Abstract
Candida albicans is an opportunistic human fungal pathogen and a causative agent of oropharyngeal candidiasis (OPC), the most frequent opportunistic infection among patients with AIDS. Fluconazole and other azole antifungal agents have proven effective in the management of OPC; however, with increased use of these agents treatment failures have occurred. Such failures have been associated with the emergence of azole-resistant strains of C. albicans. In the present study we examined changes in the genome-wide gene expression profile of a series of C. albicans clinical isolates representing the stepwise acquisition of azole resistance. In addition to genes previously associated with azole resistance, we identified many genes whose differential expression was for the first time associated with this phenotype. Furthermore, the expression of these genes was correlated with that of the known resistance genes CDR1, CDR2, and CaMDR1. Genes coordinately regulated with the up-regulation of CDR1 and CDR2 included the up-regulation of GPX1 and RTA3 and the down-regulation of EBP1. Genes coordinately regulated with the up-regulation of CaMDR1 included the up-regulation of IFD1, IFD4, IFD5, IFD7, GRP2, DPP1, CRD2, and INO1 and the down-regulation of FET34, OPI3, and IPF1222. Several of these appeared to be coordinately regulated with both the CDR genes and CaMDR1. Many of these genes are involved in the oxidative stress response, suggesting that reduced susceptibility to oxidative damage may contribute to azole resistance. Further evaluation of the role these genes and their respective gene products play in azole antifungal resistance is warranted.
The human fungal pathogen Candida albicans is a cause of mucosal, cutaneous, and systemic infections, including oropharyngeal candidiasis (OPC), the most frequent opportunistic infection among AIDS patients (11, 21). The azole antifungal agents, particularly fluconazole, have proven effective for the management of OPC. The repetition and lengthy duration of therapy for OPC in this patient population has led to an increased incidence of treatment failures secondary to the emergence of azole resistance in this pathogenic fungus (33, 42, 55; S. L. Kelly, D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele, Letter, Lancet 348:1523-1524, 1996).
The azole antifungal agents exert their activity by inhibiting the biosynthesis of ergosterol, the major membrane sterol of fungi. By binding to the heme in the active site of the cytochrome P450 enzyme, lanosterol demethylase, azoles inhibit its activity (19). Several mechanisms of resistance to azole antifungal agents in C. albicans have been described (12, 13, 28, 43-45, 53, 54). Point mutations in the gene encoding lanosterol demethylase (ERG11) may result in conformational changes that prevent effective binding between azoles and their target, while not affecting the function of the enzyme in ergosterol biosynthesis (44, 54). Increased expression of the ERG11 gene has also been associated with azole resistance (13, 37, 53). This presumably results in increased production of lanosterol demethylase, which exceeds the capacity of the azole antifungal agent.
In addition to reducing the synthesis of ergosterol, the inhibition of lanosterol demethylase leads to the accumulation of the toxic sterol, 14α-methylergosta-8,24(28)-dien-3β,6α-diol, which inhibits fungal cell growth (20). Alterations in ergosterol biosynthesis enzymes that result in the accumulation of alternate, nontoxic sterols in place of toxic sterols would allow the organism to circumvent this effect and lead to azole resistance. Indeed, inactivation of the ERG3 gene product, Δ5,6 desaturase, results in elevated membrane fecosterol content and fluconazole resistance (20, 36; Kelly et al., letter).
Two classes of efflux pumps have been implicated in azole resistance: the ATP binding cassette (ABC) transporters and the major facilitators (12, 13, 28, 45, 53). The genes encoding two of the ABC transporters in C. albicans, CDR1 and CDR2, as well as that encoding a major facilitator, CaMDR1 (BMR1), have been shown to be overexpressed in resistant isolates. Overexpression of these efflux pumps is presumed to prevent accumulation of effective concentrations of the azole antifungal agent in the fungal cell. While all of the mechanisms described here can impart reduced azole susceptibility, high-level azole resistance appears to accumulate gradually over time and to involve multiple mechanisms (37). These mechanisms individually contribute only partially to the overall high-level, clinically relevant, resistant phenotype.
The constitutive overexpression of the ERG11, CDR1, CDR2, and MDR1 genes in azole-resistant isolates of C. albicans is suggestive of mutations in the cis or trans regulatory elements of these genes. When the promoter sequences of CaMDR1 alleles of two azole-resistant isolates of C. albicans were compared to those of their respective matched susceptible counterparts, no mutations were found that might be responsible for altered CaMDR1 activation. Using a green fluorescence protein fused to the CaMDR1 promoter from an azole susceptible strain, these investigators observed increased fluorescence when the construct was introduced into an azole-resistant strain that constitutively overexpresses CaMDR1 (56). Other investigators using a Renilla luciferase reporter system fused to CDR1 and CDR2 promoters cloned from azole-susceptible C. albicans isolates showed that their expression was increased in an azole-resistant strain in which these genes are constitutively up-regulated. These investigators also identified a common unique drug response element (DRE) in the promoters of CDR1 and CDR2 (6). The trans and cis regulatory elements responsible for the regulation of genes associated with azole resistance in C. albicans have yet to be identified.
Recently DNA microarray analysis has been applied to the study of azole resistance in C. albicans in both laboratory (4) and clinical (41) isolates. In the former study, azole resistance was experimentally induced by passage of the organism in increasing concentrations of fluconazole (4). The expression profiles of multiple isolates in these series were then compared using a DNA microarray representing a portion of the C. albicans genome. In addition to CDR2 and MDR1, these investigators identified a number of genes that were differentially expressed in experimental azole resistance, including the dehydrogenases/oxidoreductases YPL88, YPX98 (IFD6), and YPR127W. Other genes found to be differentially expressed included the glutathione peroxidase gene HYR1 (GPX1), ADH4, GRE99, YOR49 (RTA2), HSP12, and URE2. Using a similar DNA microarray, we recently compared the partial expression profiles of a well-characterized matched pair of azole-susceptible and -resistant clinical isolates (41). In addition to CDR1, genes found to be differentially expressed included the oxidative stress response genes CRD2, GPX1 (as previously reported in experimental azole resistance), and IFD5 (similar to IFD6). Other genes found to be differentially expressed in this study included the ergosterol biosynthesis pathway gene ERG2 and the drug resistance gene RTA3 (similar to RTA2). In the study by Cowen et al., microarray experiments were limited to three sets of isolates where fluconazole resistance (MIC ≥ 64 μg/ml) was experimentally induced, used DNA arrays inclusive of only a portion of the C. albicans genome, and included evaluation of only 10 genes by Northern blot analysis in unmatched azole-susceptible and -resistant clinical isolates. Likewise, our previous study (41) was limited to the initial and final isolates in a series of clinical isolates representing the stepwise acquisition of azole resistance (29, 39, 40, 53). Furthermore, this previous study also used DNA microarray analysis inclusive of only a portion of the genome (3,529 annotated genes) and only included genes for which differential expression was confirmed by RT-PCR.
In the present study, we expand on our previous work by using a genome-wide DNA microarray (representing 6,039 unique open reading frames [ORFs]) to examine changes in the gene expression profile during the stepwise acquisition of the azole-resistant phenotype in this series of clinical isolates. In addition to the novel identification of a number of genes that are differentially expressed in azole resistance, we have found groups of genes that are differentially expressed across this set of isolates coordinately with either CDR1 and CDR2, CaMDR1, or genetic changes in the ERG11 gene. These findings suggest that these sets of coordinately regulated genes may be controlled by common regulatory systems and lend insight into the elucidation of their regulatory elements.
MATERIALS AND METHODS
C. albicans isolates and growth conditions.
C. albicans isolates 2-79, 2-80, 8-46, and 12-99 were obtained from Spencer W. Redding and Theodore C. White. A compilation of MICs, alternate names, and previously described resistance mechanisms for this set of isolates is presented in Table 1. An aliquot of glycerol stock from each isolate was diluted in YPD broth (1% yeast extract, 2% peptone, 1% dextrose) and grown overnight at 30°C in an environmental shaking incubator. Two independent sets of cultures were diluted to an optical density at 600 nm (OD600) of 0.2 in fresh YPD and grown as before to the early logarithmic phase to the equivalent OD (0.4 to 0.5) for subsequent RNA isolation.
TABLE 1.
Compilation of previously reported and verified MICs and described resistance mechanisms for C. albicans isolates used in this study
| Isolate | Alternate isolate namea | MIC (μg/ml)a
|
Previously described resistance mechanism(s)a | |
|---|---|---|---|---|
| Previously determineda | This study | |||
| 2-79 | 2 | 2 | 0.25 | |
| 2-80 | 3 | 8 | 4 | Increased expression of MDR1 |
| 8-46 | 15 | 32 | 16 | Increased expression of MDR1 and ERG11, loss of allelic variation in ERG11, point mutation in ERG11 |
| 12-99 | 17 | >64 | >64 | Increased expression of MDR1 and ERG11, loss of allelic variation in ERG11, point mutation in ERG11, increased expression of CDR1 and CDR2 |
As reported in reference 55.
Susceptibility testing of C. albicans isolates.
The MICs of fluconazole were determined using broth microdilution techniques as described by the National Committee for Clinical Laboratory Standards (22). The cells were cultured in RPMI 1640 buffered at pH 7.0 with MOPS (morpholinepropanesulfonic acid) with the starting inoculum at approximately 0.5 × 103 to 2.5 × 103 CFU/ml. Microtiter trays were incubated at 35°C in a moist, dark chamber, and the MICs were recorded after 48 h of incubation. The susceptibility endpoint was defined as the lowest concentration of antifungal which resulted in visible growth that was reduced by 80% compared with growth of the control. The determinations of the MICs for the isolates were performed prior to experimental use.
Total RNA isolation.
RNA was isolated using the hot phenol method (13). Briefly, cells were collected by centrifugation and snap-frozen in liquid nitrogen. Frozen cells were resuspended in 12 ml of AE buffer (50 mM sodium acetate [pH 5.2], 10 mM EDTA) at room temperature, after which 800 μl of 25% sodium dodecyl sulfate and 12 ml of acid phenol (Fisher Scientific, Houston, Tex.) were added. The cell lysate was then incubated 10 min at 65°C with vortexing each minute, cooled on ice 5 min, and subjected to centrifugation for 15 min at 11,952 × g. Supernatants were transferred to new tubes containing 15 ml of chloroform, mixed, and subjected to centrifugation at 200 × g for 10 min. RNA was precipitated from the resulting aqueous layer by transferring that portion to new tubes containing 1 volume isopropanol and 0.1 volume 2 M sodium acetate, pH 5.0; mixing well; and subjecting the mixture to centrifugation at 17,211 × g for 35 min at 4°C. The supernatants were removed, the pellet was resuspended in 10 ml of 70% ethanol, and the RNA was collected by centrifugation at 17,211 × g for 20 min at 4°C. Supernatants were again removed, and RNA was resuspended in diethyl pyrocarbonate-treated water. The OD260 and OD280 were measured, and the integrity of the RNA was visualized by subjecting 2 to 5 μl of the sample to electrophoresis through a 1% agarose-MOPS gel.
Microarray design and preparation.
The C. albicans microarray was manufactured by Eurogentec SA (Ivoz-Ramet, Belgium) in collaboration with the European Galar Fungail Consortium (www.pasteur.fr/recherche/unites/Galar_Fungail/). Primers for each of the 6,039 putative ORFs in the C. albicans genome were designed to amplify a specific region of each ORF. Both the forward and reverse primers for each ORF contained a 15-bp universal sequence at the 5′ end to generate 5′ amino-modified product for covalent attachment to aldehyde-coated glass slides. Amplicons were an average length of 300 bp and were spotted in duplicate, along with 27 control genes, using a ChipWriter Pro (Virtek Vision Intl., Waterloo, Ontario, Canada) robotic array printer.
Probe preparation.
Ten micrograms of total RNA sample was added to a mixture of 1 pmol of T20VN and oligo(dT) (18- to 21-mer) primer mix; 0.5 mM (each) dATP, dGTP, and TTP; 20.5 μM dCTP; 37.5 μM Cy3- or Cy5-dCTP (NEN Life Sciences, Boston, Mass.); and 10 mM dithiothreitol in a buffer containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, and 3 mM MgCl2. The reaction mixture was denatured at 65°C for 5 min and incubated at 42°C for 5 min, after which 1 μl of RNasin (Promega, Madison, Wis.) and 200 U of Superscript II reverse transcriptase (LifeTechnologies/Invitrogen, Carlsbad, Calif.) were added to the mixture. The reaction proceeded at 42°C for 1 h, after which an additional 200 U of Superscript II reverse transcriptase was added, and the reaction mixture was incubated at 42°C for an additional hour. To stop the reaction, EDTA (pH 8.0) and sodium hydroxide were added to a final concentration of 5 mM and 0.4 N, respectively, and the mixture was incubated at 65°C for 20 min. Finally, acetic acid was added to achieve a final concentration of 0.37 M.
To purify the labeled cDNA probes, the volume of the cDNA reaction was adjusted to 100 μl with water and mixed with 0.5 ml of buffer PB (Qiagen, Valencia, Calif.) and 13 μl of 3 M sodium acetate (pH 5.2). The sample was applied to a Qia-quick column (Qiagen) and centrifuged at 17,982 × g for 60 s. The flowthrough was discarded, and two washes of 750 μl of buffer PE (Qiagen) each were applied to the column, with a 60-s centrifugation after each wash. After an additional centrifugation step to dry the column, 50 μl of prewarmed (42°C) water was applied to the column and incubated for 3 min at room temperature. To elute the sample, the column was centrifuged as before, and then an additional elution step was performed. Eluates were pooled, applied to a Microcon-30 filter (Millipore, Bedford, Mass.), and centrifuged as before for 3 min, and the membrane unit was vortexed briefly and centrifuged for an additional 60 s. The final eluate volume was adjusted to 5 μl with water.
Microarray hybridization.
Fluorescently labeled cDNA probes were generated from duplicate sets of RNA isolated from isolates 2-79, 2-80, 8-46, and 12-99. Five microliters each of the Cy3- and Cy5-labeled probes were mixed with 50 μg of heat-denatured salmon sperm DNA, incubated at 95°C for 2 min, and snap-cooled on ice. The mixture was added to 40 μl of hybridization buffer and applied to the array slides under glass coverslips. Hybridization was performed at 37°C overnight in a humidified chamber (Corning Life Sciences, Acton, Mass.). To wash the slides, the coverslip was removed, and the slide was incubated at room temperature in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate for 5 min with agitation, rinsed at room temperature with 0.2× SSC for 5 min with agitation, and spin-dried at 500 rpm for 5 min. Slides were scanned using a ChipReader microarray scanner (Virtek Vision Intl.).
Data analysis.
GenePix 1.0 software (Axon Instruments, Inc.) was used for image analysis and data visualization. The local background values were calculated from the area surrounding each spot and subtracted from the total spot signal values. These adjusted values were used to determine differential gene expression (Cy3/Cy5 ratio) for each spot. A normalization factor was applied to account for systematic differences in the probe labels by using the differential gene expression ratio to balance the Cy5 signals. In the present study, only spots with a mean balanced differential expression ratio greater than or equal to 1.5 or less than or equal to 0.67 (−1.5) for both spots representing a given cDNA on the array in two independent experiments were considered to be differentially expressed. DNA sequences were annotated on the basis of results of BLASTn searches using GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi), the Stanford University (Palo Alto, Calif.) sequencing database (http://www-sequence.stanford.edu/group/candida), and the CandidaDB database (http://www.pasteur.fr/Galar_Fungail/CandidaDB/). Using the software of Eisen at al. (11), genes were clustered into groups on the basis of their coordinate regulation with either CDR genes or CaMDR1.
cDNA synthesis and reverse transcription-PCR (RT-PCR).
To synthesize cDNA, approximately 2 μg of total RNA was placed in a 0.5-ml reaction tube with 1 μg of oligo(dT) primer stock (ResGen/Invitrogen Corp., Carlsbad, Calif.), and the volume was adjusted to 15.5 μl using diethyl pyrocarbonate-treated water. The mixture was incubated 10 min at 70°C and chilled on ice 1 min, after which the remainder of the reaction mixture was added from a master mix to the reaction tube in order for each reaction to contain a 1.25 mM concentration (each) dATP, dCTP, dGTP, and TTP; 40 U of RNase Inhibitor (Stratagene, La Jolla, Calif.); and 25 U of Moloney murine leukemia virus reverse transcriptase (Gibco/Invitrogen Corp.) in a buffer consisting of 0.05 M Tris-HCl (pH 8.3), 75 mM KCl, and 3 mM MgCl2. After brief mixing, the reaction was incubated for 10 min at room temperature, followed by incubation at 37°C for 1 h. Finally, the reaction mixture was heated at 90°C for 5 min and either cooled on ice for 10 min or immediately stored at 4°C until use.
PCR was performed by mixing 1 μl of the appropriate dilution of cDNA (empirically determined for each gene to give product in the linear range), 0.5 μg each of forward and reverse primer, 2.5 U of Taq polymerase (Sigma, St. Louis, Mo.), and 0.1% Triton X-100 in EasyStart Micro50 PCR tubes and subjecting the reaction mixture to the following reaction conditions: one repetition of 94°C for 5 min; 32 repetitions of 94°C for 1 min, gene-specific annealing temperature for 1 min, and 72°C for 2 min; and 1 repetition of 72°C for 5 min. Equivalent volumes of PCR product were applied to a 3% agarose gel and separated by gel electrophoresis in 1× TAE (0.02 M Tris, 0.02 M acetate, 0.001 M EDTA). Primer sequences used for amplification of specific genes by RT-PCR are shown in Table 2.
TABLE 2.
Sequences and melting temperatures (Tm) of primers used in RT-PCR
| Primer | Sequence | Tm (°C) |
|---|---|---|
| RTA3-F | 5′-CGAAAGCAAACCAAGTCCAT-3′ | 54.9 |
| RTA3-R | 5′-TACCAATCATTGCTGCATCC-3′ | 54.3 |
| IPF3121-F | 5′-TTGCCACCACCTAATCATCA-3′ | 55.3 |
| IPF3121-R | 5′-CAACAGGTTCCGTTTGTGTG-3′ | 55.6 |
| DDR48-F | 5′-TTTCGGTTTCGGTAAAGACG-3′ | 54.1 |
| DDR48-R | 5′-TGAGTCGGTCTTGGAGGAAC-3′ | 57.2 |
| EBP1-F | 5′-GCTTATGTCCCAACCACCAG-3′ | 56.7 |
| EBP1-R | 5′-CTGATGGCGCAATAAGAGGT-3′ | 56.1 |
| CDR1-F | 5′-GGCAATTAGTCAAGACTCTTCTTCAG-3′ | 57.2 |
| CDR2-F | 5′-CAGCTAGACGAAAAGCCATGG-3′ | 57.1 |
| CDR-Ra | 5′-CACCTGGTCTCATAATGGCATC-3′ | 56.7 |
| IFD1-F | 5′-CGACAATGGCTTCCGTACTT-3′ | 56.2 |
| IFD4-F | 5′-GATTGGGATCCAGTTGGAGA-3′ | 55.0 |
| IFD5-F | 5′-GGGAAGATTTTGATCCTGTTGATAG-3′ | 55.2 |
| LPG20-Rb | 5′-CACCAGCAATTGGAATCACA-3′ | 54.7 |
| IFD7-F | 5′-AGATGGTTTGGGTGAAGTCG-3′ | 56.1 |
| IFD7-R | 5′-CACCGGCAATTGGGACAACTCC-3′ | 62.1 |
| GRP2-F | 5′-ACATACCGCATCTCCATTCC-3′ | 55.6 |
| GRP2-R | 5′-TGGAACCAACATAACCAGCA-3′ | 55.7 |
| DPP1-F | 5′-TCAACCATTTGCACGACAAT-3′ | 54.6 |
| DPP1-R | 5′-TCAGGAGTTCCCTTTTGTGG-3′ | 55.7 |
| CRD2-F | 5′-GCTTGTTCTGCTGCTCAATG-3′ | 55.8 |
| CRD2-R | 5′-TTTCACCATCTCTAGTACCATCACA-3′ | 57.1 |
| INO1-F | 5′-GATTTCAAGGCCAAAAACGA-3′ | 53.0 |
| INO1-R | 5′-TTTGATACCAGCATCGACCA-3′ | 55.2 |
| IPF1222-F | 5′-CTCCAAAGGCAGTCCATCAT-3′ | 55.9 |
| IPF1222-R | 5′-TTGTGTTAGCATTGCCGTGT-3′ | 56.8 |
| OPI3-F | 5′-GGCACAAAATTTCCCACAAT-3′ | 53.4 |
| OPI3-R | 5′-GCAAACAATGCAATGGCTAA-3′ | 53.7 |
| MDR1-F | 5′-TTTTTGGGTGGATTCTTTGC-3′ | 53.0 |
| MDR1-R | 5′-CGGTGATGGCTCTCAATCTT-3′ | 55.8 |
| 18S rRNA-F | 5′-GCC AGC GAG TAT AAG CCT TG-3′ | 56.7 |
| 18S rRNA-R | 5′-AGG CCT CAC TAA GCC ATT CA-3′ | 57.3 |
The CDR-R primer is used with both the CDR1-F and CDR2-F primers.
The LPG20-R primer is used with IFD1-F, IFD4-F, and IFD5-F primers.
RESULTS AND DISCUSSION
Experimental design and global findings.
The development of microarray technology and the sequencing of the C. albicans genome makes possible a more complete identification of genes associated with drug resistance in C. albicans (Stanford C. albicans genome project, Stanford University [http://www-sequence.stanford.edu/group/candida]). In the present study, changes in the gene expression profile throughout a set of isogenic clinical isolates of fluconazole-susceptible and -resistant C. albicans were identified using a DNA array that is inclusive of virtually the entire C. albicans genome. The four isolates used in this study are from a set of 17 isogenic clinical isolates that have been extensively characterized (29, 39, 40, 53, 54). These isolates were chosen because they each represent previously identified molecular changes that correlate with increases in azole MIC (Table 1). The complete data set for all microarray experiments performed may be found at http://dcp.utmem.edu/drogers/. It is important to note that, while the genes identified in the present study are differentially expressed in association with the acquisition of azole resistance, some of these changes in gene expression may be unique to the specific isolate and not necessarily relevant to the azole resistance phenotype. By our criteria, we identified a total of 32 genes to be up-regulated and 14 genes to be down-regulated between isolates 2-79 and 12-99 (Fig. 1A). These included genes previously shown to be up-regulated (CDR1, CDR2, CaMDR1, ERG2, GPX1, RTA3, IFD5, IPF5987, and CRD2) and down-regulated (FET34) in this series (29, 41, 53). Selected genes were chosen for confirmation of differential expression by RT-PCR (Fig. 1B). Consistent with our previous findings, we did not observe a change in expression for ERG11 between isolates 2-79 and 12-99 (average of 1.24-fold change in expression). As isolate 12-99 maintained its azole resistance phenotype, this finding suggests a limited role for the overexpression of ERG11 in this process. Indeed, a recent study found only a 1.2- to 2.3-fold change in expression of ERG11 between the initial and final isolates in this series (29).
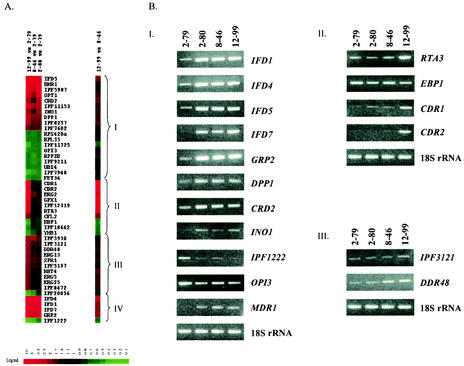
FIG. 1.
(A) Cluster images showing genes differentially expressed in this series of isolates. Red represents genes that are up-regulated, and green represents genes that are down-regulated. Genes shown in both cluster images appear in the same order. Genes are grouped as being coordinately differentially expressed with CaMDR1 (I), CDR genes (II), neither CaMDR1 nor CDR genes (III), or both CaMDR1 and CDR genes (IV). (B) Confirmation of differential expression by RT-PCR of select genes found to be differentially expressed by cDNA microarray analysis. RT-PCR of C. albicans 18S rRNA was performed as a control.
Among the up-regulated genes were seven that have homologs in Saccharomyces cerevisiae that were among those found previously to be up-regulated in the activated mutants of the transcription factors Pdr1p or Pdr3p, the PDR1-3 mutant and PDR3-7 mutant, respectively (7, 34). These were CDR1 and CDR2 (homologs of ScPDR5), IFD1, IFD4, IFD5, and IFD7 (homologs of ScYPL088w), and GRP2 (homolog of ScGRE2 and ScYLL056c). A common feature of these S. cerevisiae genes is that they contain pleiotropic DREs in their promoters which are controlled by either Pdr1p and/or Pdr3p. The promoter region of the ScRTA1 also contains a pleiotropic DRE. This is of interest as the C. albicans ScRTA1 homolog, RTA3, was up-regulated in association with azole resistance in the present study. These similarities between gene expression profiles in azole resistance in S. cerevisiae and C. albicans lend insight into the transcriptional regulation of these genes.
Genes coordinately regulated with CDR1 and CDR2.
We found seven genes to be coordinately up-regulated and three genes to be coordinately down-regulated with the up-regulation of CDR1 and CDR2 (Table 3). Two of these (GPX1 and RTA3) were identified previously as being differentially expressed in this set of isolates (41). GPX1 has also been shown to be up-regulated in experimentally induced, azole-resistant C. albicans (4).
TABLE 3.
Differentially expressed genes which exhibit coordinate regulation with CDR1 and CDR2 in isolates 2-79 through 12-99 in stepwise acquisition of azole resistance
| Gene | Description | Fold change
|
|||
|---|---|---|---|---|---|
| 2-80 vs 2-79 | 8-46 vs 2-79 | 12-99 vs 2-79 | 12-99 vs 8-46 | ||
| CDR1a,c | Multidrug resistance protein | 1 | 0.9 | 9.2 | 11 |
| CDR2a,b,c | Multidrug resistance protein | 0.9 | 0.9 | 8.4 | 9.4 |
| ERG2a | C-8 sterol isomerase | 1.1 | 1.6 | 3.7 | 2.3 |
| GPX1a,b | Glutathione peroxidase | 1.4 | 1.2 | 3.2 | 2.7 |
| IPF12319 | Unknown function | 1.1 | 1.1 | 2.7 | 2.5 |
| RTA3a,c | Unknown function | 1.1 | 1.1 | 2.6 | 2.6 |
| CFL2 | Ferric reductase | 1.2 | 1.1 | 1.8 | 1.7 |
| IPF10662 | Unknown function | 0.8 | 0.9 | 0.6 | 0.6 |
| EBP1 | NADPH dehydrogenase | 0.8 | 0.9 | 0.5 | 0.6 |
| YHB1 | Flavohemoglobin | 1 | 0.8 | 0.5 | 0.6 |
Gene previously reported to be differentially expressed in this set of isolates.
Gene previously reported to be differentially expressed in experimental azole resistance in C. albicans.
Gene with homolog in S. cerevisiae identified previously as having PDRE in its promoter.
GPX1, up-regulated 3.2-fold in isolate 12-99, encodes glutathione peroxidase, a significant component of the glutathione and glutathione-dependent enzyme system. This system protects cells from environmental toxins and has been implicated in the resistance of tumor cells to anticancer agents, particularly those whose effects are mediated by free radicals (2, 8). Increased activity of this enzyme system is often observed in conjunction with increased activity of the ABC transporter Mdr1p in drug-resistant human cancer cells (3, 26).
RTA3, up-regulated 2.6-fold in isolate 12-99, is a gene of unknown function. However, as noted earlier, it is homologous to ScRTA1, which has been implicated in resistance to 7-aminocholesterol (47). Up-regulation of RTA3 may also contribute to the azole-resistant phenotype. Of note, the RTA3 homolog RTA2 (YOR49) was reported to be up-regulated in experimentally induced, azole-resistant isolates (4).
ERG2, up-regulated 3.7-fold in isolate 12-99, encodes C-8 sterol isomerase, an enzyme in the late stages of the ergosterol biosynthesis pathway responsible for the conversion of fecosterol to episterol. A relationship between the late stages of ergosterol biosynthesis and the activity of the S. cerevisiae homolog of Cdr1p, Pdr5p, has been reported. Disruptions in ScERG6, ScERG2, ScERG3, and ScERG4 each reduced the ability of Pdr5p to confer drug resistance (18). Up-regulation of ERG2 could therefore serve to provide membrane conditions optimal for Cdr1p and Cdr2p activity in C. albicans. Furthermore, up-regulation of ERG2 could potentially compensate for a partial inhibition of lanosterol demethylase by an azole antifungal agent. Theoretically, any lanosterol successfully converted to ignosterol could more efficiently be further converted to ergosterol.
EBP1, down-regulated twofold (0.5) in isolate 12-99, is homologous to the ScOYE2 gene which encodes a flavoprotein that acts as an NADPH oxidoreductase, although its physiological role is unknown (35).
Genes coordinately regulated with CaMDR1.
We found 14 genes up-regulated and 10 genes down-regulated in coordination with the up-regulation of CaMDR1 (Table 4). Four of these (IFD5, CRD2, and FET34) were identified previously as being differentially expressed in this set of isolates (41).
TABLE 4.
Differentially expressed genes which exhibit coordinate regulation with CaMDR1 in isolates 2-79 through 12-99 in stepwise acquisition of azole resistance
| Gene | Description | Fold change
|
|||
|---|---|---|---|---|---|
| 2-80 vs 2-79 | 8-46 vs 2-79 | 12-99 vs 2-79 | 12-99 vs 8-46 | ||
| IFD4c,d | Putative aryl-alcohol dehydrogenase | 11.8 | 10.3 | 20.5 | 2 |
| IFD1c,d | Putative aryl-alcohol dehydrogenase | 17.6 | 13 | 20 | 1.6 |
| IFD5c | Putative aryl-alcohol dehydrogenase | 12.6 | 9.3 | 13.6 | 1.6 |
| IFD7c,d | Putative aryl-alcohol dehydrogenase | 5.4 | 5.6 | 8.7 | 1.6 |
| MDR1a,b | Benomyl resistance protein | 3.8 | 5.8 | 5.4 | 0.9 |
| GRP2c,d | Reductase | 2.8 | 2.5 | 4.1 | 1.7 |
| IPF5987b | Unknown function | 3.4 | 2.4 | 3 | 1.3 |
| IPF11153 | Unknown function | 2 | 2 | 2.5 | 1.3 |
| OPT1 | Oligopeptide transporter | 3 | 1.8 | 2.5 | 1.4 |
| DPP1 | Diacylglycerol pyrophos- phate phosphatase | 1.7 | 1.8 | 2.1 | 1.4 |
| IPF8257 | Unknown function | 1.6 | 1.6 | 2.1 | 1.4 |
| CRD2a | Copper-binding metallo- thionein | 2.2 | 1.4 | 2 | 1.5 |
| INO1 | myo-inositol-1-pyrophos- phate synthase | 1.9 | 1.5 | 2 | 1.4 |
| IPF7602 | Oxidoreductase | 1.6 | 1.6 | 1.7 | 1.2 |
| RPP2B | Acidic ribosomal protein | 0.6 | 0.6 | 0.6 | 1.3 |
| RPL35 | Ribosomal protein | 0.7 | 0.6 | 0.6 | 1.1 |
| RPS620a | Unknown function | 0.7 | 0.6 | 0.6 | 1.1 |
| IPF9211 | Unknown function | 0.6 | 0.6 | 0.6 | 1.1 |
| OPI3 | Methylene-fatty-acyl- phospholipid synthase | 0.7 | 0.7 | 0.6 | 1 |
| UBI4 | Polyubiquitin | 0.6 | 0.6 | 0.6 | 1.1 |
| FET34a | Multicopper oxidase | 0.6 | 0.6 | 0.5 | 0.9 |
| IPF11725 | Unknown function | 0.7 | 0.7 | 0.5 | 0.8 |
| IPF7940 | Unknown function | 0.6 | 0.5 | 0.5 | 0.9 |
| IPF1222d | Similar to superoxide dismutase | 0.5 | 0.4 | 0.3 | 0.6 |
Gene previously reported to be differentially expressed in this set of isolates.
Gene previously reported to be differentially expressed in experimental azole resistance in C. albicans.
Gene with homolog in S. cerevisiae identified previously as having PDRE in its promoter.
Gene which is further differentially expressed coordinately with the up-regulation of CDR genes.
Several genes involved in phospholipid biosynthesis and metabolism were differentially expressed concomitantly with the overexpression of CaMDR1. DPP1, up-regulated from 1.7- to almost 2.1-fold in isolates 2-80 through 12-99, encodes a diacylglycerol pyrophosphate phosphatase whose homolog is involved in phospholipids metabolism in S. cerevisiae. This enzyme catalyzes the dephosphorylation of phosphatidate, leading to the availability of diacylglycerol for the synthesis of phosphatidylethanolamine and phosphatidylcholine (50). INO1, up-regulated from 1.5- to 2-fold in isolates 2-80 through 12-99, encodes inositol-1-phosphate synthase, a key enzyme in the synthesis of inositol for phosphotidylinositol synthesis (48). OPI3, down-regulated 1.7-fold (0.6) in isolates 2-80 through 12-99, encodes phosphatidylethanolamine N-methyltransferase, an enzyme involved in the biosynthesis of phosphatidylcholine (9). Up-regulation of DPP1 and INO1 and down-regulation of OPI3 is suggestive of altered phospholipid metabolism associated with azole resistance. Indeed, alterations in phospholipid content has been associated with this phenotype (15, 16, 24, 27), however, none of these studies employed matched sets of clinical isolates. Closer examination of the phospholipid content of this and other matched sets of clinical isolates representing the acquisition of azole resistance is warranted.
FET34, down-regulated 1.7- (0.6) to 3-fold (0.5) in isolates 2-80 through 12-99, is required for FTR1-mediated iron uptake in S. cerevisiae (41). Previous studies have indicated a relationship between iron accessibility and azole resistance in C. albicans. In addition to studies that have demonstrated a synergistic effect of lactoferrin (or related compounds) and fluconazole on fungal cell killing (25, 30), Wakabayashi and colleagues showed that clinically isolated azole-resistant strains of C. albicans were more susceptible to lactoferrin treatment than azole-susceptible strains (52). Whether down-regulation of FET34 in fluconazole-resistant isolates is responsible for enhanced lactoferrin susceptibility remains uncertain. Further studies should be performed to determine what role down-regulation of these iron transport genes has in fluconazole resistance.
Interestingly, several of these genes appeared to be coordinately regulated with both the CDR genes and CaMDR1. IFD1, IFD4, IFD5, and IFD7, up-regulated from 5.4- to 20.5-fold in isolates 2-80 through 12-99, are members of a family of homologs of S. cerevisiae YPL088W, a putative alcohol dehydrogenase/oxidoreductase. YPL088W was recently shown to contain a DRE in its promoter which leads to induction of mRNA expression upon estradiol treatment of C. albicans (7). This DRE was shared by only three other genes, including CDR1 and CDR2. While these genes were coordinately differentially expressed with CaMDR1, it is possible that a trans regulatory factor for CDR1 and CDR2 contributes to their enhanced overexpression.
Likewise, GRP2, up-regulated from 2.8- to 4.1-fold in isolates 2-80 through 12-99, is a homolog of ScGRE2, which encodes a putative reductase with similarity to plant dihydroflavonol-4-reductases (14). While ScGRE2 is responsive to osmotic, ionic, oxidative, and heat stresses, its function is unknown. CRD2, up-regulated up to twofold in isolates 2-80 through 12-99, encodes a copper-binding metallothionein which also plays a role in reducing oxidative stress (51). Copper is essential for enzymes involved in a multitude of biological processes such as respiration, free radical destruction, and iron homeostasis. Like glutathione peroxidase, up-regulation of these genes in the resistant cell may compensate for azole-induced susceptibility to oxidative stress.
Miconazole and fluconazole have been shown to induce the production of endogenous reactive oxygen species (ROS) in Candida species in a dose-dependent fashion (23). Both azole-induced ROS and Candida cell death was attenuated by the antioxidant pyrrolidinedithiocarbamate. Furthermore, in multiple clinical isolates, azole-induced ROS production inversely correlated with the MIC. These data suggest a role for ROS production in the mechanism of action of the azole antifungal agents. Indeed, isolates of C. glabrata lacking the ERG11 gene have been shown to be more susceptible to oxidative effects of neutrophils and H2O2 than isolates with ERG11 intact (17). It is therefore possible that pharmacological inhibition of the ERG11 gene product, lanosterol demethylase, with an azole antifungal agent may produce a similar effect. This suggests that imparting to the fungal cell such enhanced susceptibility to oxidative damage may be part of the mechanism of action of the azole antifungals. The common theme of up-regulation of genes involved in the oxidative stress response in the present study support this hypothesis.
IPF1222, down-regulated from 2- (0.5) to 3.3-fold (0.3) in isolates 2-80 through 12-99, is a homolog of the SOD1 gene in S. cerevisiae. Three superoxide dismutase (SOD) genes have been identified in C. albicans; those encoding the copper- and zinc-containing (Cu/Zn) SOD, SOD1, and the manganese-containing (Mn) SODs, SOD2 and SOD3. IPF1222 likely encodes an additional uncharacterized Cu/Zn SOD. While S. cerevisiae Sod1p has been shown to have a role in protecting the mitochondrion against oxidative damage (51), deficiency in Sod1p appears to be beneficial for the acquisition of heat and ethanol tolerance (38).
Genes not coordinately regulated with known resistance genes.
The up-regulation of 10 genes and down-regulation of 1 gene was not correlated with the expression of either CDR1 and CDR2 nor with CaMDR1 (Table 5). IPF3121, up-regulated 1.8-fold in isolate 12-99, is homologous to the RME1 gene in S. cerevisiae which encodes a zinc finger protein involved in the negative regulation of meiosis (46). DDR48, up-regulated 1.8-fold in isolate 12-99, is homologous to a gene in S. cerevisiae that encodes a DNA damage-inducible protein. In S. cerevisiae, its expression has been shown to be responsive to cellular stresses such as cadmium exposure and osmotic stress (31, 32). Other genes in this category include the ergosterol biosynthesis genes ERG5, ERG13, and ERG25, up-regulated 1.6-, 1.6-, and 1.7-fold, respectively. Both ERG5 and ERG25 have been shown to be responsive to azole treatment in vitro in both S. cerevisiae and C. albicans (1, 5).
TABLE 5.
Differentially expressed genes not coordinately regulated with known resistance genes in stepwise acquisition of azole resistance
| Gene | Description | Fold change
|
|||
|---|---|---|---|---|---|
| 2-80 vs 2-79 | 8-46 vs 2-79 | 12-99 vs 2-79 | 12-99 vs 8-46 | ||
| IPF3930 | Unknown function | 1.4 | 2 | 1.8 | 1 |
| IPF3121 | Unknown function | 1.4 | 1.5 | 1.8 | 1.3 |
| DDR48 | Stress response protein | 1.2 | 1.4 | 1.8 | 1.4 |
| ERG13 | 3-Hydroxy-3-methylglutaryl coenzyme A synthase | 1 | 1.4 | 1.7 | 1.3 |
| ZPR1 | Zinc finger protein | 1.4 | 1.5 | 1.7 | 1.3 |
| IPF5197 | Unknown function | 1.3 | 1.4 | 1.7 | 1.2 |
| RBT4 | Repressed by TUP1 protein | 1.3 | 1.2 | 1.7 | 1.4 |
| ERG5 | C-22 sterol desaturase | 1.1 | 1.3 | 1.6 | 1.4 |
| ERG25 | C-4 sterol methyl oxidase | 1 | 1.3 | 1.6 | 1.4 |
| IPF8472 | Unknown function | 1.4 | 1.3 | 1.6 | 1.3 |
| IPF20056 | Unknown function | 0.8 | 0.5 | 0.5 | 0.9 |
Azole resistance mechanisms acquired by isolate 8-46 in this series include the acquisition of a point mutation and loss of allelic variation in the ERG11 gene. While there is no evidence that the two are directly linked, it is possible that changes in gene expression not correlating with the expression of CDR genes or CaMDR1 are in some way reflective of alterations in the ERG11 gene.
Conclusions.
This study is the first to use global microarray analysis to characterize changes in gene expression throughout the stepwise acquisition of azole antifungal resistance in clinical isolates of C. albicans. To this end we have identified the differential expression of genes previously shown to be differentially expressed in both experimentally induced and clinically acquired azole resistance, as well as genes whose differential expression was found for the first time to be associated with this process. Furthermore, by examining the expression profiles of serial isolates in a series that represents almost all known mechanisms of azole resistance, it was possible to group these genes by their coordinate regulation with that of other known resistance genes. Further study of multiple matched sets of azole-susceptible and -resistant C. albicans isolates will substantiate these associations and help pinpoint the most-common and critical mechanisms of azole resistance. Investigation into the specific contribution of these genes to the azole-resistant phenotype is warranted. Such studies will lend insight into the regulatory mechanisms that govern the coordinate expression of these genes and provide new targets for the development of pharmacological strategies to circumvent azole antifungal resistance and improve the therapeutic index of this class of antifungal agents.
Acknowledgments
This work was supported by a 2001-2002 Society of Infectious Diseases Pharmacists/Pfizer Research Award.
We thank Spencer Redding and Theodore White for kindly providing the isolates used in this study and Russ Lewis for performing susceptibility testing.
REFERENCES
- 1.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, S. M., and C. R. Wolf. 1991. The role of glutathione-dependent enzymes in drug resistance. Pharmacol. Ther. 51:139-154. [DOI] [PubMed] [Google Scholar]
- 3.Buser, K., F. Joncourt, H. J. Altermatt, M. Bacchi, A. Oberli, and T. Cerny. 1997. Breast cancer: pretreatment drug resistance parameters (GSH-system, ATPase, P-glycoprotein) in tumor tissue and their correlation with clinical and prognostic characteristics. Ann. Oncol. 8:335-341. [DOI] [PubMed] [Google Scholar]
- 4.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 7.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 8.Doroshow, J. H., S. Akman, F. F. Chu, and S. Esworthy. 1990. Role of the glutathione-glutathione peroxidase cycle in the cytotoxicity of the anticancer quinones. Pharmacol. Ther. 47:359-370. [DOI] [PubMed] [Google Scholar]
- 9.Dowd, S. R., M. E. Bier, and J. L. Patton-Vogt. 2001. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J. Biol. Chem. 276:3756-3763. [DOI] [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feigal, D. W., M. H. Katz, D. Greenspan, J. Westenhouse, W. Winkelstein, Jr., W. Lang, et al. 1991. The prevalence of oral lesions in HIV-infected homosexual and bisexual men: three San Francisco epidemiological cohorts. AIDS 5:519-525. [DOI] [PubMed] [Google Scholar]
- 12.Franz, R., M. Ruhnke, and J. Morschhäuser. 1999. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses 42:453-458. [DOI] [PubMed] [Google Scholar]
- 13.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhäuser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garay-Arroyo, A., and A. A. Covarrubias. 1999. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 15:879-892. [DOI] [PubMed] [Google Scholar]
- 15.Hitchcock, C. A., K. J. Barrett-Bee, and N. J. Russell. 1986. The lipid composition of azole-sensitive and azole-resistant strains of Candida albicans. J. Gen. Microbiol. 132:2421-2431. [DOI] [PubMed] [Google Scholar]
- 16.Hitchcock, C. A., K. J. Barrett-Bee, and N. J. Russell. 1987. The lipid composition and permeability to azole of an azole- and polyene-resistant mutant of Candida albicans. J. Med. Vet. Mycol. 25:29-37. [DOI] [PubMed] [Google Scholar]
- 17.Kan, V. L., A. Geber, and J. E. Bennett. 1996. Enhanced oxidative killing of azole-resistant Candida glabrata strains with ERG11 deletion. Antimicrob. Agents Chemother. 40:717-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur, R., and A. K. Bachhawat. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145:809-818. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, S. L., A. Arnoldi, and D. E. Kelly. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, et al. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 21.Klein, R. S., C. A. Harris, C. B. Small, B. Moll, M. Lesser, and G. H. Friedland. 1984. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N. Engl. J. Med. 311:354-358. [DOI] [PubMed] [Google Scholar]
- 22.Klepser, M. E., D. Malone, R. E. Lewis, E. J. Ernst, and M. A. Pfaller. 2000. Evaluation of voriconazole pharmacodynamics using time-kill methodology. Antimicrob. Agents Chemother. 44:1917-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, D., K. Kondo, N. Uehara, S. Otokozawa, N. Tsuji, A. Yagihashi, and N. Watanabe. 2002. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob. Agents Chemother. 46:3113-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohli, A., Smriti, K. Mukhopadhyay, A. Rattan, and R. Prasad. 2002. In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 46:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuipers, M. E., H. G. de Vries, M. C. Eikelboom, D. K. Meijer, and P. J. Swart. 1999. Synergistic fungistatic effects of lactoferrin in combination with antifungal drugs against clinical Candida isolates. Antimicrob. Agents Chemother. 43:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, W. P., C. L. Lee, and H. C. Lin. 1996. Glutathione S-transferase and glutathione peroxidase are essential in the early stage of adriamycin resistance before P-glycoprotein overexpression in HOB1 lymphoma cells. Cancer Chemother. Pharmacol. 38:45-51. [DOI] [PubMed] [Google Scholar]
- 27.Loffler, J., H. Einsele, H. Hebart, U. Schumacher, C. Hrastnik, and G. Daum. 2000. Phospholipid and sterol analysis of plasma membranes of azole-resistant Candida albicans strains. FEMS Microbiol. Lett. 185:59-63. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyons, C. N., and T. C. White. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minn, Y., E. Brummer, and D. A. Stevens. 1997. Effect of iron on fluconazole activity against Candida albicans in presence of human serum or monocyte-derived macrophages. Mycopathologia 138:29-35. [DOI] [PubMed] [Google Scholar]
- 31.Miralles, V. J., and R. Serrano. 1995. A genomic locus in Saccharomyces cerevisiae with four genes up-regulated by osmotic stress. Mol. Microbiol. 17:653-662. [DOI] [PubMed] [Google Scholar]
- 32.Momose, Y., and H. Iwahashi. 2001. Bioassay of cadmium using a DNA microarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ. Toxicol. Chem. 20:2353-2360. [DOI] [PubMed] [Google Scholar]
- 33.Morschhäuser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240-248. [DOI] [PubMed] [Google Scholar]
- 34.Nawrocki, A., S. J. Fey, A. Goffeau, P. Roepstorff, and P. M. Larsen. 2001. The effects of transcription regulating genes PDR1, pdr1-3 and PDR3 in pleiotropic drug resistance. Proteomics 1:1022-1032. [DOI] [PubMed] [Google Scholar]
- 35.Niino, Y. S., S. Chakraborty, B. J. Brown, and V. Massey. 1995. A new old yellow enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 270:1983-1991. [DOI] [PubMed] [Google Scholar]
- 36.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, et al. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, et al. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira, M. D., E. C. Eleutherio, and A. D. Panek. 2001. Acquisition of tolerance against oxidative damage in Saccharomyces cerevisiae. BMC Microbiol. 1:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaller, M. A., J. Rhine-Chalberg, S. W. Redding, J. Smith, G. Farinacci, A. W. Fothergill, and M. G. Rinaldi. 1994. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J. Clin. Microbiol. 32:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redding, S., J. Smith, G. Farinacci, M. Rinaldi, A. Fothergill, J. Rhine-Chalberg, and M. Pfaller. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin. Infect. Dis. 18:240-242. [DOI] [PubMed] [Google Scholar]
- 41.Rogers, P. D., and K. S. Barker. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans using cDNA microarray analysis. Antimicrob. Agents Chemother. 46:3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruhnke, M., A. Eigler, I. Tennagen, B. Geiseler, E. Engelmann, and M. Trautmann. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J. Clin. Microbiol. 32:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanglard, D. 2002. Resistance of human fungal pathogens to antifungal drugs. Curr. Opin. Microbiol. 5:379-385. [DOI] [PubMed] [Google Scholar]
- 44.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu, M., W. Li, P. A. Covitz, M. Hara, H. Shindo, and A. P. Mitchell. 1998. Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res. 26:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soustre, I., Y. Letourneux, and F. Karst. 1996. Characterization of the Saccharomyces cerevisiae RTA1 gene involved in 7-aminocholesterol resistance. Curr. Genet. 30:121-125. [DOI] [PubMed] [Google Scholar]
- 48.Sreenivas, A., M. J. Villa-Garcia, S. A. Henry, and G. M. Carman. 2001. Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase C. J. Biol. Chem. 276:29915-29923. [DOI] [PubMed] [Google Scholar]
- 49.Sturtz, L. A., K. Diekert, L. T. Jensen, R. Lill, and V. C. Culotta. 2001. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276:38084-38089. [DOI] [PubMed] [Google Scholar]
- 50.Toke, D. A., W. L. Bennett, J. Oshiro, W. I. Wu, D. R. Voelker, and G. M. Carman. 1998. Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 273:14331-14338. [DOI] [PubMed] [Google Scholar]
- 51.Viarengo, A., B. Burlando, N. Ceratto, and I. Panfoli. 2000. Antioxidant role of metallothioneins: a comparative overview. Cell. Mol. Biol. 46:407-417. [PubMed] [Google Scholar]
- 52.Wakabayashi, H., T. Okutomi, S. Abe, H. Hayasawa, M. Tomita, and H. Yamaguchi. 1998. Enhanced anti-Candida activity of neutrophils and azole antifungal agents in the presence of lactoferrin-related compounds. Adv. Exp. Med. Biol. 443:229-237. [DOI] [PubMed] [Google Scholar]
- 53.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14alpha demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wirsching, S., S. Michel, G. Kohler, and J. Morschhäuser. 2000. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]