option is to assess user skills with bioinformatics resources, rather than self-identified needs that are limited by a user's current knowledge. One way to do that is to present problem-based questions relevant to the users' area of study and assess their knowledge and skill in use of bioinformatics resources based on the approach they use to solve the problem. Services can then be designed that expand an individual's or group's existing skills.
Minie, for example, developed a brief pretest to assess the baseline skills of participants [58]. It presents a block of sequence data followed by five questions related to the identity and function of the sequence. A similar skills assessment (Appendix D, available only online) has been developed and administered to “NAWBIS” participants and could be adapted for use at their institutions. However, the development and implementation of this type of skills assessment requires expertise with the relevant subject and bioinformatics tools, so it is most feasible to do after a bioinformatics specialist has been hired. It is also more effectively administered in an instructional setting, such as in the context of a workshop, to tailor the lecture and training to the needed areas of emphasis.
GOAL SETTING IN THE ESTABLISHMENT OF NEW SERVICES
Before setting goals in the establishment or expansion of library-based services, it is helpful to identify current service providers on campus and determine which user groups and needs are already being met. Although most universities do not offer bioinformatics workshops, when they are offered, they can be found in such places as bioinformatics centers, information technology (IT) centers, and continuing education programs [1].
Even universities with bioinformatics centers do not always provide these services. Among those that do, it is generally not possible for them to address all the user groups and needs. In such cases, developing a collaborative, not competitive, relationship with the bioinformatics center or other service provider to develop complementary services will result in the best possible range of services to the university community.
For example, Figures 2 and 3 provide a model of the possible distribution of bioinformatics educational and end-user support services for research and clinical/ consumer health communities, respectively. Various borders are used to delineate user groups and needs that can potentially be served by the library and/or bioinformatics center, depending on the expertise of the staff in each unit and its mission or primary focus—service, research, or both. Many variants of the model are possible depending on an institution's organizational structure. For example, libraries could be natural collaborators with a university's computing or bioinformatics center to establish and manage DSpace-like programs [64], and library staff could and sometimes already do engage in research collaborations on data retrieval and analysis projects. Lyon et al. [65] provide examples of collaborations that have been established at four universities.
Partnerships between libraries and bioinformatics centers can also be beneficial to support staff as they refer end users to each other. This increases the visibility of the services each one provides and frees up staff time in each center, enabling them to focus on their primary mission.
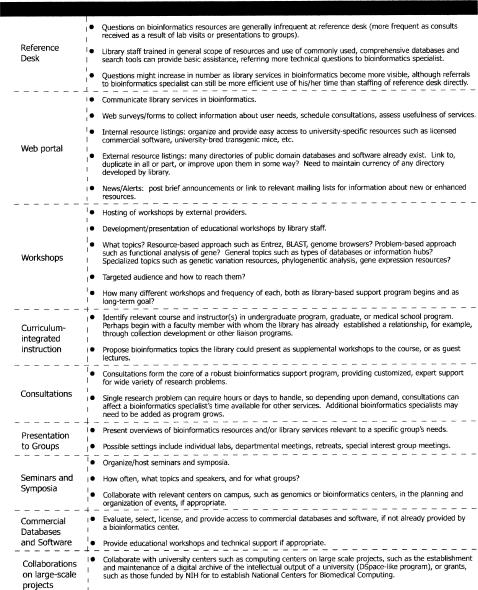
When a library has identified the user groups and needs that will initially be addressed, services can be implemented in a variety of ways. Figure 6 provides examples of support services to consider, ranging from the development of a Web portal and workshops to collaborations with other university centers on large scale projects, and summarizes some issues to consider in regard to each service. The planning process for support services can be guided by an intersection of information from Figures 2, 3, and 6; the results of local needs assessments; and dialog with other service providers on campus that can lead to a synergy of efforts.
Figure 6.
Bioinformatics support services to consider in establishing a library-based program
Finally, as new services are implemented, it is important to develop methods of gauging their success. Examples of feedback mechanisms include surveys, tracking of service use, comment or feedback links on a Web portal, formation of user groups, and open dialog among the bioinformatics specialists, users, and management to identify successes as well as challenges. In combination, these can be used to refine the existing services and justify the development of additional services.
OUTREACH: BRIDGING THE COMMUNICATION GAP BETWEEN THE LIBRARY AND RESEARCH COMMUNITY
The development of services, unfortunately, does not guarantee their use. A communication gap currently exists between end users of bioinformatics resources and libraries with staff that can help them. Users often do not think of the library as a place to receive assistance with these resources. In addition, users are unlikely to be familiar with the full extent of resources and tend to frame their questions only in terms of familiar resources. Conversely, many library staff think that users of bioinformatics resources are already highly skilled with those tools and do not need help, or the staff are not yet aware of the specific types of help various users might need.
The 2001 AAHSL RFI mentioned above provided some evidence of this communication gap. While fourteen (39%) of the institutions responding to the RFI said they provide user services support for nonbibliographic molecular biology databases and tools, only two (6%) of institutions responding to the survey said they handled an average of at least five user questions per week on molecular biology databases and tools (excluding bibliographic search questions). Ten (28%) of the institutions said they had no plans to begin or increase molecular biology support services. Several specifically explained that they have not been asked for such support by their clients and the faculty did not see the library as a resource for nonbibliographic databases.
However, a number of libraries have developed popular and successful bioinformatics end-user support programs. One fundamental element of their success has been the implementation of active outreach programs to assess user needs and communicate relevant library services [54, 57–59]. In each case, contact with the research and clinical communities predictably increased the library's visibility and credibility in the area of bioinformatics support.
SUMMARY AND CONCLUSIONS
The fundamental role of bioinformatics resources in biological research and medicine brings both opportunities and challenges. The resources provide powerful ways to establish connections between biological data and bring new insights to basic research and clinical medicine. At the same time, the rapid growth in the number and complexity of bioinformatics resources seems to outpace most researchers' ability to keep up with the fast-moving field. Because most researchers primarily focus on laboratory experiments, they have limited time for exploration of bioinformatics resources. So they necessarily use the resources they know, get what they (think they) need, and move on. In the process, they might be unaware of advanced techniques that could help them to more effectively retrieve or analyze their data or they might be missing other, more appropriate resources for their information needs. If, for example, users seldom employ features in a commonly used search system such as Entrez, it is reasonable to assume their overall knowledge of the plethora of bioinformatics resources, some of them quite complicated, is more limited.
To make optimal use of these tools, users can benefit from outside assistance in increasing their awareness of the resources that are available, learning how to use them effectively, and learning how to view the data critically and use them judiciously. Based on the relative scarcity of educational and end-user support services among most universities [1], libraries definitely have opportunities to take the lead in bioinformatics support.
This paper and others in the symposium focus on bioinformatics educational and end-user support services that are available to the general university community who might need to use bioinformatics resources as part of their work, rather than to students and researchers specializing in the study of bioinformatics as a discipline or who are being trained as bioinformatics researchers [66]. The latter will be primarily served by relevant academic programs.
Additionally, as each academic setting is unique, this paper does not attempt to provide a prescribed approach for libraries considering involvement in bioinformatics but hopefully provides a process for building a program that is appropriate for an individual institution. Such programs may be very diverse in their implementation. Nevertheless, additional library involvement in bioinformatics education and end-user support can undoubtedly lead to the more effective use of biological data in the scientific discovery process.
Acknowledgments
The author gratefully acknowledges Barbara Rapp, for leading the NLM Associate project in 1994 that provided a foundation for the work described here; Mark Frisse, Bob Engeszer, and the staff of the Bernard Becker Medical Library at Washington University School of Medicine for providing early experiences that demonstrated the role libraries could play in bioinformatics; and Dick West, posthumously, for his mentorship. The author is also grateful to Robert Yates, for his graphics work; Nancy Roderer, AHIP, and Jayne Campbell of the Welch Medical Library at Johns Hopkins University for posting and summarizing the 2001 RFI to AAHSL; David Osterbur and Diane Rein for their editorial assistance; and Dennis Benson, Lewis Geer, and Betsy Humphreys for many helpful discussions and their ongoing support of this work.
APPENDIX A
Assessing user needs
Sample survey tool developed for adaptation and use by participants in the “National Center for Biotechnology Information (NCBI) Advanced Workshop for Bioinformatics Information Specialists (NAWBIS)” <http://www.ncbi.nlm.nih.gov/Class/NAWBIS/Modules/OutreachCommunication/user_needs_survey.html>: Designed to initiate communication with labs and other groups in your institution to find out what type of bioinformatics help they might need and to let them know that help is available. Edit, format, or distribute the survey in any way that is optimal for you.
Bioinformatics Support Program: “Mining Molecular Databases”
Bioinformatics specialist: Cameron Smith, Hart University Library, Bldg. 41; telephone: 987.123.4567; csmith@hart.edu.
To provide our university with an effective bioinformatics support program that meets your needs, please complete the brief survey below and return by email to csmith@hart.edu (or by mail to the address above). All of the information will be kept confidential and will be used only to tailor our services to your lab's needs.
If you would like a lab demo of any of the resource categories and/or a consultation, please indicate that below. Please note that our support program is designed to assist faculty, staff, and students with the selection and use of a wide range of molecular databases and retrieval and analysis software. We do not currently provide support with management of laboratory data or programming needs.
About you
 |
Resources of interest to you
Please circle your frequency of use for each category of resources below, and indicate whether you would like a lab demo on the resources in any category. If numerous labs request demos, it might be necessary to offer a brief course (1-4 hours, depending on topic) in the library's computer lab to accommodate all the interested staff.
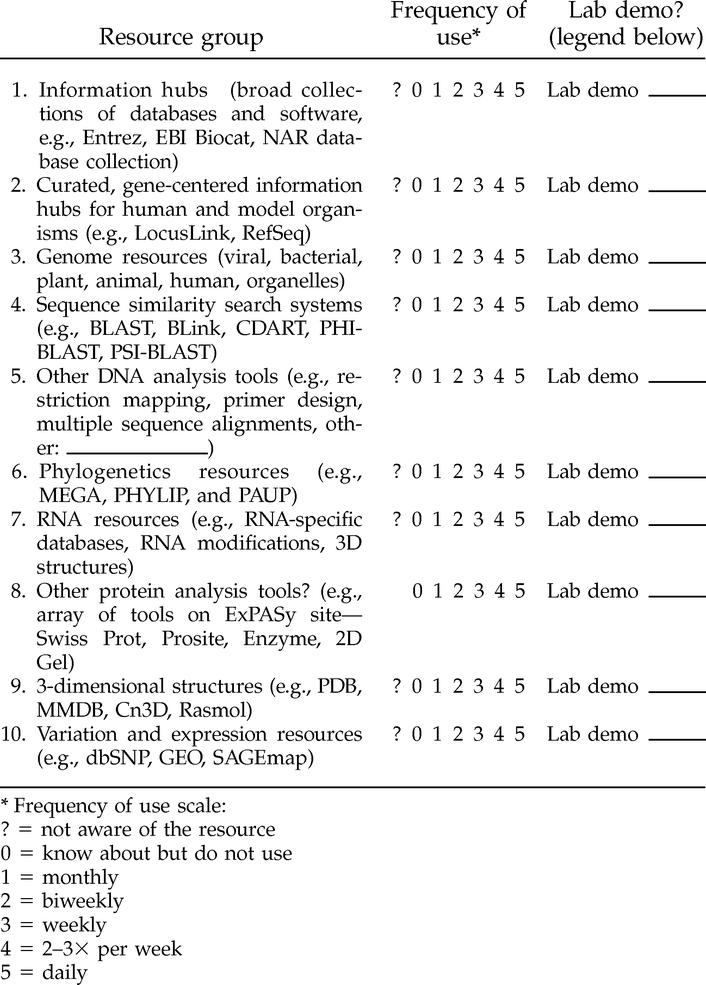 |
Lab visit
We provide consultations to individual labs on the selection and use of various resources specific to your research problems. If you are interested in this service, please summarize the problem with which you need assistance. We will call you to arrange a consultation in your lab or our office, as you prefer. Problem summary: _______________________________________________________________
APPENDIX B
Questionnaire posted on August 5, 1994, to MEDLIB-L about molecular biology services in medical libraries
Molecular biology researchers are creating and using a new array of information resources from which many of us are currently removed. In the near future, it would perhaps be helpful to form a Molecular Biology Special Interest Group in MLA. We could then assist each other in the use of these important resources and in identifying the types of services we could provide to students, researchers, and clinicians in need of molecular biology information.
The purpose of this questionnaire is to determine:
the current familiarity of medical librarians with molecular biology information resources
how many medical libraries provide molecular biology information services
what types of services are being provided
Your completion of this questionnaire is greatly appreciated. There are twenty brief questions. Even if your library currently does not provide molecular biology information services or you are not aware of the services and databases discussed, that is important to know as well.
Please send your responses directly to Renata McCarthy <renata@ncbi.nlm.nih.gov>. Results will be summarized and posted to the list. Thank you for your valuable input.
Institution: __________________
City, state: __________________
Country, if not US: __________________
Molecular biology resources
Questions 1–9: Please enter the letter a, b, c, or d, from the scale below, that best represents your level of familiarity with each database or software tool listed.
Scale:
a. = not familiar
b. = familiar with scope or content but do not use
c. = use occasionally (few times per year)
d. = use regularly (monthly or more often)
GenBank ____________
PIR v ____________
SwissProt ____________
GDB ____________
OMIM ____________
Entrez (Which version? CD-ROM ____________ client/server ____________)
Retrieve email server ____________
BLAST email server ____________
Others? (Please list any other resources and your level of familiarity with them): ____________
Services provided
Questions 10–16: Please place an X beside the molecular biology information services your library provides. If your library provides services 10 through 12, for example, but not the others, questions 13 through 16 will be left blank.
Search services
10. a. Search bibliographic databases for articles related to molecular biology. ____________
10. b. Which databases are used most frequently? ____________
11. a. Search sequence databases by text word (searching header/descriptive information). ____________
11. b. Which databases are used most frequently? ____________
12. a. Conduct similarity searches of DNA or protein sequences (sequence analysis). ____________
12. b. Which databases are used most frequently? ____________
12. c. Which software or sequence analysis algorithm do you use most frequently? ____________
End-user instruction
13. a. For searching MEDLINE and/or other bibliographic databases (instruction provided to molecular biology, genetics, or related departments). ____________
13. b. Comments? ____________
14. a. For searching sequence databases. ____________
14. b. Comments? ____________
15. a. For use of molecular biology-related software such as Entrez, BLAST, GCG, Intelligenetics, etc. ____________
15. b. Comments? ____________
16. a. Assistance with sequence submission. ____________
16. b. Comments? ____________
End-user workstations
17. a. Are there any end-user workstations in your library that provide local or remote access to sequence databases? Yes ____________ No ____________
17. b. If yes, how many workstations? ____________
17. c. What software is used? ____________
Staff and training
18. a. How many reference librarians are there in your library? ____________
18. b. Do you have a specific reference librarian(s) who provides molecular biology info services? Yes ____________ No ____________
18. c. If yes, how many? ____________
19. If the answer to 18b is yes, how did that person(s) receive her/his training? (e.g., self-trained by experimenting with systems, read documentation, attended workshops). ____________
Special interest group
20. Would you be interested in joining a special interest group (SIG) for molecular biology information services if one was formed? Its purpose would be to share information about molecular biology resources, skills in their use, and strategies for expanding library services to include them. Yes ____________ No ____________
If yes, please provide your name, postal address, phone, and Internet address below:
Name: ________________________
Address: ________________________
Telehone: ________________________
Internet: ________________________
Thank you again for your time and input. Please send responses directly to Renata McCarthy at renata@ncbi.nlm.nih.gov.
APPENDIX C
Questionnaire posted on May 8, 2001, to the Association of Academic Health Sciences Libraries (AAHSL) email discussion list about molecular biology services in medical libraries
Colleagues,
Earlier this year staff from several Association of Academic Health Sciences Libraries (AAHSL) libraries convened at the National Center for Biotechnology Information (NCBI) to learn about the databases and services that NCBI offers to support molecular biologists. A next step is to determine what information services academic health sciences libraries collectively provide to the molecular biology community. Your assistance in this effort is greatly appreciated. The results will be shared with NCBI and, of course, distributed to all of you.
By May 14, please send your responses to Jayne M. Campbell at [email address].
–Nancy Roderer.
Name: ________________________
Institution: ________________________
-
Does your library provide user services support for nonbibliographic molecular biology databases and tools? Yes ____________ No ____________
If yes, please continue. If no, skip to # 6.
Does your library handle an average of at least five user questions per week on molecular biology databases and tools (excluding bibliographic search questions)? Yes ____________ No ____________
Does your library provide training to users in molecular biology databases and tools? Yes ____________ No ____________
-
Has your library developed training materials for molecular biology information resources? Yes ____________ No ____________
If yes, what audiences are they intended to reach? what content/databases do they cover? what format are they? ____________
-
Does your library have staff whose time is officially dedicated to these services? Yes ____________ No ____________
If yes, how many staff and what percent of their time is allocated to molecular biology services? ____________
What plans, if any, does your library have to begin or increase molecular biology user support services? ____________
-
If off-site training courses were offered in molecular biology databases and tools, would you send a staff member if the course took
3 days? Yes ____________ No ____________
5 days? Yes ____________ No ____________
10 days? Yes ____________ No ____________
If an off-site training course were offered to your staff what, if any, times during the academic year would be most favorable? ____________
-
Does your library have an active collaboration with a bioinformatics center in your institution? Yes ____________ No ____________
If yes, please describe. ____________
APPENDIX D
Assessing user skills: pretest given to participants of the “National Center for Biotechnology Information (NCBI) Advanced Workshop for Bioinformatics Information Specialists (NAWBIS)” 2005 course
Gauge your current familiarity with National Center for Biotechnology Information (NCBI) resources …an informal pretest <http://www.ncbi.nlm.nih.gov/Class/NAWBIS/Modules/InfoHubs/infohubs3a.html>. Below is a sample human sequence, followed by twelve questions about it, to gauge your current familiarity with some of NCBI's bioinformatics resources. Just answer the questions you can and do not worry about the others. By the end of the module, you should be able to answer all of the questions in ten to fifteen minutes or less. The questions apply some key concepts and skills covered in the three-day introductory course, which are briefly summarized in this information hubs module.
>gi|904118 Sample human sequence for NAWBIS/InfoHubs module
TGGGACAGGCAGCTCCGGGGTCCGCGGTTTCACATCGGAAACAAAACAGCGGCTGGTCTGGAAGGAACCT
GAGCTACGAGCCGCGGCGGCAGCGGGGCGGCGGGGAAGCGTATACCTAATCTGGGAGCCTGCAAGTGACA
ACAGCCTTTGCGGTCCTTAGACAGCTTGGCCTGGAGGAGAACACATGAAAGAAAGAACCTCAAGAGGCTT
TGTTTTCTGTGAAACAGTATTTCTATACAGTTGCTCCAATGACAGAGTTACCTGCACCGTTGTCCTACTT
CCAGAATGCACAGATGTCTGAGGACAACCACCTGAGCAATACTGTACGTAGCCAGAATGACAATAGAGAA
CGGCAGGAGCACAACGACAGACGGAGCCTTGGCCACCCTGAGCCATTATCTAATGGACGACCCCAGGGTA
ACTCCCGGCAGGTGGTGGAGCAAGATGAGGAAGAAGATGAGGAGCTGACATTGAAATATGGCGCCAAGCA
TGTGATCATGCTCTTTGTCCCTGTGACTCTCTGCATGGTGGTGGTCGTGGCTACCATTAAGTCAGTCAGC
TTTTATACCCGGAAGGATGGGCAGCTAATCTATACCCCATTCACAGAAGATACCGAGACTGTGGGCCAGA
GAGCCCTGCACTCAATTCTGAATGCTGCCATCATGATCAGTGTCATTGTTGTCATGACTATCCTCCTGGT
GGTTCTGTATAAATACAGGTGCTATAAGGTCATCCATGCCTGGCTTATTATATCATCTCTATTGTTGCTG
TTCTTTTTTTCATTCATTTACTTGGGGGAAGTGTTTAAAACCTATAACGTTGCTGTGGACTACATTACTG
TTGCACTCCTGATCTGGAATTTTGGTGTGGTGGGAATGATTTCCATTCACTGGAAAGGTCCACTTCGACT
CCAGCAGGCATATCTCATTATGATTAGTGCCCTCATGGCCCTGGTGTTTATCAAGTACCTCCCTGAATGG
ACTGCGTGGCTCATCTTGGCTGTGATTTCAGTATATGATTTAGTGGCTGTTTTGTGTCCGAAAGGTCCAC
TTCGTATGCTGGTTGAAACAGCTCAGGAGAGAAATGAAACGCTTTTTCCAGCTCTCATTTACTCCTCAAC
AATGGTGTGGTTGGTGAATATGGCAGAAGGAGACCCGGAAGCTCAAAGGAGAGTATCCAAAAATTCCAAG
TATAATGCAGAAAGCACAGAAAGGGAGTCACAAGACACTGTTGCAGAGAATGATGATGGCGGGTTCAGTG
AGGAATGGGAAGCCCAGAGGGACAGTCATCTAGGGCCTCATCGCTCTACACCTGAGTCACGAGCTGCTGT
CCAGGAACTTTCCAGCAGTATCCTCGCTGGTGAAGACCCAGAGGAAAGGGGAGTAAAACTTGGATTGGGA
GATTTCATTTTCTACAGTGTTCTGGTTGGTAAAGCCTCAGCAACAGCCAGTGGAGACTGGAACACAACCA
TAGCCTGTTTCGTAGCCATATTAATTGGTTTGTGCCTTACATTATTACTCCTTGCCATTTTCAAGAAAGC
ATTGCCAGCTCTTCCAATCTCCATCACCTTTGGGCTTGTTTTCTACTTTGCCACAGATTATCTTGTACAG
CCTTTTATGGACCAATTAGCATTCCATCAATTTTATATCTAGCATATTTGCGGTTAGAATCCCATGGATG
TTTCTTCTTTGACTATAACCAAATCTGGGGAGGACAAAGGTGATTTTCCTGTGTCCACATCTAACAAAGT
CAAGATTCCCGGCTGGACTTTTGCAGCTTCCTTCCAAGTCTTCCTGACCACCTTGCACTATTGGACTTTG
GAAGGAGGTGCCTATAGAAAACGATTTTGAACATACTTCATCGCAGTGGACTGTGTCCCTCGGTGCAGAA
ACTACCAGATTTGAGGGACGAGGTCAAGGAGATATGATAGGCCCGGAAGTTGCTGTGCCCCATCAGCAGC
TTGACGCGTGGTCACAGGACGATTTCACTGACACTGCGAACTCTCAGGACTACCGGTTACCAAGAGGTTA
GGTGAAGTGGTTTAAACCAAACGGAACTCTTCATCTTAAACTACACGTTGAAAATCAACCCAATAATTCT
GTATTAACTGAATTCTGAACTTTTCAGGAGGTACTGTGAGGAAGAGCAGGCACCAGCAGCAGAATGGGGA
ATGGAGAGGTGGGCAGGGGTTCCAGCTTCCCTTTGATTTTTTGCTGCAGACTCATCCTTTTTAAATGAGA
CTTGTTTTCCCCTCTCTTTGAGTCAAGTCAAATATGTAGATTGCCTTTGGCAATTCTTCTTCTCAAGCAC
TGACACTCATTACCGTCTGTGATTGCCATTTCTTCCCAAGGCCAGTCTGAACCTGAGGTTGCTTTATCCT
AAAAGTTTTAACCTCAGGTTCCAAATTCAGTAAATTTTGGAAACAGTACAGCTATTTCTCATCAATTCTC
TATCATGTTGAAGTCAAATTTGGATTTTCCACCAAATTCTGAATTTGTAGACATACTTGTACGCTCACTT
GCCCCCAGATGCCTCCTCTGTCCTCATTCTTCTCTCCCACACAAGCAGTCTTTTTCTACAGCCAGTAAGG
CAGCTCTGTCRTGGTAGCAGATGGTCCCATTATTCTAGGGTCTTACTCTTTGTATGATGAAAAGAATGTG
TTATGAATCGGTGCTGTCAGCCCTGCTGTCAGACCTTCTTCCACAGCAAATGAGATGTATGCCCAAAGCG
GTAGAATTAAAGAAGAGTAAAATGGCTGTTGAAGC
Questions
This is the sequence data from what human gene?
Did the data come from a primary (archival) or derivative (curated) database? How can you tell?
If it is archival, how can you find a curated mRNA record for this human gene, or vice versa?
What is the official gene symbol? By what other gene symbols has it been known?
What is the location of this gene on a cytogenetic map, and what is its base pair location on the assembled human genome sequence?
How many transcript variants is it known to have?
On what mouse and rat chromosomes are the homologs found?
What phenotypes are associated with this gene?
How many allelic variants are documented in OMIM for this gene?
Name a clinical laboratory in the United States that offers genetic testing for one of the phenotypes.
Does NCBI offer a software tool for making a restriction map of the query sequence? How can you find out?
If a user has the genomic DNA for the gene and wants to identify putative transcription factor binding sites, what are some databases and/or software tools that could potentially be useful? How or where did you find these?
Footnotes
This article has been approved for the Medical Library Association's Independent Reading Program.
* Based in part on a presentation titled, “Medical Information in the Genomic Era: Beyond Literature” given at the Swedish Conference for Medical Librarians; Stockholm, Sweden; November 6, 2000; and at the inaugural meeting of the National Center for Biotechnology Information Educational Collaborators, “Educollab,” at the National Library of Medicine on January 12, 2001.
† An archival set of the position descriptions is available by request from the author.
REFERENCES
- Messersmith DJ, Benson DA, and Geer RC. A Web-based assessment of bioinformatics end-user support services at US universities. J Med Libr Assoc. 2006 Jul; 94(3):299–305. E-156–88. [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, and Wheeler Dl. GenBank. Nucleic Acids Res. 2005 Jan 1; 33database issue. D34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenBank sequence database, release 149.0: distribution release notes. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2005. [rev. 15 Aug 2005; cited 15 Sep 2005]. <ftp://ftp.ncbi.nih.gov/genbank/release.notes/gb149.release.notes>. [Google Scholar]
- Sprinzl M, Gauss DH.. Compilation of tRNA sequences. Nucleic Acids Res. 1984;(12 Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Gauss DH.. Compilation of sequences of tRNA genes. Nucleic Acids Res. 1984;(12 Suppl):r59–131. doi: 10.1093/nar/12.suppl.r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann VA, Wolters J, Huysmans E, Vandenberghe A, De Wachter R.. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1984;(12 suppl)::r133–66. doi: 10.1093/nar/12.suppl.r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ.. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 Suppl:r167–204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. The molecular biology database collection: 2005 update. Nucleic Acids Res. 2005 Jan; 33(Database Issue:):D5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Web server issue [editorial] Nucleic Acids Res. 2003;31(13):3289. [Google Scholar]
- Web server issue [editorial] Nucleic Acids Res. 2005;33(Web server issue):W1. doi: 10.1093/nar/gkq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct; 215(3):403–10. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 Sep; 25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twenty years of citation superstars. Science Watch 2003; 14(5). [Google Scholar]
- Top 10 papers published. The Scientist. 2005;19(20):26. [Google Scholar]
- Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, and Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov; 63(4):851–9. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Searching for drug targets in microbial genomes. Curr Opin Biotechnol. 1999 Dec; 10(6:):571–8. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Greenhut SF, Grouse LH, Schaefer CF, and Buetow KH. In silico analysis of cancer through the Cancer Genome Anatomy Project. Trends Cell Biol. 2001 Nov; 11(11):S66–71. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE. Collins FS: Genomic medicine—a primer. N Engl J Med. 2002 Nov; 347(19):1512–20. [DOI] [PubMed] [Google Scholar]
- Gibbs R. Deeper into the genome. Nature. 2005 Oct; 437(7063):1233–4. [DOI] [PubMed] [Google Scholar]
- Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, Beahler C, Bird TD, Popovich B, Nesbitt C, Dolan C, Marymee K, Hanson NB, Neufeld-Kaiser W, McCullough Grohs G, Kicklighter T, Abair C, Malmin A, Barclay M, and Dharani Palepu R. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002 May; 19(5):501–9. [DOI] [PubMed] [Google Scholar]
- Pagon RA. Molecular genetic testing for inherited disorders. Expert Rev Mol Diagn. 2004 Mar; 4(2):135–40. [DOI] [PubMed] [Google Scholar]
- Thomas FJ, McLeod HL, Watters JW.. Pharmacogenomics: the influence of genomic variation on drug response. Curr Top Med Chem. 2004;4(13):1399–409. doi: 10.2174/1568026043387638. [DOI] [PubMed] [Google Scholar]
- Thorn CF, Klein TE, Altman RB.. PharmGKB. the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol. 2005;311:179–91. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]
- MacMullen WJ, Denn SO. Information problems in molecular biology and bioinformatics. J Am Soc Inf Sci Tech. 2005 Mar; 56(5):447–56. [Google Scholar]
- Bartlett JC, Toms EG. Developing a protocol for bioinformatics analysis: an integrated information behavior and task analysis approach. J Am Soc Inf Sci Tech. 2005 Mar; 56(5:):469–82. [Google Scholar]
- Stevens R, Goble C, Baker P, and Brass A. A classification of tasks in bioinformatics. Bioinformatics. 2001 Feb; 17(2):180–8. [DOI] [PubMed] [Google Scholar]
- Baker PG, Goble CA, Bechhofer S, Paton NW, Stevens R, and Brass A. An ontology for bioinformatics applications. Bioinformatics. 1999 Jun; 15(6):510–20. [DOI] [PubMed] [Google Scholar]
- Ohles JA. Clinical resources. [Web document]. 2002. [rev. 31 Jul 2005; cited 15 Sep 2005]. <http://www.ncbi.nlm.nih.gov/Class/NAWBIS/Modules/ClinicalResources/clin1.html>. [Google Scholar]
- Campbell AM. Public access for teaching genomics, proteomics, and bioinformatics. Cell Biol Educ. 2003 Summer; 2(2):98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A field guide to NCBI resources. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information. [rev. 10 Aug 2005; cited 4 Oct 2005]. <http://www.ncbi.nlm.nih.gov/Class/FieldGuide/>. [Google Scholar]
- A decade of data at NCBI: integrated approaches to managing the information explosion. NCBI News. 1999;Summer::1–4. [Google Scholar]
- Maglott DR, Katz KS, Sicotte H, and Pruitt KD. NCBI's LocusLink and RefSeq. Nucleic Acids Res. 2000 Jan; 28(1):126–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, and Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000 Jan; 28(1):33–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostell J. Chapter 14: the Entrez search and retrieval system. In: The NCBI handbook. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2002. [rev. 13 Aug 2003; cited 15 Sep 2005]. <http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=handbook.chapter.588>. [Google Scholar]
- Paranthaman P. Entrez query statistics for nucleotide, protein, gene. Email message to: Renata Geer. 2005 Sep 28, 12:15 p.m. [Google Scholar]
- Geer RC, Sayers EW. Entrez: making use of its power. Brief Bioinform. 2003 Jun; 4(2):179–84. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, and Maglott DR. NCBI Reference Sequence(RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005 Jan; 33(database issue):D501–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV.. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1(1):55–67. [PubMed] [Google Scholar]
- Minie ME. Introduction: expression resources. [Web document]. 2003. [rev. 25 Jul 2005; cited 15 Sep 2005]. <http://www.ncbi.nlm.nih.gov/Class/NAWBIS/Modules/Expression/exp1.html>. [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002 Dec; 32(suppl):490–5. [DOI] [PubMed] [Google Scholar]
- NCBI at a glance: our mission. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2005. [rev. 21 May 2004; cited 11 Oct 2005]. <http://www.ncbi.nlm.nih.gov/About/glance/ourmission.html>. [Google Scholar]
- NCBI at a glance: programs and activities. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2005. [rev. 21 May 2004; cited 11 Oct 2005]. <http://www.ncbi.nlm.nih.gov/About/glance/programs.html>. [Google Scholar]
- Geer RC. MLA CE course manual: molecular biology information resources. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 1999. [rev. 12 Nov 2002; cited 5 Oct 2005]. <http://www.ncbi.nlm.nih.gov/Class/MLACourse/Original8Hour/>. [Google Scholar]
- Geer RC, Messersmith DJ. Introduction to molecular biology resources. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2002. [rev. 12 Jul 2005; cited 5 Oct 2005]. <http://www.ncbi.nlm.nih.gov/Class/MLACourse/>. [Google Scholar]
- Geer RC, Messersmith DJ, Alpi K, Bhagwat M, Chattopadhyay A, Gaedeke N, Lyon J, Minie ME, Morris RC, Ohles JA, Osterbur DL, and Tennant MR. NCBI advanced workshop for bioinformatics information specialists. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2002. [rev. 16 Aug 2005; cited 6 Oct 2005]. <http://www.ncbi.nlm.nih.gov/Class/NAWBIS/>. [Google Scholar]
- Alpi KM, Hendler GY, and Ohles JA. Making sense of molecular medicine: New York librarians' involvement with bioinformatics. [Web document]. 2001. [cited 6 Oct 2005]. <http://library.med.cornell.edu/Library/nahsl/molbioposter.pdf>. [Google Scholar]
- Delwiche FA. Introduction to resources in molecular genetics. Med Ref Serv Q. 2001 Summer; 20(2):33–50. [DOI] [PubMed] [Google Scholar]
- Tennant MR, Miyamoto MM. The role of medical libraries in undergraduate education: a case study in genetics. J Med Libr Assoc. 2002 Apr; 90(2):181–93. [PMC free article] [PubMed] [Google Scholar]
- Osterbur DL, Alpi K, Canevari C, Corley PM, Devare M, Gaedeke N, Jacobs DK, Kirlew P, Ohles JA, Vaughan KTL, Wang L, Wu Y, and Geer RC. Vignettes: diverse library staff offering diverse bioinformatics services. J Med Libr Assoc. 2006 Jul; 94(3):306– E-188–91. [PMC free article] [PubMed] [Google Scholar]
- Lyon J. Beyond the literature: bioinformatics training for medical librarians. Med Ref Serv Q. 2003 Spring; 22(1):67–74. [DOI] [PubMed] [Google Scholar]
- Alpi K. Bioinformatics training by librarians and for librarians: developing the skills needed to support molecular biology and clinical genetics information instruction. Issues Sci Tech Lib 2003 Spring;37. [Google Scholar]
- Lyon J, Giuse NB, Williams A, Koonce T, and Walden R. A model for training the new bioinformationist. J Med Libr Assoc. 2004 Apr; 92(2):188–95. [PMC free article] [PubMed] [Google Scholar]
- Yarfitz S, Ketchell DS. A library-based bioinformatics services program. Bull Med Libr Assoc. 2000 Jan; 88(1):36–48. [PMC free article] [PubMed] [Google Scholar]
- Tennant MR.. Bioinformatics librarian: meeting the information needs of genetics and bioinformatics researchers. Ref Serv Rev. 2005;33(1):12–9. [Google Scholar]
- Geer RC. Molecular biology and genomics SIG of the Medical Library Association. [Web document]. 1998. [rev. 9 Aug 2005; cited 12 Oct 2005]. <http://medicine.wustl.edu/∼molbio/>. [Google Scholar]
- MacMullen WJ, Vaughan KTL, and Moore ME. Planning bioinformatics education and information services in an academic health sciences library. Coll Res Libr. 2004 Jul; 65(4:):320–33. [Google Scholar]
- Chattopadhyay A, Tannery NH, Silverman DAL, Bergen P, and Epstein BA. Design and implementation of a library-based information service program in molecular biology and genetics at the University of Pittsburgh. J Med Libr Assoc. 2006 Jul; 94(3):307–13. E-192. [PMC free article] [PubMed] [Google Scholar]
- Minie M, Bowers S, Tarczy-Hornoch P, Roberts E, James RA, Rambo N, and Fuller S. The University of Washington Health Sciences Library BioCommons: an evolving northwest biomedical research information support infrastructure. J Med Libr Assoc. 2006 Jul; 94(3):321–9. [PMC free article] [PubMed] [Google Scholar]
- Rein DC. Developing library bioinformatics services in context: the Purdue University Libraries bioinformationist program. J Med Libr Assoc. 2006 Jul; 94(3):314–20. E-193–7. [PMC free article] [PubMed] [Google Scholar]
- Davidoff F, Florance V. The informationist: a new health profession? Ann Intern Med. 2000 Jun; 132(12):996–8. [DOI] [PubMed] [Google Scholar]
- Florance V, Giuse NB, and Ketchell DS. Information in context: integrating information specialists into practice settings. J Med Libr Assoc. 2002 Jan; 90(1):49–58. [PMC free article] [PubMed] [Google Scholar]
- PowerTools NCBI technical workshop series. [Web document]. Bethesda, MD: US National Library of Medicine, National Center for Biotechnology Information, 2004. [rev. 2 Feb 2005; cited 13 Oct 2005]. <http://www.ncbi.nlm.nih.gov/Class/PowerTools/>. [Google Scholar]
- Epstein BA. A management case study: challenges of initiating an information service in molecular biology and genetics [editorial]. J Med Libr Assoc. 2006 Jul; 94(3):245–7. [PMC free article] [PubMed] [Google Scholar]
- Smith M, Barton M, Branschofsky M, McClellan G, Walker JH, Bass M, Stuve D, and Tansley R. DSpace: an open source dynamic digital repository. D-Lib Magazine 2003; 9(1). [Google Scholar]
- Lyon JA, Tennant MR, Messner KR, and Osterbur DL. Carving a niche: establishing bioinformatics collaborations. J Med Libr Assoc. 2006 Jul; 94(3):330–5. [PMC free article] [PubMed] [Google Scholar]
- Pevzner PA. Educating biologists in the 21st century: bioinformatics scientists versus bioinformatics technicians. Bioinformatics. 2004 Sep; 20(14):2159–61. [DOI] [PubMed] [Google Scholar]