Abstract
Objectives: The paper describes collaborations and partnerships developed between library bioinformatics programs and other bioinformatics-related units at four academic institutions.
Methods: A call for information on bioinformatics partnerships was made via email to librarians who have participated in the National Center for Biotechnology Information's Advanced Workshop for Bioinformatics Information Specialists. Librarians from Harvard University, the University of Florida, the University of Minnesota, and Vanderbilt University responded and expressed willingness to contribute information on their institutions, programs, services, and collaborating partners. Similarities and differences in programs and collaborations were identified.
Results: The four librarians have developed partnerships with other units on their campuses that can be categorized into the following areas: knowledge management, instruction, and electronic resource support. All primarily support freely accessible electronic resources, while other campus units deal with fee-based ones. These demarcations are apparent in resource provision as well as in subsequent support and instruction.
Conclusions and Recommendations: Through environmental scanning and networking with colleagues, librarians who provide bioinformatics support can develop fruitful collaborations. Visibility is key to building collaborations, as is broad-based thinking in terms of potential partners.
Highlights
This paper discusses four cases of bioinformatics collaborations between libraries and nonlibrary bioinformatics support groups.
The authors compare collaborative efforts in very different institutional settings and identify commonalities in areas of knowledge management, instruction, and electronic resource support.
Implications for practice
Librarians are uniquely situated to develop bioinformatics collaboration networks and provide users with assistance in finding the right provider.
In these four cases, librarians have focused on supporting free electronic resources rather than supporting commercial, fee-based resources.
Knowledge of the institution, visibility, and creativity in choosing partners are important to the development of collaborations.
INTRODUCTION
Because of its broad scope and relative newness, the field of bioinformatics attracts individuals from a wide range of backgrounds and has engendered the development of bioinformatics-related support services in multiple pockets of biomedical institutions, including the medical library. This evolution of service providers reflects the interdisciplinary nature of the field, the large amount of data it generates, and the complexity of organizations involved. Bioinformatics clients and service providers come from many subpopulations (faculty, postdoctoral associates and students, clinicians and researchers, patients, and others), adding another dimension of complexity that creates both challenges and opportunities for medical librarians as they seek to position themselves as vital information providers, while building collaborations with other interested units of the institution.
To date, several accounts have been published of the early development of library-based bioinformatics programs [1–9]. Most of these initial efforts focused on creating a direct service relationship between librarians and end users, including biotechnologists, molecular biology researchers, undergraduate and graduate students, academic faculty, and even other medical librarians. These efforts assessed the clients' needs, detailed the types of services provided, and evaluated success for the particular client group; most focused on training, such as the development of workshops, courses, or other educational media for bioinformatics resources. Other areas of interest include collection development and resource access, particularly for electronic resources, and expansion of reference consultations to include more specialized types of queries beyond literature searching.
Bell proposes that academic librarians in bioinformatics assume roles as “scouts, instructors, and advocates” [10] and be knowledgeable about not only the resources available in the electronic world at-large, but also intra-institutional electronic resources and expertise. The ability of the librarian to make internal connections in the organization is vital.
Little published discussion has addressed library collaboration with other bioinformatics support units on campus or identified the most efficient demarcation of responsibilities between libraries and those other units. To explore such issues, two investigators (Lyon and Tennant) solicited feedback via email from information specialists who had participated in the “National Center for Biotechnology Information Advanced Workshop for Bioinformatics Information Specialists” <http://www.ncbi.nlm.nih.gov/Class/NAWBIS/>. Librarians from Harvard University and the University of Minnesota self-identified as having developed such collaborations and were subsequently queried concerning their institutions, programs, services, and collaborating partners.
This paper summarizes the institutional collaborations identified by all four librarians and explores the development of library-provided bioinformatics support and collaborations with other members of institutional bioinformatics support networks. Both similarities in approach and differences related to the individual institutional environments, as well as recommendations for beginning such collaborations, are discussed.
THE INSTITUTIONS, LIBRARIES, AND BIOINFORMATICS-RELATED CLIENTS SERVED
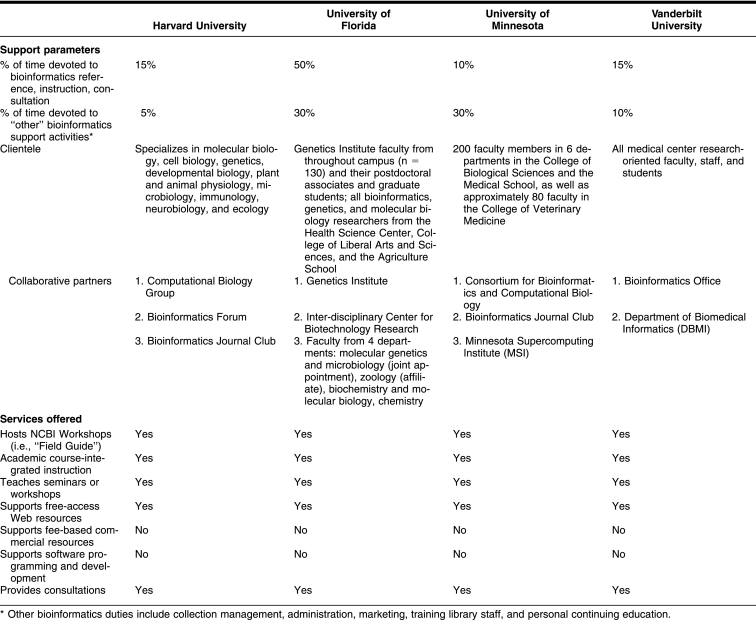
The four institutions and the roles of the library and librarian in each have a number of differences that may affect the type of services provided, as well as the time and abilities librarians have to create collaborations with other campus units. It is important for librarians to understand and work in the context and needs of their institution. The case examples include two private universities, Harvard University and Vanderbilt University, and two public institutions, University of Florida and University of Minnesota. The sizes and organization of the institutions vary, as well as the position of the librarians in the institutions and the types of patrons served (Table 1).
Table 1 Librarian positions
Library-based bioinformatics services
Although the percentages of time devoted to bioinformatics services differ greatly—ranging from 20% to 80%—the libraries at all four institutions have developed unique bioinformatics support programs (Table 2). Librarians provide workshops on bioinformatics-related tools, course-integrated instruction, and reference and consultation. A detailed description of library-based bioinformatics services at the University of Florida is available in the literature [8]. Vanderbilt's Eskind Biomedical Library offers a Research Informatics Consult Service (RICS), providing training seminars and individualized consultations to researchers, combined with extensive provision of electronic resources through a customized BioResearch digital library and an electronic search-advice service called BioSearchDoc. The librarian at Harvard provides hands-on training sessions on a number of different resources as well as private consultation with researchers. At the University of Minnesota, the molecular biosciences librarian offers seminars and workshops, academic course–integrated instruction, and research consultation services.
Table 2 Service parameters by institution
Institutional bioinformatics support partners
Collaboration can occur at a variety of levels: college, institution and center, department, educational program, and even with individual faculty members. The University of Florida Genetics Institute (UFGI) funds the bioinformatics librarian position housed in the Health Science Center Library. Partnerships involve teaching faculty from individual departments (molecular genetics and microbiology, biochemistry and molecular biology, zoology, and chemistry) and programs (first-year medical student and doctoral student education) who work closely with the librarian to develop bioinformatics education services. The University of Florida's Interdisciplinary Center for Biotechnology Research, a core facility providing fee-based laboratory and computational education and services, has divided responsibilities with the library as described in the “Fee versus Free” section.
At Vanderbilt University Medical Center (VUMC), collaborators include the Bioinformatics Office in the Program of Human Genetics (PHG) and the Department of Biomedical Informatics (DBMI). The Bioinformatics Office licenses commercial software programs and specialized computer programming applications. The librarian refers patron inquiries to the Bioinformatics Office when users require licensed software, while the Bioinformatics Office assists in advertising library-sponsored seminars. Additional collaborative opportunities have arisen for the librarian given the library's integration in VUMC's Informatics Center and the Informatics Center's strategic interest in increasing VUMC's bioinformatics capabilities [11]. Such positioning provides opportunities to work closely with the faculty, staff, and students in the DBMI as well as other parts of the informatics enterprise at VUMC.
The Computational Biology Group at the Center for Genomics Research at Harvard University provides three-hour, hands-on workshops in a wide range of bioinformatics programs, from microarray analysis to perl programming. Librarian-taught classes are part of this group's offerings and concentrate on free, Web-based resources. This teaching provides an opportunity to demonstrate some of the advantages of having a librarian's assistance when searching. Further, the collaboration includes cross-referring individuals based on their various areas of expertise.
The collaborative partners at the University of Minnesota include the Center for Computational Genomics and Bioinformatics (CCGB), the Minnesota Supercomputing Institute (MSI), and an interdisciplinary bioinformatics graduate minor program. The librarian provides lectures on bioinformatics topics for undergraduate and graduate courses in the biosciences, while CCGB and MSI focus on building new bioinformatics tools and providing licensed software and hardware and resources for high-performance computing. The librarian refers patron inquires related to those specialties to the centers, while the centers can refer basic bioinformatics questions, especially concerning the use of Web-based resources, to the librarian. The librarian participates in the minor program's journal club, bioinformatics interest email list, and annual on-campus bioinformatics symposium cosponsored with the MSI.
DEVELOPING COLLABORATIONS
In all cases, visibility appears to be the key to developing collaborative relationships. All four libraries host the “National Center for Biotechnology Information Field Guide” class [12] and assist with the teaching during those sessions.
The UFGI director agreed to fund the new bioinformatics librarian position based on the work an existing librarian had performed and the strong client connections she had forged in her previous six years as liaison to the University of Florida College of Medicine's basic science departments. Other connections were made with units and faculty via librarian participation on campus- and health center–wide grant proposal and program development committees. The librarian also made initial contacts through the bioinformatics database workshops she teaches. She now makes connections through the UFGI seminar series she organizes and moderates. Each of these opportunities educates potential collaborators about the role that librarians can take in bioinformatics instruction and services.
At the University of Minnesota, the librarian participates in the Bioinformatics Journal Club, an important tool in building connections to the research and learning communities in the field. The librarian has presented on open access publishing and multimedia resources available for molecular biology teaching and attends other presentations, occasionally providing “just in time” information during those sessions. This presence has led to many positive interactions with faculty and students, bringing a more personal face to library services for this part of the university community.
At Harvard, the librarian participates in a weekly journal club sponsored by the Computational Biology Group and in semi-monthly meetings attended by various groups across Harvard interested in bioinformatics. Coordinated by the Computational Biology Group, the meetings are held on a rotating basis at various venues across Harvard, with various groups from each venue being responsible for the presentations. This structure facilitates the librarian's staying informed about the areas of expertise in the community and the interests of each of the participating groups.
At Vanderbilt, the bioinformatics specialist at the Bioinformatics Office and the librarian have developed an agreement to support each other's training efforts and to delineate areas of responsibility (see “Fee versus Free”). Continuing cooperation between the library and the DBMI on other projects has also created partnership opportunities. The librarian has been consulted on larger projects including wide-scale research grants and has been given the opportunity to participate in VUMC's research enterprise planning. Finally, teaching well-publicized seminars and workshops has demonstrated to research faculty that the librarian can speak their language and has knowledge and skills relevant to their needs.
MAJOR AREAS OF COLLABORATION
At all four institutions, three major areas of collaboration are observed: knowledge management and information dissemination, resource provision and licensing, and instruction and consultation.
Knowledge management
All four librarians serve as “scouts” and “advocates,” as defined by Bell [10]. Although they perform these roles in a variety of ways, they all know their institutions, the research performed in bioinformatics and related fields, the stakeholders, and the resources and local expertise available at their institutions. They also know how to put people together with information and each other. Librarians advise students about which academic classes cover which bioinformatics tools, identify which researchers own various pieces of laboratory equipment, and point out which units license particular bioinformatics tools. In more formal ways, they collaborate with other units on campus reciprocally by advertising seminars and events, providing Websites with faculty research interests and contact information, and publicizing received grants and important papers published by their researchers.
All four librarians work with multiple divisions and various patron groups in their institutions, allowing them to serve as connections between those groups. By understanding various collaborative resources and faculty research interests, the librarians are able to triage patron questions and needs to the appropriate support providers in their institutions. In all cases, the librarian acts as a first line of support, directing users to available resources including specific databases and their collaborative partners depending on the nature of the request. The ability to triage questions effectively is based on the librarian's understanding of the entire institution, the available bioinformatics support units, and the types of clients presenting with bioinformatics-related questions at the library. These librarians serve as clearinghouses for information on the state of bioinformatics at their institutions, providing direct instruction as well as referrals to other experts; they are true “information hubs.”
Fee versus free
Another important commonality is the demarcation of responsibilities between the librarian and other bioinformatics units in terms of the types of resources supported. In these four cases, the librarian provides instruction, consultation, and support for noncommercial, Web-based resources and databases that do not require paying fees and licensing. Patrons needing assistance with purchasing commercial software or support for such software are referred to another bioinformatics unit. Consequently, an important role for the librarians has been to match patrons to information resources provided by various campus units, regardless of whether their libraries directly provide the resource.
It is important to note that, although the four institutions profiled in this paper approach the fee versus free issue in the same way, such is not the case for all libraries that provide bioinformatics support and resources. Examples of libraries that license fee-based bioinformatics resources can be found elsewhere in this issue [13–15]. Each library should evaluate its specific situation and resources to determine whether it has the expertise and capability to provide software licensing.
ENVIRONMENT PROMOTES SUCCESS
The uniqueness of these four university environments strongly impacts the success of each librarian. Harvard's decentralization has fostered the librarian's creation of his own niche, focused specifically on supporting the faculty and students in the bioscience departments that he supports, and allows him to set priorities for managing his own time. He has managed a Website and server space, allowing him to provide twenty-four-hour access to teaching resources. UFGI funding for the bioinformatics librarian position (full salary and the costs of one conference per year) has allowed her to concentrate on bioinformatics, genetics, and related clientele almost exclusively.
In the complex environment of the University of Minnesota, the primary task faced by the biosciences librarian has been to carve an appropriate, sustainable niche. The multiple bioinformatics support units across campus provide many opportunities for collaboration but also create challenges in coordination. However, because the librarian has been affiliated with multiple libraries serving the medical, agricultural, and other user groups, he has been able to serve as a connector and communication point for bioinformatics support efforts.
At Vanderbilt, the librarian has a small portion of her time devoted to bioinformatics support; the remainder of her time is focused on supporting clinical departments (emergency medicine and internal medicine). Though she has limited time for networking, marketing, and course development, the full support of the library's director for cross-training among library staff provides a unique advantage [3]. Most librarians providing bioinformatics services are the sole providers in their libraries; in their absence, no support is available. Because Vanderbilt librarians and some support staff are trained in the basics of molecular biology and genetics and related resources, incoming bioinformatics questions are triaged so that desk staff handle simple questions, other senior librarians handle mid-difficulty questions, and the more advanced questions are referred to the specialist librarian [3, 16]. This approach is presently unique to Vanderbilt among the four examples.
CONCLUSIONS AND RECOMMENDATIONS
Even with the diverse nature of the universities described in this paper, their approaches have commonalities that have led these programs to both define unique roles at their institutions and develop fruitful collaborations. Bioinformatics encompasses a broad, multidisciplinary area of interest. With many information and educational needs, it is unlikely that any one unit on campus can provide all necessary services. The following recommendations may help librarians in establishing or refining bioinformatics support services:
Perform an environmental scan to determine bioinformatics information needs, identify stakeholders, learn about services that are already available at the institution and their providers, and identify information and educational gaps to be filled.
Find initial advocates on the faculty and research staff who are library supporters, particularly those who champion bioinformatics education (e.g., at the undergraduate level). They can serve as an entry point into the research community.
Be visible and advocate for the service. Let potential clients and collaborators see that the library and librarians can be useful partners in bioinformatics. Host the “National Center for Biotechnology Information Field Guide” course [12], become involved in campus-wide committees, participate in journal clubs, attend seminars and research days, etc.
Seek out collaborations once library services are developed, and be broad in thinking of potential partners. Terms such as bioinformatics, computational biology, biotechnology, genetics, genomics, proteomics, pharmacogenetics/genomics, and molecular biology generally indicate potential partners; however, some potential clients will not be as obvious. A department in the dental school may be interested in bioinformatics tools or genetic techniques to treat dental caries; a surgery department may be interested in microarray techniques to study wound healing.
Use the other authors who appear in this issue and colleagues as contacts for advice and suggestions. Scanning the bioinformatics-focused literature should also yield service ideas.
As illustrated in this paper, the burgeoning field of bioinformatics provides numerous opportunities for librarians to provide bioinformatics-related services, resources, and instruction. Developing partnerships and collaborations with appropriate nonlibrary units throughout the institution can increase the likelihood of success in these endeavors and produce results that are more comprehensive than any unit could individually provide.
Contributor Information
Jennifer A. Lyon, Email: jennifer.lyon@Vanderbilt.Edu.
Michele R. Tennant, Email: michele@library.health.ufl.edu.
Kevin R. Messner, Email: messn006@tc.umn.edu.
David L. Osterbur, Email: dosterbu@mcb.harvard.edu.
REFERENCES
- Yarfitz S, Ketchell DS. A library-based bioinformatics services program. Bull Med Libr Assoc. 2000 Jan; 88(1):36–48. [PMC free article] [PubMed] [Google Scholar]
- Delwiche FA. Introduction to resources in molecular genetics. Med Ref Serv Q. 2001 Summer; 20(2):33–50. [DOI] [PubMed] [Google Scholar]
- Lyon J, Giuse NB, Williams A, Koonce T, and Walden R. A model for training the new bioinformationist. J Med Libr Assoc. 2004 Apr; 92(2):188–95. [PMC free article] [PubMed] [Google Scholar]
- MacMullen WJ, Vaughan KTL, and Moore ME. Planning bioinformatics education and information services in an academic health science library. Coll Res Libr. 2004 Jul; 65(4:):320–33. [Google Scholar]
- Moore ME, Vaughan KTL, and Hayes BE. Building a bioinformatics community of practice through library education programs. Med Ref Serv Q. 2004 Fall; 23(3):71–9. [DOI] [PubMed] [Google Scholar]
- Pratt GF. A health sciences library liaison project to support biotechnology research. Bull Med Libr Assoc. 1990 Jul; 78(3):302–3. [PMC free article] [PubMed] [Google Scholar]
- Pratt GF. Liaison services for a remotely located biotechnology research center. Bull Med Libr Assoc. 1991 Oct; 79(4:):394–401. [PMC free article] [PubMed] [Google Scholar]
- Tennant MR. Bioinformatics librarian: meeting the information needs of genetics and bioinformatics researchers. Ref Serv Rev. 2005 Jan; 33(1):12–9. [Google Scholar]
- Tennant MR, Miyamoto MM. The role of medical libraries in undergraduate education: a case study in genetics. J Med Libr Assoc. 2002 Apr; 90(2):181–93. [PMC free article] [PubMed] [Google Scholar]
- Bell JW. Our genome in common: genomic data release policies and the academic librarian. Libraries and the Academy. 2003 Apr; 3(2):293–306. [Google Scholar]
- Vanderbilt University Medical Center Informatics Center. Strategic plan for VUMC informatics & roadmap to 2010. [Web document]. Nashville, TN: The Center, 2005. [cited 7 Feb 2006]. <http://www.mc.vanderbilt.edu/infocntr/IC_Strategic_Plan_05.pdf>. [Google Scholar]
- National Center for Biotechnology Information. A field guide to NCBI resources. [Web document]. Bethesda: MD. The Center, 2005. [rev. 27 Dec 2005; cited 20 Jan 2006]. <http://www.ncbi.nlm.nih.gov/Class/FieldGuide/>. [Google Scholar]
- Chattopadhyay A, Tannery NH, Silverman DAL, Bergen P, and Epstein BA. Design and implementation of a library-based information service program in molecular biology and genetics at the University of Pittsburgh. J Med Libr Assoc. 2006 Jul; 94(3):307–13. E-192. [PMC free article] [PubMed] [Google Scholar]
- Minie M, Bowers S, Tarczy-Hornoch P, Roberts E, James RA, Rambo N, and Fuller S. The University of Washington Health Sciences Library BioCommons: an evolving northwest biomedical research information support infrastructure. J Med Libr Assoc. 2006 Jul; 94(3):321–9. [PMC free article] [PubMed] [Google Scholar]
- Osterbur D, Canevari C, Kirlew P, Jacobs DK, Ohles JA, Alpi K, Vaughan KTL, Wang L, Devare M, Corley PM, Wu Y, and Geer RC. Vignettes: diverse library staff offering diverse bioinformatics services. J Med Libr Assoc. 2006 Jul; 94(3):306– E-188–91. [PMC free article] [PubMed] [Google Scholar]
- Lyon J. Beyond the literature: bioinformatics training for medical librarians. Med Ref Serv Q. 2003 Spring; 22(1):67–74. [DOI] [PubMed] [Google Scholar]