Abstract
Recently, evidence was presented that in a collection of fluconazole-resistant strains of Candida albicans there was a much higher proportion of homozygotes for the mating type locus (MTL) than in a collection of fluconazole-sensitive isolates, suggesting the possibility that when cells become MTL homozygous they acquire intrinsic drug resistance. To investigate this possibility, an opposite strategy was employed. First, drug susceptibility was measured in a collection of isolates selected for MTL homozygosity. The majority of these isolates had not been exposed to antifungal drugs. Second, the level of drug susceptibility was compared between spontaneously generated MTL-homozygous progeny and their MTL-heterozygous parent strains which had not been exposed to antifungal drugs. The results demonstrate that naturally occurring MTL-homozygous strains are not intrinsically more drug resistant, supporting the hypotheses that either the higher incidence of MTL homozygosity previously demonstrated among fluconazole-resistant isolates involved associated homozygosity of a drug resistance gene linked to the MTL locus, or that MTL-homozygous strains may be better at developing drug resistance upon exposure to the drug than MTL-heterozygous strains. Furthermore, the results demonstrate that a switch by an MTL-homozygous strain from the white to opaque phenotype, the latter functioning as the facilitator of mating, does not notably alter drug susceptibility.
The great majority of Candida albicans strains are heterozygous at the mating type-like locus MTL, carrying MTLa1 on one chromosome and MTLα1 and MTLα2 on the homologous chromosome (7, 14). Fusion has been demonstrated to occur both in vivo (8) and in vitro (16, 17) between engineered homozygous MTLa (a/−) and MTLα (α/−) cells, but not between parental MTL-heterozygous (a/α) cells. Recently, Miller and Johnson (17) discovered an extraordinary and unexpected relationship between mating and white-opaque switching. They demonstrated that while cells of the MTL-heterozygous parent strain CAI4 (a/α) did not undergo white-opaque switching, derivative homozygous MTLa and homozygous MTLα cells underwent white-opaque switching at high frequency, and that when homozygous cells were in the opaque phase, the efficiency of mating increased 106-fold. Lockhart et al. (14) subsequently demonstrated that all white-opaque switchers obtained from a collection of clinical isolates were MTL homozygous and, conversely, that the majority of MTL homozygotes obtained from a collection of clinical isolates underwent white-opaque switching. Lockhart et al. (15) subsequently demonstrated that fusion occurred only between naturally occurring homozygous MTLa and homozygous MTLα strains in the opaque phase. Finally, Lockhart et al. (15) described for the first time the cell biology of C. albicans mating, including shmooing, chemotropism of conjugation tubes, fusion of tubes, nuclear association, vacuole expansion and nuclear separation in the conjugation bridge, asynchronous nuclear division in the zygote, daughter cell growth, nuclear migration in the daughter cell, septation, and daughter cell budding.
Recently, Rustad et al. (27) presented evidence suggesting a link between homozygosity at the MTL locus and fluconazole resistance in C. albicans. An analysis of 46 fluconazole-sensitive and 50 fluconazole-resistant strains revealed that while only 2% of the former were homozygous for the MTL locus (i.e., either a/a or α/α), 22% of the latter were homozygous. Rustad et al. (27) hypothesized that the association of fluconazole resistance and MTL homozygosity could be due to two possible mechanisms. First, drug resistance could be the direct result of the loss of either MTLa1 or MTLα1/MTLα2 (i.e., the direct result of MTL homozygosity), which in turn suggests that MTL-homozygous strains may be intrinsically resistant. Alternatively, they hypothesized that drug resistance could be the result of homozygosity of an allele linked to MTL on chromosome 5, through mitotic crossing over. Rustad et al. (27) demonstrated that homozygous MTLa and homozygous MTLα cells, engineered from the heterozygous MTLa/α strain CAI4 by Hull et al. (8), were as susceptible to fluconazole as the parent MTL-heterozygous strain, which supported the hypothesis that the high level of MTL homozygosity among fluconazole-resistant strains was not due to the immediate acquisition of drug resistance through MTL alleles becoming homozygous without drug exposure.
To investigate further the relationship between fluconazole resistance and MTL homozygosity, we have used a strategy opposite that of Rustad et al. (27). Rather than measure the proportion of MTL homozygotes among identified drug-resistant strains, we measured drug susceptibility among identified MTL-homozygous strains, the majority of which were collected from patients prior to drug therapy. In addition, we tested the susceptibility of the MTL-homozygous progeny of three MTL-heterozygous strains that spontaneously formed MTL-homozygous progeny at high frequency. If resistance is directly associated with the loss of either MTLa1 or MTLα1/MTLα2, then MTL homozygotes randomly selected from a large collection of clinical isolates should be intrinsically drug resistant without exposure to drugs, and MTL homozygotes spontaneously generated by naturally occurring, unstable MTL heterozygotes should be more drug resistant than their heterozygous parent strains without exposure to drugs. The results presented here demonstrate that in the absence of previous antifungal drug exposure, naturally occurring MTL-homozygous strains of C. albicans are not intrinsically more drug resistant.
MATERIALS AND METHODS
C. albicans isolates.
A general collection of over 300 clinical isolates representing the five major clades distinguished by the complex DNA fingerprinting probe Ca3 (5, 14, 23, 25) were tested for MTL zygosity by PCR methods (14, 15). A total of 16 MTL-homozygous strains were identified, including 7 MTLa-homozygous and 9 MTLα-homozygous strains. The majority of isolates were previously described (14). Two additional isolates, P37037 and P37039, were both collected from healthy individuals in Wisconsin and New Jersey, respectively. To this collection was added strain WO-1, which is MTLα homozygous. This population represents a mixed collection of isolates from the United States (nine isolates), Canada (one isolate), and South Africa (five isolates) from diverse clinical origins (14). With the exception of 19F and WO-1, these isolates were not exposed to any antifungal agent prior to collection. 19F was exposed to terconazole, and WO-1 was exposed to 5-fluorcytosine and amphotericin B prior to collection. Isolates were stored in 20% glycerol at 80°C prior to experimental use.
MIC testing.
Antifungal susceptibility testing was performed by the reference broth microdilution method as described in the National Committee for Clinical Laboratory Standards document M27-A (18). Caspofungin (Merck Research Laboratories, Rahway, N.J.), amphotericin B (Sigma, St. Louis, Mo.), flucytosine (5-FC; Sigma), fluconazole (Pfizer Inc., New York, N.Y.), voriconazole (Pfizer), itraconazole (Janssen, Beerse, Belgium), posaconazole (Schering-Plough, Kenilworth, N.J.), and ravuconazole (Bristol-Myers Squibb, Wallingford, Conn.) were obtained from their respective manufacturers. Serial dilutions were made in RPMI 1640 broth medium buffered to pH 7.0 with 0.165 M morpholinepropanesulfonic acid. Microdilution trays were prepared in a single lot as previously described (21), with the addition of an array of caspofungin at final dilutions of 0.007 to 8 μg/ml. The trays were stored at −70°C prior to use.
Prior to testing, each isolate was passaged on supplemented Lee's agar medium (4, 11) to assess colony phenotypes and once on potato dextrose agar (Remel, Lenexa, Kans.) to ensure optimal growth characteristics. One hundred microliters of a suspension of cells varying between 0.5 × 103 and 2.5 × 103 cells per ml was added to each well of a microdilution tray. The trays were incubated in air at 35°C, and MIC endpoints were read after 48 h. Drug- and yeast-free controls were included in each tray. Following incubation, the growth in each well was compared with that in the control wells. The MICs of each of the triazoles, caspofungin, and 5-FC were defined as the lowest concentration resulting in approximately 50% growth inhibition. The MIC of amphotericin B was defined as the lowest concentration resulting in 100% growth inhibition. Quality control of the susceptibility assays was performed on the reference isolates Candida parasilosis ATCC 22091 and Candida krusei ATCC 6258 (3, 18).
DNA fingerprinting.
The complex DNA fingerprinting probe Ca3 (1, 13, 24, 28) was used to confirm the identity of the progeny isolates from the parental strains P37037par and P37039par and to exclude the possibility of contaminations. DNA was extracted from cells (31), digested with EcoRI, and electrophoresed in a 0.8% agarose gel at 50 V according to methods previously described (12, 25, 32, 34). The DNA was then transferred to a Hybond N+ membrane (Amersham, Piscataway, N.J.) by capillary blotting. Prehybridization and hybridization with 32P-labeled Ca3 probe were performed as previously described (32). Southern blots were then autoradiographed.
PCR analysis of the MTL locus.
The PCR protocol used to analyze the MTL locus was previously described (14). The presence of MTLa was ascertained by amplifying MTLa1 and the associated gene OBPa (7). The presence of MTLα was ascertained by amplifying MTLα1 and MTLα2 and the associated gene OBPα. MTLα1, MTLα2, and MTLa1 are the C. albicans homologues of the Saccharomyces cerevisiae mating type genes MATα1, MATα2, and MATa1, respectively (7).
RESULTS
Drug susceptibility of a and α cells.
To test whether naturally occurring MTL-homozygous strains are more resistant to antifungal drugs than naturally occurring MTL-heterozygous strains, we first compared the susceptibility of 7 a strains and 10 α strains identified in a large collection of clinical isolates with the susceptibility of an independent set of unselected isolates from two large collections of naturally occurring isolates previously analyzed (6, 22). Since Lockhart et al. recently demonstrated that approximately 97% of naturally occurring C. albicans isolates are heterozygous at the MTL locus (14), we assumed here that the large collections previously analyzed for drug susceptibility were predominantly MTL heterozygous. In Table 1, the MICs of fluconazole, ravuconazole, posaconazole, voriconazole, itraconazole, caspofungin, amphotericin B, and 5-FC are presented for each of the 17 homozygous strains. In addition, the MIC50 (the concentration that inhibited 50% of isolates) and MIC90 (the concentration that inhibited 90% of isolates) of the entire collection of 17 MTL-homozygous strains and of the previously studied collections of predominantly MTL-heterozygous strains are provided in Table 1.
TABLE 1.
Antifungal susceptibility of 17 C. albicans strains homozygous at the mating type-like locus (MTL)
| MTL genotype | Strain | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Flu. | Ravu. | Posa. | Vori. | Itra. | Caspo. | Ampho. B | 5-FC | ||
| a | L26 | 2.00 | 0.060 | 0.120 | 0.030 | 0.500 | 0.12 | 1.0 | 2.00 |
| 12C | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.06 | 0.5 | 0.25 | |
| G106 | 0.50 | ≤0.007 | 0.030 | ≤0.007 | 0.120 | 0.06 | 1.0 | 0.25 | |
| OKP90 | ≤0.12 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.12 | 1.0 | 16.00 | |
| P60 | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.060 | 0.06 | 1.0 | 0.25 | |
| P37005 | 0.25 | ≤0.007 | 0.030 | ≤0.007 | 0.060 | 0.06 | 1.0 | 1.00 | |
| P75063 | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.12 | 1.0 | 0.12 | |
| α | WO-1 | ≤0.12 | ≤0.007 | 0.030 | 0.015 | 0.060 | 0.12 | 1.0 | 0.12 |
| 19F | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.12 | 1.0 | ≥64.00 | |
| GC75 | ≤0.12 | ≤0.007 | ≤0.007 | ≤0.007 | 0.030 | 0.25 | 1.0 | 0.25 | |
| P87 | 0.25 | ≤0.007 | 0.030 | ≤0.007 | 0.060 | 0.12 | 1.0 | 0.12 | |
| P37035 | 0.50 | 0.015 | 0.015 | ≤0.007 | 0.060 | 0.06 | 1.0 | 0.25 | |
| P37037 | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.25 | 1.0 | 0.25 | |
| P37039 | ≤0.12 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.06 | 1.0 | 0.25 | |
| P57072 | 0.25 | ≤0.007 | ≤0.007 | ≤0.007 | 0.015 | 0.06 | 1.0 | 0.12 | |
| P78048 | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.06 | 1.0 | 0.25 | |
| P80001 | 0.50 | ≤0.007 | 0.030 | ≤0.007 | 0.120 | 0.06 | 1.0 | 0.12 | |
| All isolates | |||||||||
| MIC50b | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.06 | 1.0 | 0.25 | |
| MIC90b | 0.50 | ≤0.007 | 0.030 | ≤0.007 | 0.120 | 0.12 | 1.0 | 2.00 | |
| a/α (97%)c | |||||||||
| MIC50b | 0.25 | ≤0.007 | 0.015 | ≤0.007 | 0.030 | 0.12 | 1.0 | 0.25 | |
| MIC90b | 0.50 | 0.015 | 0.060 | 0.015 | 0.060 | 0.25 | 1.0 | 1.00 | |
Flu., fluconazole; Ravu., ravuconazole; Posa., posaconazole; Vori., voriconazole; Itra., itraconazole; Caspo., caspofungin; Ampho.B, amphotericin B.
The MIC50 and MIC90 values represent the concentration of each antifungal agent that inhibited 50% and 90%, respectively, of the tested isolates.
Data were obtained from Pfaller et al. (22) for a collection of 486 isolates of C. albicans tested for caspofungin and from Diekema et al. (6) for a collection of 148 isolates of C. albicans tested for the remaining antifungals. The collections used as controls were assumed to be 97% a/α based on the results of Lockhart et al. (14).
Of the 17 MTL-homozygous strains tested, only one strain, L26, exhibited increased resistance to fluconazole (Table 1). This strain also exhibited increased resistance to the other tested azoles and a slight increase in 5-FC resistance, but no resistance to caspofungin or amphotericin B. Two strains, OKP90 and 19F, exhibited increased resistance to 5-FC but normal susceptibility to all other drugs. With the exception of resistance to 5-FC observed for 19F, the two MTL-homozygous strains that had been exposed to drug therapy prior to collection, strains WO-1 and 19F, exhibited no increases in resistance to the tested drugs. These results suggest that becoming homozygous in and of itself is not sufficient to confer drug resistance in C. albicans.
Drug susceptibility of spontaneous MTL-homozygous progeny of MTL-heterozygous strains.
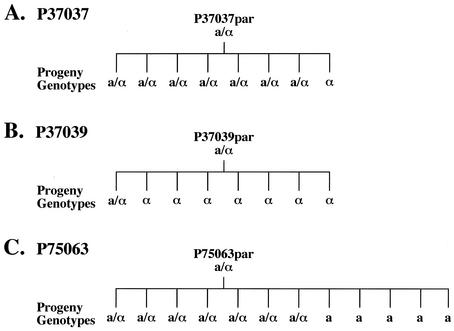
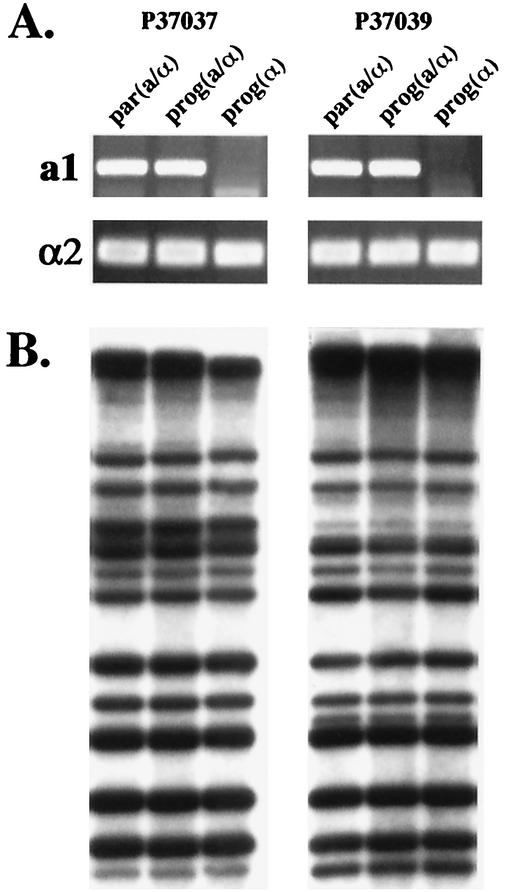
We next tested whether MTL heterozygotes gave rise to drug-resistant MTL-homozygous progeny. In an analysis of 30 MTL-heterozygous isolates, 3 were identified that formed opaque-phase sectors at high frequency. Since white-opaque switching is suppressed in MTL-heterozygous strains (14, 17), it appeared likely that these particular strains gave rise spontaneously to MTL-homozygous progeny at high frequency. To test this possibility, randomly selected progeny of the three such strains were tested for MTL zygosity by PCR analysis of mating type genes. For strains P37037 and P37039, MTLa/α and MTLα progeny were identified and, for strain P75063, MTLa/α and MTLa progeny were identified (Fig. 1). In Fig. 2A, PCR amplification patterns of MTLa1 and MTLα2 are presented for the MTL-heterozygous parental strains P37037par and P37039par and both MTL-heterozygous and MTL-homozygous progeny of each. A similar analysis of strain P75063 was previously reported (14). To demonstrate that MTL-heterozygous and MTL-homozygous isolates were progeny of the respective parent strains and not contaminants, isolates were DNA fingerprinted by Southern blot hybridization with the complex probe Ca3 (32, 34). The Southern blot hybridization patterns of the parent strain and the progeny of each strain were identical (Fig. 2B).
FIG. 1.
Pedigrees demonstrating spontaneous in vitro loss of heterozygosity at the MTL locus. Selected progeny of three MTL-heterozygous parental strains, P37037par, P37039par, and P75063par, that formed opaque-phase sectors were analyzed by PCR amplification of MTL genes. Their MTL genotypes are coded a/α for MTL heterozygotes, a for MTLa homozygotes, and α for MTLα homozygotes.
FIG. 2.
Demonstration by PCR of spontaneous in vitro loss of heterozygosity at the MTL locus by strains P37037 and P37039. (A) PCR analysis of the MTL locus demonstrating MTL heterozygosity of the parent strains [par(a/α)] and MTL homozygosity of the progeny isolate (α/α). The MTLa1 and MTLα2 genes of each test isolate were amplified by PCR and run in a 1% agarose gel. (B) Southern blot analysis with the complex fingerprinting probe Ca3 confirmed that, in both strains, the heterozygous and homozygous isolates were progeny of the original parental strain. EcoRI-digested DNA was run in a 0.8% agarose gel, Southern blotted, and hybridized with radiolabeled Ca3 fingerprinting probe.
The drug susceptibilities of the three selected MTL-heterozygous parent strains and a single MTL-homozygous progeny of each were compared. For strains P37039 and P75063, there was no significant difference (i.e., more than twofold increase) between the MICs for the parental MTL heterozygotes and their MTL-homozygous progeny. For strain P37037, there was no significant difference between the MTL-heterozygous parental isolates and MTL-homozygous progeny for seven of the eight antifungal drugs. In the case of 5-FC, however, the MTL heterozygote was 16-fold less susceptible than the MTL homozygote. Their respective 5-FC MICs were 4.00 and 0.25 μg per ml. Together, these data demonstrate that spontaneous MTL-homozygous progeny that have not been exposed to antifungal drugs are not more resistant to fluconazole or the other tested drugs than their MTL-heterozygous parents.
Drug susceptibility and the white-opaque transition.
It has been demonstrated that while MTL-homozygous strains of C. albicans undergo white-opaque switching, MTL-heterozygous strains do not. Since high-frequency phenotypic switching has been shown to alter drug susceptibility levels (38), the possibility was entertained that the higher proportion of fluconazole-resistant strains in the collection of MTL-homozygous strains analyzed by Rustad et al. (27) may have been due to expression of the opaque-phase phenotype (9, 26, 33, 36, 37). To test this possibility, we compared the susceptibility of white- and opaque-phase cells of six MTL-homozygous strains to the eight antifungal drugs. For five of the six strains, there was no significant difference between white- and opaque-phase cells in their susceptibility to the eight antifungal drugs. One strain, 19F, exhibited increased resistance to 5-FC in both white- and opaque-phase cells but showed relatively low MICs of the seven other antifungal drugs. In this one strain, white-phase cells were significantly more resistant than opaque-phase cells to 5-FC. Their 5-FC MICs were ≥64.00 and 16.00 μg per ml, respectively. Together, these results indicate that, on average, white- and opaque-phase cells exhibit similar drug susceptibility patterns. Hence, the increase in the proportion of homozygous strains among fluconazole-resistant strains in the collection analyzed by Rustad et al. (27) was not due to increased expression of the opaque-phase phenotype.
DISCUSSION
Although it does not appear that specific drug-resistant strains of C. albicans have established themselves as significant components of the general colonizing populations of C. albicans (19, 20, 30, 35), drug-resistant strains have emerged in specific cases of candidiasis. In recent years, a number of genes involved in drug resistance have been identified and cloned (29, 39). Among these, genes of the ABC transporter family have been implicated in clinical resistance to the azoles. Two of these genes, CDR3 and CDR4, are regulated by white-opaque switching (2; D. Sanglard, F. Ischer, M. Monod, S. Dogra, R. Prasad, and J. Bille, Abstr. 5th ASM Conf. Candida Candidiasis, abstr. C27, 1999). In addition, a number of other putative drug resistance genes have been recently shown to be differentially regulated by the white-opaque transition (10). The report by Rustad et al. (27) that 22% of fluconazole-resistant strains were MTL homozygous while only 2% of fluconazole-susceptible strains were MTL homozygous was interesting in light of the recent discovery that only MTL-homozygous strains of C. albicans undergo white-opaque switching (14, 17).
Rustad et al. (27) proposed that the increase in the proportion of MTL-homozygous strains among fluconazole-resistant strains was either a direct result of becoming homozygous at the MTL locus or homozygous at a gene linked to MTL. To distinguish between these alternatives, we used a strategy opposite that of Rustad et al. (27) in which we tested whether strains selected for MTL homozygosity were more resistant to antifungal drugs. Because homozygosity in natural populations is relatively rare in C. albicans (14), the MTL-homozygous isolates analyzed here were obtained from different geographical locales and different clinical settings. This collection was distinct from the collection analyzed by Rustad et al. (27) in that isolates of their collection were selected solely for fluconazole resistance. If resistance in C. albicans isolates was a direct consequence of MTL homozygosity, it would be expected that naturally occurring MTL-homozygous isolates that have not been exposed to fluconazole or other antifungal drugs would nevertheless show intrinsic resistance. None of the 17 isolates had been exposed to fluconazole prior to collection, while two were exposed to antifungal drugs other than fluconazole. We found no elevated levels of resistance to fluconazole and very limited resistance to the seven additional drugs among the 17 MTL-homozygous isolates. The two MTL-homozygous isolates that had been exposed to drug therapy prior to collection were not resistant to the drugs they had been exposed to. These results suggest that the relation between MTL homozygosity and fluconazole resistance is not direct, and they are consistent with the alternative hypothesis proposed by Rustad et al., namely, that a gene linked to MTL, which becomes homozygous when MTL becomes homozygous, is responsible for the acquisition of drug resistance (27). An analysis of the region of chromosome 5 harboring the MTL locus in the Stanford genome database, however, failed to reveal a possible gene candidate (i.e., potential drug resistance gene). ERG11, which is the only known gene involved in fluconazole resistance on chromosome 5, does not appear to be a viable candidate since it is not closely linked to the MTL locus and has been found to be heterozygous in some MTL-homozygous strains (27).
To further test whether MTL homozygosity leads to drug resistance in the absence of drug exposure, we analyzed MTL-homozygous progeny of three MTL-heterozygous strains that produce MTL-homozygous progeny at high frequency. In no case did we observe fluconazole resistance or general drug resistance in the MTL-homozygous progeny. Finally, we tested the possibility that the increase in MTL-homozygous isolates among drug-resistant isolates was the result of their MTL-homozygous-associated characteristic of switching from white to opaque (14, 17). A comparison of white- and opaque-phase cells (9, 26, 33, 36, 37) from six MTL-homozygous strains revealed no opaque-phase-specific resistance, thus eliminating this possibility as an explanation of the results reported by Rustad et al. (27).
Among the 17 MTL-homozygous isolates analyzed, one was resistant to 5-FC and a second one exhibited intermediary resistance to this drug. In addition, significant differences in susceptibility to 5-FC were observed between white- and opaque-phase cells of one strain and between the MTL-heterozygous parent and the MTL-homozygous progeny of another strain. These changes, however, did not support the acquisition of drug resistance as a result of becoming homozygous.
Together, our results demonstrate that MTL homozygosity in and of itself is not sufficient to confer fluconazole or general drug resistance. In addition, MTL-homozygous progeny formed spontaneously from MTL-heterozygous strains exhibit the same levels of susceptibility as their MTL-heterozygous parent strains. These results are consistent with an alternative hypothesis proposed by Rustad et al. (27) that the higher incidence of MTL-homozygous strains among fluconazole-resistant strains is the result of associated homozygosity of an unidentified gene linked to the MTL locus. These results are also consistent with the possibility that MTL-homozygous strains develop drug resistance more rapidly than MTL-heterozygous strains when exposed to antifungal drugs, a hypothesis now under investigation.
Acknowledgments
We are grateful to S. R. Lockhart for valuable suggestions.
This research was supported by NIH grant AI2392 to D.R.S.
REFERENCES
- 1.Anderson, J., T. Srikantha, B. Morrow, S. H. Miyasaki, T. C. White, N. Agabian, J. Schmid, and D. R. Soll. 1993. Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J. Clin. Microbiol. 31:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balan, I., A. M. Alarco, and M. Raymond. 1997. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J. Bacteriol. 179:7210-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, A. L., M. A. Pfaller, S. D. Brown, A. Espinel-Ingroff, M. A. Ghannoum, C. Knapp, R. P. Rennie, J. H. Rex, and M. G. Rinaldi. 2000. Quality control limits for broth microdilution susceptibility tests of ten antifungal agents. J. Clin. Microbiol. 38:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedell, G. W., and D. R. Soll. 1979. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and zinc-sensitive pathways for mycelium formation. Infect. Immun. 26:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blignaut, E., C. Pujol, S. Lockhart, S. Joly, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals new clade in South Africa. J. Clin. Microbiol. 40:826-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diekema, D. J., S. A. Messer, A. B. Brueggemann, S. L. Coffman, G. V. Doern, L. A. Herwaldt, and M. A. Pfaller. 2002. Epidemiology of candidemia: three-year results from the Emerging Infections and the Epidemiology of Iowa Organisms study. J. Clin. Microbiol. 40:1298-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 8.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 9.Kvaal, C., S. A. Lachke, T. Srikantha, K. Daniels, J. McCoy, and D. R. Soll. 1999. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67:6652-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan, C. Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 12.Lockhart, S., C. Pujol, S. Joly, and D. R. Soll. 2001. Development and use of complex probes for DNA fingerprinting the infectious fungi. Med. Mycol. 39:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Lockhart, S., J. J. Fritch, A. S. Sturdevant Meier, K. Schroppel, T. Srikantha, R. Galask, and D. R. Soll. 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockhart, S. R., C. Pujol, K. Daniels, M. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart, S. R., K. Daniels, R. Zhao, D. Wessels, and D. R. Soll. 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 2:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 17.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Pfaller, M. A., and D. J. Diekema. 2002. Role of sentinel surveillance of candidemia: trends in species distribution and antifungal susceptibility. J. Clin. Microbiol. 40:3551-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., D. J. Diekema, R. N. Jones, S. A. Messer, R. J. Hollis, and the SENTRY Participant Group. 2002. Trends in antifungal susceptibility of Candida spp. from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 40:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaller, M. A., D. J. Diekema, S. A. Messer, L. Boyken, H. Huynh, and R. J. Hollis. 2002. Clinical evaluation of a frozen commercially prepared microdilution panel for antifungal susceptibility testing of seven antifungal agents, including the new triazoles posaconazole, ravuconazole, and voriconazole. J. Clin. Microbiol. 40:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller, M. A., S. A. Messer, K. Mills, A. Bolmstrom, and R. N. Jones. 2001. Evaluation of E-test method for determining caspofungin (MK-0991) susceptibilities of 726 clinical isolates of Candida species. J. Clin. Microbiol. 39:4387-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pujol, C., M. Pfaller, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans bloodstream isolates from the United States, Canada, South Africa, and Europe reveals a European clade. J. Clin. Microbiol. 40:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pujol, C., S. Joly, B. Nolan, T. Srikantha, and D. R. Soll. 1999. Microevolutionary changes in Candida albicans identified by the complex Ca3 fingerprinting probe involve insertions and deletions of the full-length repetitive sequence RPS at specific genomic sites. Microbiology 145:2635-2646. [DOI] [PubMed] [Google Scholar]
- 25.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rikkerink, E. H. A., B. B. Magee, and P. T. Magee. 1988. Opaque-white phenotypic transition: a programmed morphological transition in Candida albicans. J. Bacteriol. 170:895-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rustad, T. R., D. A. Stevens, M. A. Pfaller, and T. C. White. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061-1072. [DOI] [PubMed] [Google Scholar]
- 28.Sadhu, C., M. J. McEachern, E. P. Rustchenko-Bulgac, J. Schmid, D. R. Soll, and J. B. Hicks. 1991. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J. Bacteriol. 173:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard, D. 2002. Resistance of human fungal pathogens to antifungal drugs. Curr. Opin. Microbiol. 5:379-385. [DOI] [PubMed] [Google Scholar]
- 30.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 31.Scherer, S., and D. A. Stevens. 1987. Application of DNA fingerprinting methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid, J., E. Voss, and D. R. Soll. 1990. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J. Clin. Microbiol. 28:1236-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soll, D. R. 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13:332-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soll, D. R. 2002. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 81:101-110. [DOI] [PubMed] [Google Scholar]
- 36.Soll, D. R. Candida albicans. In A. Craig and A. Scherf (ed.), Antigenic variation, in press. Academic Press, London, United Kingdom.
- 37.Srikantha, T., and D. R. Soll. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53-60. [DOI] [PubMed] [Google Scholar]
- 38.Vargas, K., S. A. Messer, M. Pfaller, S. R. Lockhart, J. T. Stapleton, J. Hellstein, and D. R. Soll. 2000. Elevated phenotypic switching and drug resistance of Candida albicans from human immunodeficiency virus-positive individuals prior to first thrush episode. J. Clin. Microbiol. 38:3595-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]