Abstract
The heat shock response, which is accompanied by a rapid and robust upregulation of heat shock proteins (Hsps), is a highly conserved protection mechanism against protein-damaging stress. Hsp induction is mainly regulated at transcriptional level by stress-inducible heat shock factor 1 (HSF1). Upon activation, HSF1 trimerizes, binds to DNA, concentrates in the nuclear stress granules, and undergoes a marked multisite phosphorylation, which correlates with its transcriptional activity. In this study, we show that HSF1 is modified by SUMO-1 and SUMO-2 in a stress-inducible manner. Sumoylation is rapidly and transiently enhanced on lysine 298, located in the regulatory domain of HSF1, adjacent to several critical phosphorylation sites. Sumoylation analyses of HSF1 phosphorylation site mutants reveal that specifically the phosphorylation-deficient S303 mutant remains devoid of SUMO modification in vivo and the mutant mimicking phosphorylation of S303 promotes HSF1 sumoylation in vitro, indicating that S303 phosphorylation is required for K298 sumoylation. This finding is further supported by phosphopeptide mapping and analysis with S303/7 phosphospecific antibodies, which demonstrate that serine 303 is a target for strong heat-inducible phosphorylation, corresponding to the inducible HSF1 sumoylation. A transient phosphorylation-dependent colocalization of HSF1 and SUMO-1 in nuclear stress granules provides evidence for a strictly regulated subnuclear interplay between HSF1 and SUMO.
The heat shock response is an evolutionarily well-conserved cellular stress response mechanism that is characterized by the elevated synthesis and accumulation of heat shock proteins (Hsps). Hsps are molecular chaperones involved in protein folding and maintenance of protein homeostasis, and a robust increase in Hsp levels is essential for survival when cells are exposed to various protein-damaging stresses (for reviews see references 15 and 34). The stress-inducible expression of Hsp-encoding genes is regulated by a family of heat shock transcription factors (HSFs), which bind to the heat shock element (HSE) in the promoter of heat shock genes and stimulate their transcription (for reviews see references 37 and 62). Three members of the HSF family, HSF1, HSF2, and HSF4, have been identified in mammalian species (41, 47, 54, 56). HSF1, the vertebrate homologue of the single HSF found in the yeast and the fly, is the major stress-responsive family member, since no other HSF is able to functionally substitute for HSF1 or to rescue the heat shock response in HSF1-deficient cells or mice (35, 44, 64).
The elevated synthesis of Hsps upon heat shock is caused by a multistep activation of HSF1 (for a review see reference 45). Under normal growth conditions, inactive HSF1 is distributed throughout the cell as a monomer that is constitutively phosphorylated on certain serine residues. Upon activation, HSF1 undergoes several modifications, such as trimerization and localization to specific nuclear structures, called nuclear stress granules (for a review see reference 25). Independently of the monomer-to-trimer transition, which is required for DNA binding, phosphorylation of HSF1 is strongly induced during heat shock. Although the level of HSF1 bulk phosphorylation correlates well with its transcriptional activity, the data on phosphorylation-mediated regulation of HSF1 have remained relatively scattered. So far, four phosphorylated serine residues (S230, S303, S307, and S363) have been identified. In reporter assays, the constitutive phosphorylation of S303, S307, and S363 has been shown to repress the transactivation capacity of HSF1 under normal growth conditions (5, 8, 31, 32). However, kinetic analysis of the phosphorylation status of these sites during the HSF1 activation-deactivation cycle is missing. In contrast to the repressive role of S303, S307, and S363, S230 was recently demonstrated to be inducibly phosphorylated upon heat shock and to positively contribute to the transcriptional activity of HSF1 (17). Since the majority of HSF1 phosphorylation sites are still unknown, the biological relevance of HSF1 phosphorylation in regulating various aspects of HSF1 functions remains to be established.
Posttranslational modification by sumoylation has been reported for a variety of proteins, including several transcriptional regulators, such as p53, c-Jun, c-Myb, AP-2, androgen receptor, promyelocytic leukemia protein (PML), and IκBα (3, 11, 46; for a review see reference 36). This modification involves covalent conjugation of a small ubiquitin-like modifier 1 (SUMO-1, also called GMP-1, PIC1, sentrin, Ubl1, Smt3c, and hSmt3) to one or several lysines of the substrate by an isopeptide bond (for a review see reference 36). SUMO-1 conjugation utilizes a unique E1-activating enzyme complex termed SAE1/SAE2 (Aos1/Uba2 [24]) and an E2-conjugating enzyme, Ubc9 (22), as well as a group of recently characterized E3 ligases, a family of protein-inhibitor-of-activated-STAT (PIAS) proteins (21, 23, 29, 33, 50, 55, 60), and nucleoporin RanBP2 (43). SUMO-1 modification has been indicated to have diverse substrate-specific functions. Sumoylation has been shown to act antagonistically to ubiquitination by enhancing protein stability, as exemplified by sumoylation of the NF-κB inhibitor IκBα (9). In addition to regulation of protein half-life, SUMO-1 modification may alter other properties of its targets and consequently their activities. In the case of p53, SUMO-1 modification has been reported to affect the transactivation capacity of p53 as well as its intracellular localization (12, 13, 39, 49, 55). SUMO-1 has also been shown previously to localize into PML nuclear bodies where it is covalently bound to the PML protein (10), which appears to be essential for the structural and functional integrity of PML nuclear bodies (20, 65). In addition to SUMO-1, the SUMO protein family has two other members, SUMO-2 and SUMO-3, which are highly homologous to each other and share about 50% sequence identity with SUMO-1 (for a review see reference 36). Although there is evidence that endogenous SUMO-2/3 would have, at least partially, distinct sets of substrates compared to those of SUMO-1 (51), the specificity of the individual SUMO family members remains to be elucidated.
Considering the many functions attributed to SUMO proteins, understanding the biological relevance of sumoylation requires further elucidation of the mechanisms that regulate SUMO conjugation. Phosphorylation has been proposed as an important regulatory mechanism of ubiquitination and acetylation (1, 2). Interestingly, phosphorylation of SUMO substrates, such as c-Jun, p53, IκBα, and PML, has been reported previously to inhibit their sumoylation (9, 39, 40). In this study, we examined whether inducible phosphorylation of human HSF1 is accompanied by other posttranslational modifications. Our results show that lysine 298, located in the regulatory domain of HSF1, functions as an in vivo sumoylation site, and importantly, this sumoylation occurs in conjunction with phosphorylation of nearby serine 303. Phosphorylation on S303 and sumoylation on K298 were markedly enhanced in response to stress stimuli, and HSF1 and SUMO-1 were transiently colocalized in the nuclear stress granules in a phosphorylation-dependent manner. This strict cooperativity provides, to our knowledge, the first evidence of phosphorylation as a positive regulator of sumoylation.
MATERIALS AND METHODS
Plasmid constructs.
The green fluorescent protein (GFP)-fused expression plasmids SUMO-1 WT and GA (46), His-SUMO-1 (kindly provided by Anne Dejean, Institut Pasteur, Paris, France [39]), and His-SUMO-2 (a kind gift from Hetti Poukka, University of Helsinki, Helsinki, Finland), as well as the Myc epitope-tagged human HSF1 expression plasmid (17), have been described previously. The point mutations were made by using the QuickChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing.
Cell culture and transient-transfection experiments.
Human K562 erythroleukemia cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and antibiotics (penicillin and streptomycin) in a humidified 5% CO2 atmosphere at 37°C. Mouse embryonic fibroblasts (MEFs), derived from a hsf1−/− mouse (hsf1−/− MEFs [35]), were maintained in Dulbecco's modified Eagle's medium containing 10% FCS, 10 mM minimal essential medium nonessential amino acids, 0.96 μl of β-mercaptoethanol per 100 ml, and antibiotics (penicillin and streptomycin). HeLa cells were grown in Dulbecco's modified Eagle's medium containing 5% FCS with supplements. A total amount of 20 or 30 μg of DNA was transfected into cells by electroporation, 250 V (975 μF) for K562 cells, 220 V (975 μF) for HeLa cells, and 280 V (975 μF) for hsf1−/− MEFs, with a Gene Pulser electroporator (Bio-Rad Laboratories). Cells were incubated at 37°C for 24 to 36 h prior to the experimental treatments. Heat shock was performed at 42°C for indicated time periods. The proteasome inhibitor MG132 (Peptide Institute, Inc.) was used at 20 μM for 5 h.
Immunoprecipitation and immunoblotting assays.
For in vivo immunoprecipitation experiments, the cell pellet from transiently transfected cells was resuspended in 200 μl of lysis buffer (25 mM HEPES, 100 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 20 mM β-glycerophosphate, 20 mM paranitrophenylphosphate, 100 μM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 1× Complete mini-protease inhibitor cocktail [Roche Diagnostics]) supplemented with 20 mM N-ethylmaleimide and 5 μM MG132, followed by centrifugation for 25 min at 15,000 × g at 4°C. Subsequent to protein extraction, 500 μg of total cell proteins was preincubated with 40 μl of a 50% slurry of protein G-Sepharose (Amersham) in TEG buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol) containing 150 mM NaCl and 0.1% Triton X-100 for 30 min at 4°C followed by brief centrifugation. The precleared cellular lysate was incubated with anti-c-Myc (9E10; Sigma), anti-Flag M2 (Sigma), or monoclonal anti-human HSF1 (Neomarkers) antibodies at room temperature for 30 min under rotation, after which 40 μl of a 50% slurry of protein G-Sepharose was added to the reaction mixture and incubated under rotation for 1 h or overnight at 4°C. After centrifugation, the Sepharose beads were washed four times with supplemented TEG buffer, and immunoprecipitated proteins were run on a sodium dodecyl sulfate (SDS)-8% polyacrylamide gel, transferred to nitrocellulose membrane (Protran nitrocellulose; Schleicher & Schuell), and immunoblotted with anti-SUMO-1 (anti-GMP-1) antibody (Zymed Laboratories, Inc.), anti-HSF1 (18), or phosphopeptide-specific antibody anti-p-S303/7 (this study; see below). For the input control, membranes were blotted with anti-HSF1 antibodies, and for controlling the protein levels, the membranes were reblotted with anti-Hsc70 SPA-815 or anti-Hsp90 SPA-835 antibodies (StressGen). For analysis of Hsp70 levels anti-Hsp70 antibody SPA-810 (StressGen) was used. Horseradish peroxidase-conjugated secondary antibodies were purchased from Promega and Amersham, and the immunocomplexes were visualized by enhanced chemiluminescence (ECL; Amersham).
Gel mobility shift assays and antibody perturbation.
Whole-cell extracts (15 to 30 μg) were incubated with a 32P-labeled oligonucleotide representing the proximal HSE of the human hsp70 promoter (38). The protein-DNA complexes were analyzed on a native 4% polyacrylamide gel as described previously (38). For the perturbation assay, antibodies were incubated in indicated dilutions with the cell lysates at room temperature for 15 min before the gel mobility shift assay.
In vivo 32P labeling and phosphopeptide mapping.
Plasmids encoding wild-type Myc-tagged HSF1 or mutants were transfected by electroporation 40 h prior to starting the labeling. For the labeling, cells were incubated in phosphate-free minimal essential medium supplemented with 10% dialyzed FCS and 0.3 mCi of [32P]orthophosphate/ml for 3 h before heat shock. After treatments cells were washed once with ice-cold phosphate-buffered saline (PBS) and lysed with 500 μl of lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 0.1% sodium deoxycholate, 1 mM EDTA, 100 μM sodium orthovanadate, 2 mM sodium fluoride, 1× Complete mini-protease inhibitor cocktail [Roche Diagnostics]). The lysates were cleared by centrifugation (15,000 × g, 10 min, 4°C). HSF1 was immunoprecipitated from the supernatants for 2 h by using 10 μg of anti-Myc antibody (9E10; Sigma) together with protein G-Sepharose (Amersham). After the Sepharose beads were washed six times with the lysis buffer, the precipitates were boiled with 3× Laemmli sample buffer and resolved on an SDS-8% polyacrylamide gel. Tryptic digestion and two-dimensional phosphopeptide mapping were performed as described previously (17). Briefly, the labeled HSF1 band was excised from the dried gel and digested with l-1-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma). The tryptic digest was applied on a cellulose thin-layer chromatography plate (Merck) and separated in two dimensions by electrophoresis and thin-layer chromatography. The phosphopeptides were visualized by autoradiography.
Generation of the S303/7 phosphopeptide antibody and Western blotting.
For preparation of the phosphoantibody, a peptide, KEEPPpSPPQpSPRVE, was synthesized, conjugated to hemocyanin, and injected into a rabbit. The antiserum was positively affinity purified by using the KEEPPpSPPQpSPRVE peptide immobilized on a HiTrap-N-hydroxysuccinimide column (Amersham). For immunoblot analysis nitrocellulose membranes were blocked in 5% milk-25 mM MOPS (morpholinepropanesulfonic acid)-125 mM NaCl (pH 7.3) for 2 h. After blocking membranes were rinsed with MOPS buffer supplemented with 0.3% Tween 20 (MOPS-TW) and incubated in 5% bovine serum albumin-MOPS-TW for 30 min. The primary antibody was added to the bovine serum albumin solution in a dilution of 1:1,000 and incubated overnight at 4°C with gentle shaking. The excess of primary antibody was rinsed with water for 10 min followed by three 10-min washes with MOPS-TW. The horseradish peroxidase-conjugated anti-rabbit antibody (Promega) was diluted 1:20,000 in 3% milk-MOPS-TW and incubated for 1 h, followed by three 10-min washes with MOPS-TW and detection by ECL (Amersham).
SUMO-1 conjugation in vitro.
Recombinant glutathione S-transferase (GST)-SUMOGG-1, GST-Ubc9, GST-SAE1, and GST-SAE2 (SAE1 and SAE2 plasmids kindly provided by Ronald Hay, University of Dundee, Dundee, United Kingdom) were produced and purified as described previously (33). In vitro translations of HSF1 mutants were performed with the TNT-coupled transcription-translation system (Promega) in the presence of [35S]methionine. The conjugation assay was performed as described previously (33). Briefly, the in vitro-translated product was incubated with GST-SUMOGG-1, GST-Ubc9, and the mixture of GST-SAE1 and GST-SAE2 in the reaction buffer (50 mM HEPES [pH 7.7], 100 mM NaCl, 10 mM MgCl2, 2 mM ATP, 1 mM dithiothreitol) at 30°C for 1 h. The reaction was terminated by addition of 3× Laemmli sample buffer and boiling. Samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by fluorography.
Immunofluorescence microscopy.
For immunolabeling, HeLa cells were grown on coverslips. After treatments the cells were immediately fixed and permeabilized by incubating them with ice-cold methanol for 6 min. After the fixation cells were washed three times for 5 min with PBS-0.05% Tween 20 (PBST), followed by blocking with 20% boiled normal goat serum (GS) in PBST for 1 h. Primary antibody incubations were made in 5% GS-PBST with dilutions of 1:500 for anti-Myc (Sigma), 1:500 for anti-HSF1 (Neomarkers), and 1:40 for anti-p-S303/7. The excess of primary antibodies was washed off for three washes of 5 min each with PBST. Secondary antibodies (goat anti-mouse Alexa 488 and Alexa 568 for anti-Myc or goat anti-rabbit Alexa 546 for anti-p-S303/7) were diluted 1:500 in 5% GS-PBST, and cells were incubated for 1 h followed by three washes and mounting with Vectashield supplemented with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories). Immunofluorescence was analyzed with a Leica DMR fluorescence microscope equipped with a digital Hamamatsu ORCA charge-coupled device camera.
RESULTS
HSF1 undergoes stress-inducible SUMO modification.
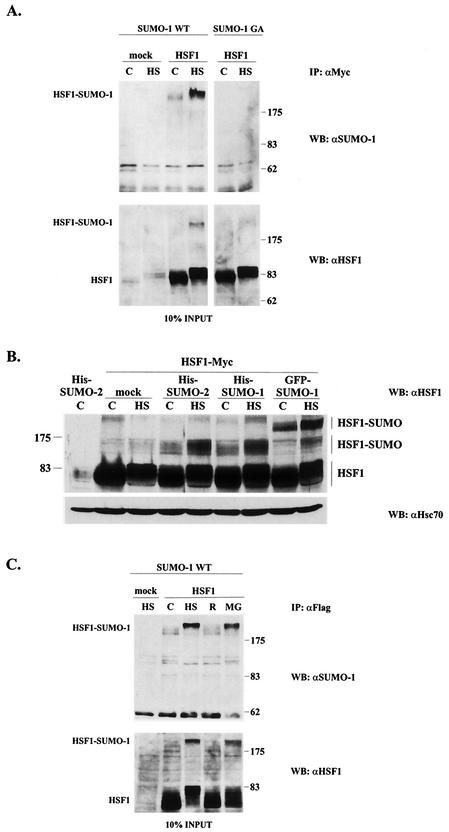
To investigate whether HSF1 undergoes sumoylation, Myc-tagged HSF1 together with GFP-fused SUMO-1 was transfected into K562 cells, whereafter the samples were immunoprecipitated with anti-Myc antibody and resolved by SDS-PAGE. Immunoblotting with anti-SUMO-1 antibody to detect SUMO-1-conjugated HSF1 revealed a band of approximately 200 kDa (Fig. 1A). SUMO-1-modified HSF1 was found both in untreated and in heat-shocked samples, but a 1-h heat shock at 42°C clearly increased the sumoylation of HSF1 (Fig. 1A). It is notable that the slower migration of HSF1 and HSF1-SUMO-1 upon heat shock was due to increased phosphorylation as previously shown (17, 53). As anticipated, upon coexpression of HSF1 with a conjugation-defective form of SUMO-1 (SUMO-1 GA), the HSF1-SUMO-1 conjugates were absent (Fig. 1A). The input controls in the lower panels were blotted with anti-HSF1 antibody to verify equal expression of HSF1. The high-molecular-weight band representing sumoylated HSF1 could also be detected from the input lysates.
FIG. 1.
HSF1 undergoes a heat-inducible modification by SUMO-1 and SUMO-2. (A) Human K562 erythroleukemia cells were transiently transfected with either empty vector (mock) or Myc-tagged human HSF1 (HSF1) together with the GFP-fused wild-type or conjugation-defective SUMO-1 (SUMO-1 WT and SUMO-1 GA, respectively), and after 24 h the cells were either left untreated (C) or exposed to heat shock at 42°C for 1 h (HS). Immunoprecipitation was performed with anti-Myc antibody, and the samples were analyzed by immunoblotting with anti-SUMO-1 antibody. The input was blotted with polyclonal anti-human HSF1 antibody. (B) HSF1 WT was cotransfected to K562 cells with either empty vector (mock), His-tagged SUMO-2 and SUMO-1, or GFP-fused SUMO-1. Cells were left untreated (C) or exposed to a 15-min heat shock at 42°C (HS). Samples were analyzed by anti-HSF1 Western blotting. Hsc70 was used as a loading control. (C) K562 cells overexpressing Flag-tagged HSF1 and wild-type GFP-SUMO-1 were subjected to a 1-h heat shock alone (HS), heat shock followed by a 3-h recovery (R), or a 5-h treatment with 20 μM proteasome inhibitor MG132 (MG). Immunoprecipitation was performed with anti-Flag antibody, and the samples were further analyzed as described for panel A. IP, immunoprecipitation; WB, Western blot. Numbers on right of panels A and C and on left of panel B are molecular masses in kilodaltons.
Since many SUMO-1 substrates can also be modified by SUMO-2 (30, 50, 52), we tested whether coexpressed SUMO-2 would be able to conjugate to HSF1. A similar slower-migrating HSF1-reactive broad band was detected in cells where His-tagged SUMO-1 or His-tagged SUMO-2 was coexpressed with HSF1 (Fig. 1B). Even without SUMO overexpression (mock), we were able to detect an HSF1-reactive band, corresponding to the mobility of His-SUMO-conjugated HSF1 (Fig. 1B). However, due to a low stoichiometry of the modification, the putative endogenous SUMO conjugate could not be further examined with the currently available antibodies. In the case of His-tagged SUMO-1 and SUMO-2, the modified HSF1 was detected at the molecular mass region of 120 to 170 kDa, corresponding to a >50-kDa decrease in the mobility, which is considerably more than the calculated molecular mass of a SUMO molecule (11 kDa). The mobility of sumoylated HSF1 was similar in several different cell lines (data not shown) and consistent with earlier studies of other SUMO substrates, showing a substantially bigger mobility shift than 11 kDa (33, 52, 59). Furthermore, the differences in electrophoretic mobilities between His-SUMO and GFP-SUMO-1 conjugates were bigger than anticipated based on the molecular mass of GFP (28 kDa) (Fig. 1B). We believe that the large increase in the apparent molecular mass is likely to derive from the presence of the conjugated SUMO polypeptide in the central region of HSF1, forming a branched protein structure and thereby leading to aberrant electrophoretic migration. Notably, migration of HSF1 on SDS-polyacrylamide gels is also strongly affected by phosphorylation, a mobility change that cannot be explained by the altered molecular weight (Fig. 1) (17, 53). In addition, gel conditions could also affect the mobility.
To examine whether the heat-induced increase in HSF1 sumoylation would persist after a return to normal temperature, heat-shocked cells were subjected to a recovery period of 3 h at normal temperature. As shown in Fig. 1C, the amount of HSF1 sumoylation returned to the control levels upon recovery, indicating that the heat-induced HSF1 sumoylation is a reversible event. Treatment of K562 cells with the proteasome inhibitor MG132 is known to induce the heat shock response, including phosphorylation and activation of HSF1 (18, 44). To study the effect of proteasome inhibition on HSF1 sumoylation, K562 cells coexpressing HSF1 and SUMO-1 were treated with MG132. As shown in Fig. 1C, MG132 treatment induced a similar SUMO-1 modification as did heat shock, indicating that SUMO modification of HSF1 is not unique to heat stress.
Lysine 298 is an in vivo SUMO modification site of HSF1.
In most proteins, SUMO modification occurs at a consensus sequence, ψKxE, including the target lysine and certain surrounding amino acids (ψ represents a hydrophobic residue and “x” represents any amino acid). Additional sequences may be required for optimal modification, and in fact, the presence of a nuclear localization signal has been reported to be of importance, as SUMO modification predominantly occurs in the nucleus (48). We analyzed the amino acid sequence of HSF1 and found three SUMO consensus sequences, i.e., K91, K126, and K298, and three sites with partial consensus, i.e., K150, K162, and K381 (Fig. 2A). After mutation of the lysines to arginines, the HSF1 mutants were coexpressed together with SUMO-1 in K562 cells and analyzed by immunoprecipitation. All mutants but one, K298R, were capable of forming SUMO-1 conjugates (Fig. 2B), suggesting that K298 is a major site for SUMO conjugation in vivo. Identical results were obtained with SUMO-2 (data not shown). As illustrated in Fig. 2C, the sumoylation target sequence on human HSF1 is conserved among vertebrates including the frog Xenopus laevis and the zebra fish Danio rerio, whereas HSF of the fruit fly Drosophila melanogaster does not contain a target sequence in this region.
FIG. 2.
Lysine 298 is a major in vivo sumoylation site on HSF1. (A) The amino acid sequence of HSF1 was analyzed in order to identify possible sumoylation sites corresponding to the consensus SUMO-1 modification sequence. Sequence motifs with perfect consensus are marked with open boxes (K91, K126, and K298), and sites with partial consensus are shaded (K150, K162, and K381). (B) The K→R mutants were coexpressed with wild-type GFP-SUMO-1 in K562 cells and subjected to a 1-h heat shock followed by immunoprecipitation analysis as described for Fig. 1A. The variation in the intensities of sumoylation of the mutants reflects differences between individual experiments. WT, wild type; IP, immunoprecipi- tation; WB, Western blot. Numbers on right are molecular masses in kilodaltons. (C) Alignment of the human HSF1 (hHSF1) SUMO-1 modification sequence in comparison to mouse HSF1 (mHSF1), rat HSF1 (rHSF1), chicken HSF1 (cHSF1), zebra fish D. rerio HSF1 (zHSF1), frog X. laevis HSF1 (XHSF1), and fruit fly D. melanogaster HSF (DrHSF). The SUMO-1 consensus sequence is boxed in the respective species. The alignment was made by using ClustalW multiple sequence alignment.
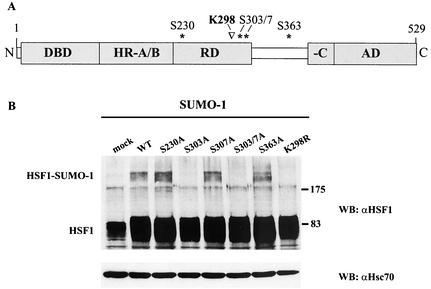
Sumoylation of HSF1 is inhibited by mutation of the phosphorylation site S303.
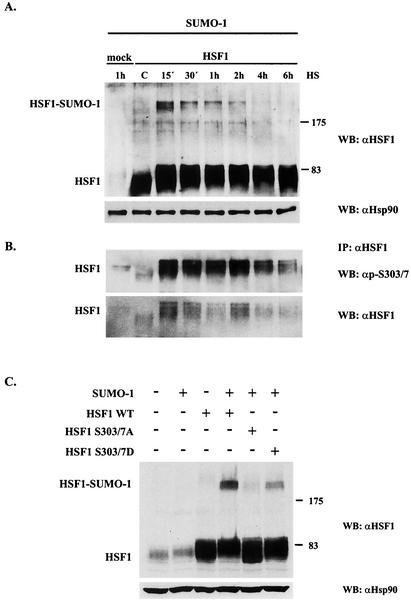
Since the HSF1 sumoylation site is located in the regulatory domain, which is essential for controlling the transcriptional activity of HSF1 and where several critical phosphorylation sites have been identified (Fig. 3A) (5, 14, 17, 31, 32, 42, 58, 66), we were prompted to study whether any cooperativity between phosphorylation and sumoylation of HSF1 existed. Mutants for all characterized HSF1 phosphorylation sites (S230A, S303A, S307A, and S363A) were tested for in vivo sumoylation by cotransfecting them together with SUMO-1 into K562 cells. Both S303A and S303/7A mutations efficiently prevented HSF1 sumoylation, since no sumoylated form of HSF1 was detected, whereas S307A and other mutants were sumoylated equally with the wild-type HSF1 (Fig. 3B). Also SUMO-2 conjugation was inhibited by the S303A mutation (data not shown). These results indicate that S303 is likely to be a critical determinant for HSF1 sumoylation.
FIG. 3.
Mutation of a phosphorylation site at serine 303 to alanine prevents HSF1 sumoylation. (A) Schematic presentation showing the location of K298 (▿) in the regulatory domain of HSF1 in the proximity of several critical phosphorylation sites (asterisks). The DNA-binding domain (DBD), N-terminal leucine zipper (HR-A/B), regulatory domain (RD), C-terminal leucine zipper (-C), and activation domain (AD) are indicated. (B) Myc-tagged S→A mutants of HSF1 phosphorylation sites were cotransfected with GFP-SUMO-1 into K562 cells followed by a 1-h heat shock, and cell lysates were resolved by SDS-PAGE. Western blot analysis was performed with anti-HSF1 antibody to detect the presence of a high-molecular-weight HSF1 band corresponding to sumoylated HSF1. Equal loading was confirmed by anti-Hsc70 immunoblotting. WT, wild type; WB, Western blot. Numbers on right are molecular masses in kilodaltons.
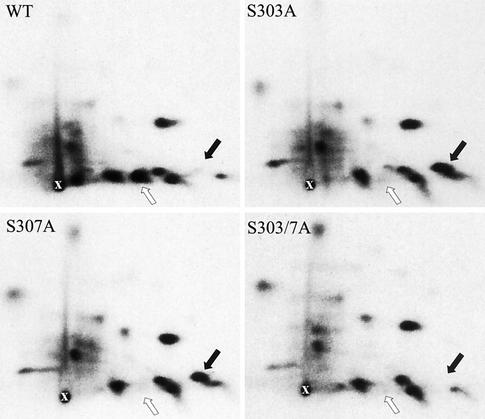
To further investigate whether phosphorylation of S303 was essential to prime HSF1 sumoylation, we proceeded to examine the phosphorylation status of S303 in K562 cells. This was particularly important, since cooperativity between sites S303 and S307 has previously been demonstrated by the finding that S303 can be phosphorylated by glycogen synthase kinase-3, but only when S307 is first phosphorylated by another kinase (4, 5). S→A mutants of serines 303 and 307 were transfected into K562 cells, in vivo 32P labeled, and analyzed by tryptic phosphopeptide mapping (Fig. 4). In the wild-type HSF1, the majority of phosphorylated molecules were phosphorylated on both S303 and S307 (white arrow), since the phosphopeptide corresponding to the single phosphorylated state was undetectable (black arrow). S303 was strongly phosphorylated in the S307A mutant, as demonstrated by the appearance of a spot corresponding to a single phosphorylated peptide (black arrow). Phosphorylation of S307 seems not to be needed to prime S303 phosphorylation, since the spot representing the single phosphorylated peptide (black arrow) in the S307A map was as strong as the respective spot in the S303A map. As expected, this spot disappeared when both S303 and S307 were mutated to alanine. Taken together, the inhibition of S303 phosphorylation in the S303A and S303/7A mutants (Fig. 4) corresponded to the lack of sumoylation in these mutants (Fig. 3B). Moreover, S303 was potently phosphorylated in the S307A mutant (Fig. 4), which also was a good substrate for SUMO-1 modification (Fig. 3B).
FIG. 4.
S303 is strongly phosphorylated in the wild-type and S307A mutant HSF1. To obtain two-dimensional tryptic phosphopeptide maps, wild-type (WT), S303A, S307A, or S303/7A HSF1 was transfected into K562 cells, after which the cells were in vivo labeled with [32P]orthophosphate and exposed to a heat shock (42°C, 1 h). The labeled HSF1 was digested with trypsin and resolved by two-dimensional mapping. The white arrow indicates phosphopeptide phosphorylated on both S303 and S307. The black arrow indicates phosphopeptides phosphorylated either on S303 or on S307. The starting points are indicated as “x.”
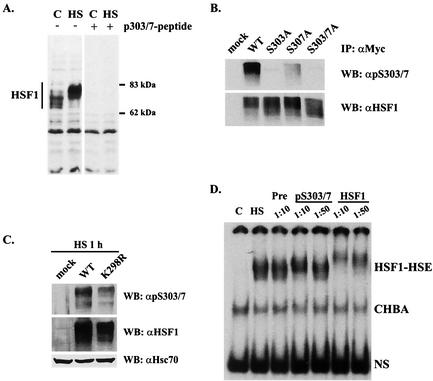
Characterization of S303/7 phosphoantibody.
To better characterize the dynamics and interplay of S303 phosphorylation and HSF1 sumoylation, we generated an antibody specific to the S303/7 phosphoepitope. This antibody recognized protein bands from control and heat-treated K562 lysates corresponding to the size of normal and hyperphosphorylated HSF1, respectively, which could be competed out by preincubating the antibody with the phosphopeptide antigen (Fig. 5A). To further evaluate the specificity of the phosphoantibody, the immunoprecipitated S→A mutants of S303 and S307 were analyzed by Western blotting. The phosphoantibody specifically recognized molecules that were phosphorylated on both S303 and S307, since only the wild-type HSF1 was efficiently detected with the phosphoantibody, whereas the anti-HSF1 antibody recognized the wild-type HSF1 and all the mutants equally well (Fig. 5B). Furthermore, the phosphoantibody recognized the K298R mutant transfected into hsf1−/− MEFs, indicating that the mutation of K298 did not inhibit phosphorylation of S303 and S307 (Fig. 5C). To elucidate whether HSF1 molecules phosphorylated on S303/7 could display binding activity to the HSE upon heat shock, an antibody perturbation assay was performed with K562 cells. As shown in Fig. 5D, with a 1:10 dilution of the phosphoantibody the HSF1-HSE complex was supershifted, indicating that HSF1 phosphorylated on S303/7 is capable of binding to HSE.
FIG. 5.
Characterization of an antibody specific to phosphorylated S303/7. (A) K562 cells were subjected to a heat shock (HS; 1 h at 42°C) or left untreated (C), and cell lysates were resolved by SDS-PAGE and blotted with anti-p-S303/7-HSF1 antibody. The specificity was verified by preincubating the antibody with a 500 M excess of free phosphopeptide (right panel). (B) K562 cells were transfected with Myc-tagged wild-type HSF1, S303A, S307A, or S303/7A, and the cells were subjected to a 1-h heat shock at 42°C. HSF1 was immunoprecipitated with anti-Myc antibody, and the immunoprecipitates were resolved by SDS-PAGE and blotted with anti-p-S303/7-HSF1 antibody. Equal amounts of HSF1 were controlled by blotting the membranes with anti-HSF1 antibody. (C) hsf1−/− MEFs were transfected with an empty plasmid (mock), wild-type HSF1, or K298R mutant. Cells were exposed to a 1-h heat shock, and cell lysates were resolved by SDS-PAGE and blotted with anti-p-S303/7, anti-HSF1, and anti-Hsc70 antibodies. (D) K562 cells were treated as indicated, and the cell lysates were preincubated with preimmune serum (Pre) or antibodies specific to phospho-S303/7-HSF1 or total HSF1. The HSE-binding HSF1 complex was analyzed by gel mobility shift assay. CHBA, constitutive HSE-binding activity; NS, nonspecific protein-DNA interaction; WT, wild type; IP, immunoprecipitation; WB, Western blot.
Sumoylation of HSF1 corresponds to heat-inducible phosphorylation of S303/7.
For a more detailed analysis of HSF1 sumoylation in relation to phosphorylation of S303/7, we performed a heat shock time course assay at 42°C. A prominent increase in HSF1 sumoylation was observed already at the 15-min time point, after which the level of sumoylation gradually decreased and returned to the control level by 4 h (Fig. 6A). Interestingly, phosphorylation on S303/7 also displayed a strong and rapid induction in response to heat stress as revealed by increased immunoreactivity of S303/7 phosphoantibody in the Western blot (Fig. 6B). The attenuation of HSF1 sumoylation, however, occurred more rapidly than the attenuation of S303/7 phosphorylation, which started to decrease only after 2 h and was still above the control level at 6 h of heat shock. It should be noted that the signal intensities of S303/7 phosphorylation and HSF1 sumoylation varied in different experiments but consistently displayed a close correlation. Further analysis with shorter time points indicated that inducible HSF1 sumoylation and S303/7 phosphorylation were induced already at 6 to 10 min and reached the maximal levels at 10 to 15 min of heat shock (data not shown).
FIG. 6.
Phosphorylation of S303/7 occurs concomitantly with sumoylation of HSF1. (A) K562 cells were cotransfected with Myc-tagged wild-type HSF1 and GFP-SUMO-1 and heat shocked for the indicated time periods. Cell lysates were resolved by SDS-PAGE and analyzed by immunoblotting with anti-HSF1 antibody to determine the level of HSF1 sumoylation. Equal loading was confirmed by anti-Hsp90 immunoblotting. (B) HSF1 was immunoprecipitated from the same samples as for panel A with a monoclonal anti-HSF1 antibody, and the immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-p-S303/7-HSF1 and anti-HSF1 antibodies. (C) Myc-tagged wild-type HSF1, S303/7A, or S303/7D was cotransfected with GFP-SUMO-1 into K562 cells followed by a 15-min heat shock at 42°C. Cell lysates were resolved by SDS-PAGE and analyzed by immunoblotting with anti-HSF1 antibody. Equal loading was confirmed by anti-Hsp90 immunoblotting. WT, wild type; WB, Western blot; IP, immunoprecipitation. Numbers on right of panels A and C are molecular masses in kilodaltons.
The inhibitory effect of the S303/7A mutant on HSF1 sumoylation was further tested with a negatively charged S303/7D mutant, the charge of which resembles that of phosphorylated S303/7. Unlike the S303/7A mutant, the S303/7D mutant was well modified by SUMO-1, although it did not reach the sumoylation level of wild-type HSF1 (Fig. 6C), perhaps due to the inability of aspartate residues to fully mimic the conformation of phosphorylated serine residues.
HSF1 S303/7D is preferentially sumoylated in vitro.
To better understand the mechanisms involved in the phosphorylation-mediated regulation of HSF1 sumoylation, we examined whether mimicking of S303 phosphorylation would affect HSF1 sumoylation also in a cell-free system. In vitro sumoylation assays were performed by incubating in vitro-translated 35S-labeled HSF1 mutants together with ATP and purified GST fusions of SAE1/SAE2 (E1), Ubc9 (E2), and SUMO-1. As shown in Fig. 7, HSF1 was sumoylated in the presence of SAE1/SAE2, Ubc9, and SUMO-1, but not if Ubc9 was absent or if lysine 298 was mutated. The mobility shift caused by attachment of the GST-tagged SUMO-1 was comparable to that detected for androgen receptor in vitro (Fig. 7) (46) and to that caused by GFP-SUMO-1 in vivo (Fig. 1). Intriguingly, mimicking of S303/7 phosphorylation by S303/7D mutation increased sumoylation of HSF1 in vitro. These results indicate that phosphorylation of serine 303 mediates its sumoylation-promoting effect by changing HSF1 conformation and thereby exposing the sumoylation site, although we cannot exclude the possibility of a phosphospecific binding partner in the reticulocyte lysate.
FIG. 7.
HSF1 S303/7D is preferentially sumoylated in vitro. In vitro-translated, 35S-labeled HSF1 mutants and androgen receptor (AR) were incubated with purified GST-SUMOGG-1 (SUMO-1) or GST-SAE1/GST-SAE2 (SAE1/SAE2) in the presence or absence of GST-Ubc9 as indicated. Reaction products were resolved by SDS-PAGE, and the signal was visualized by fluorography. Sumoylation is detected as the appearance of slower-migrating bands. WT, wild type.
Nonsumoylated HSF1 is capable of binding to HSE and inducing Hsp70 expression.
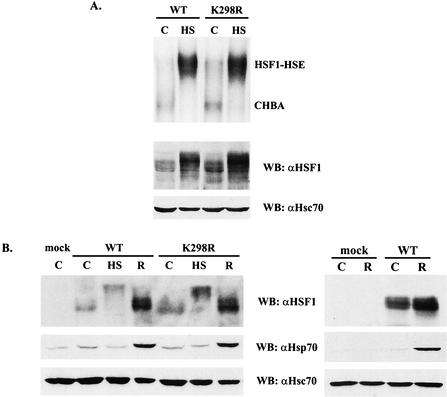
To establish a possible role of sumoylation on HSF1 DNA-binding activity, untreated or heat-shocked hsf1−/− MEFs transfected with either the wild-type HSF1 or the K298R mutant were analyzed by gel mobility shift assay. The K298R mutant was able to bind HSE upon heat shock similarly to the wild-type HSF1 (Fig. 8A), indicating that sumoylation is not essential for HSF1 DNA-binding activity. Since hsf1−/− MEFs do not display induction of Hsp70 in response to heat shock (Fig. 8B, right panel) (17, 35, 44), we transfected either the wild-type HSF1 or the K298R mutant into these cells and analyzed the rescue of Hsp70 induction upon heat shock. First, we confirmed that HSF1 was similarly sumoylated in hsf1−/− MEFs as in K562 cells and that the sumoylation was prevented by K298R mutation (data not shown). As shown in Fig. 8B, the wild-type and K298R HSF1, which were equally expressed and hyperphosphorylated in response to heat shock, were able to rescue the Hsp70 induction to the same extent.
FIG. 8.
The wild-type and K298R HSF1 rescue the heat shock response equally well in hsf1−/− MEFs. (A) Gel mobility shift assay (upper panel) was performed on hsf1−/− MEFs transfected with the wild-type or K298R mutant HSF1 and left untreated (C) or subjected to a 1-h heat shock (HS). The input lysates were resolved by SDS-PAGE and blotted with anti-HSF1 and anti-Hsc70 antibodies (lower panels). (B) hsf1−/− MEFs were transfected with empty vector (mock) or the wild-type or K298R mutant HSF1 and left untreated (C) or subjected to a 1-h heat shock (HS) or a 1-h heat shock followed by a 3-h recovery (R). Induction of the heat shock response was analyzed by Western blotting with anti-HSF1 and anti-Hsp70 antibodies. Anti-Hsc70 antibodies were used for loading control. Lack of Hsp70 induction in the absence of HSF1 (mock) is shown in the right panel. WT, wild type; WB, Western blot; CHBA, constitutive HSE-binding activity.
Transient colocalization of HSF1 and SUMO-1 to the nuclear stress granules at the onset of the heat shock response.
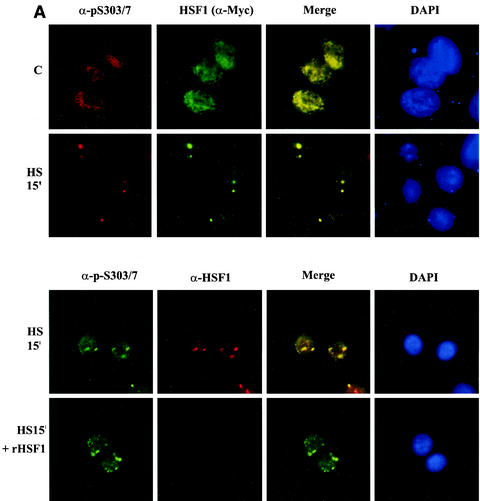
Finally, we addressed the questions whether phosphorylation on S303 has an impact on cellular localization of HSF1 and what the spatiotemporal relationship of HSF1 and SUMO-1 is in the dynamic formation of subnuclear structures upon heat stress. To study if HSF1 phosphorylated on S303/7 would form a subcellular population distinct from the total HSF1 pool, HeLa cells were transiently transfected with Myc-tagged HSF1 and double immunolabeled with antibodies specific to p-S303/7 and c-Myc. Under normal growth conditions, both the total HSF1 and the phosphoantibody-reactive HSF1 were localized diffusely in the nucleus (Fig. 9A, upper panel). Upon a 15-min heat shock, a major part of HSF1 molecules were localized to brightly stained irregular subnuclear structures called nuclear stress granules, which is in agreement with several earlier studies demonstrating stress-induced HSF1 granule formation in human cells (6, 16, 18, 26, 27, 28, 53). The phosphorylated HSF1 localized to the same granules as did the total HSF1 (Fig. 9A, upper panel), showing that HSF1 phosphorylated on S303/7 is indeed present in the nuclear stress granules. To verify the specificity of the phosphoantibody signal, we incubated recombinant human HSF1 together with the primary antibody, which blocked the function of the anti-HSF1 antibody but failed to affect anti-p-S303/7 immunoreactivity (Fig. 9A, lower panel).
FIG. 9.
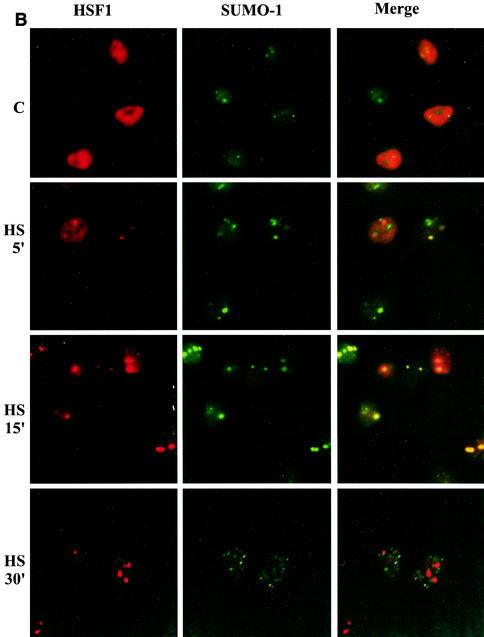
HSF1 and SUMO-1 transiently form cogranules at the onset of the heat shock response. (A) (Upper panel) HeLa cells were transiently transfected with HSF1-Myc and heat shocked for 15 min at 42°C or left untreated (C). Methanol-fixed cells were double stained by using anti-p-S303/7 antibody to detect phosphorylated HSF1 and anti-Myc antibody to localize total HSF1. Immunostaining was analyzed by fluorescence microscopy. Colocalization can be seen as yellow in the merged image. DAPI was used for nuclear staining. (Lower panel) HeLa cells were transiently transfected with HSF1-Myc and heat shocked for 15 min at 42°C. Cells were double stained with anti-p-S303/7 and anti-HSF1 antibodies in the presence or absence of 150 μg of recombinant human HSF1 (rHSF1) (17)/ml. (B) HeLa cells were transiently transfected with wild-type HSF1-Myc and GFP-SUMO-1 and heat shocked at 42°C for indicated times or left untreated (C). HSF1 was detected by using monoclonal anti-Myc antibody combined with a red fluorescent secondary antibody, and GFP-SUMO-1 was visualized through the green channel. (C) HeLa cells were transiently transfected with S303A or K298R HSF1 mutants together with GFP-SUMO-1. Cells were heat shocked at 42°C for 15 min and analyzed as described above.
In addition to HSF1, SUMO-1 has been shown to localize in distinct nuclear bodies (for a review see reference 61), which led us to study the relationship between subnuclear localization of HSF1 and that of SUMO-1. HeLa cells cotransfected with Myc-tagged HSF1 and GFP-fused SUMO-1 were subjected to heat stress for various time periods and analyzed by fluorescence microscopy. At normal temperature, SUMO-1 was partially localized to the nuclear bodies and partially diffusely distributed, being, however, predominantly nuclear (Fig. 9B). Immunolabeled HSF1 was also mainly nuclear but remained diffuse and was not concentrated in SUMO-1-containing bodies. Induction of the heat shock response led to a rapid reorganization of HSF1, since already after a 5-min exposure to heat stress, some HSF1 was localized to brightly stained nuclear stress granules. SUMO-1 displayed also a granular localization, partially overlapping with HSF1, as a mixture of colocalizing and noncolocalizing granules was observed in most cells. After 15 min, HSF1 had almost totally localized to the stress granules and the colocalization with SUMO-1 was much more prominent (Fig. 9B). To our surprise, the colocalization of HSF1 and SUMO-1 was abolished already after 30 min of heat shock; although SUMO-1 displayed a punctate localization, no overlap between HSF1 and SUMO-1 granules was observed (Fig. 9B). During longer heat treatments HSF1 remained in the stress granules, until they slowly dissociated after several hours (data not shown).
To explore whether the inhibitory effect of S303A mutation on HSF1 sumoylation was reflected in the formation of HSF1 and SUMO-1 cogranules, a similar experiment as described above was performed with the nonphosphorylatable mutant. S303A HSF1 was localized to the nuclear stress granules upon heat shock similarly to the wild type (Fig. 9C; also 9A and B). This result emphasizes that phosphorylation of S303 does not regulate HSF1 localization and excludes the possibility that S303 phosphorylation would promote HSF1 sumoylation through subcellular localization. However, no colocalization between S303A HSF1 and SUMO-1 was detected after a 15-min heat shock (Fig. 9C), indicating that HSF1 targeting to the nuclear stress granules can occur independently of SUMO-1 conjugation, whereas covalent SUMO-1 conjugation is needed for the colocalization. This finding was further supported by the K298R mutant, which also localized to the stress granules, without displaying any colocalization with SUMO-1 (Fig. 9C). As expected, under normal growth conditions both S303A and K298R displayed diffuse nuclear localization similarly to the wild-type HSF1 (data not shown). In conclusion, our immunofluorescence analysis demonstrated that stress induces a prominent but transient colocalization of HSF1 and SUMO-1 to the same subnuclear compartment at the onset of the heat shock response. The localization of HSF1 to the stress granules occurs independently of the phosphorylation status of S303, but S303 phosphorylation is required for the transient HSF1-SUMO-1 colocalization.
DISCUSSION
HSF1 has served as a typical example of a transcription factor that is under strict posttranslational control. Despite the abundant evidence for HSF1 undergoing a multistep activation-deactivation cycle, our understanding of the regulatory functions of HSF1 posttranslational modifications has remained incomplete. For example, it is well established that HSF1 is phosphorylated on several serine residues and that the induction of the bulk phosphorylation correlates positively with its transcriptional activity, but only scarce data on repressing and activating effects of single phosphorylation sites are available. More recently, sumoylation has emerged as another common posttranslational modification that targets mainly nuclear proteins, such as transcription factors. During the past several years, the number of SUMO targets has increased rapidly, but the function and regulation of SUMO modification apparently vary depending on the substrate. This study was undertaken to explore the SUMO modification and its impact on HSF1 regulation. Our results show that HSF1 is a substrate for a rapid and transient stress-inducible sumoylation on lysine 298 in the vicinity of several phosphorylation sites within the regulatory domain. Importantly, the stress-induced phosphorylation of serine 303 was found to be a prerequisite for the SUMO modification, demonstrating a dynamic cooperative action of these two modification events. A prominent but transient SUMO-1-HSF1 colocalization, which is strictly dependent on S303 phosphorylation, suggests that SUMO is required to perform certain, as yet unidentified, functions in the nuclear stress granules at the onset of heat shock response.
In vivo studies of HSF1 phosphorylation during different stages of the heat shock response are greatly challenged by the existence of multiple phosphorylation sites, the majority of which are still unknown. Another limiting factor in studying the dynamic changes of a single phosphorylation site is the poor reproducibility of the elaborate in vivo 32P-labeling and phosphopeptide mapping experiments. To avoid these technical caveats, we generated a phosphoepitope-specific antibody recognizing the phosphorylated state of serines 303 and 307. Kinetic analysis of S303/7 phosphorylation revealed a rapid and prominent increase in phosphorylation upon heat shock and gradual dephosphorylation at the attenuation phase during a continuous exposure to heat shock. This result was surprising in the light of several earlier studies that have indicated serines 303 and 307 to be constitutively phosphorylated to keep the factor inactive under normal growth conditions (4, 31, 32). Furthermore, our experiments employing pharmacological activators and inhibitors of mitogen-activated protein kinase-extracellular signal-regulated kinase signaling, which have been indicated to target S303/7 (4, 31, 32), did not show major changes in the phosphorylation status of these sites (data not shown). Therefore, it is plausible that several kinases are able to phosphorylate serines 303 and 307 in vivo, perhaps in a cell-specific manner.
During the course of this study, another report describing a strictly heat-inducible HSF1 sumoylation on K298 was published (19). Although the basic findings are similar, major controversies exist. Hong and coworkers showed a sharp ∼80-kDa SUMO-1-reactive band that appeared in HSF1 immunoprecipitates derived from heat-shocked cells and was referred to as endogenously sumoylated HSF1. Surprisingly, this band comigrates with hyperphosphorylated HSF1. Instead, we found a new HSF1-reactive band with markedly decreased electrophoretic mobility, which even upon SUMO overexpression appeared in a substoichiometric ratio. Hong and coworkers postulated that the retarded mobility on SDS-PAGE, which is characteristic of activated HSF1, is caused by heat-induced sumoylation. In our study, no difference in electrophoretic mobility between the major bands of wild-type HSF1 and K298R upon heat shock could be detected, suggesting that the heat-induced retardation around 80 kDa is caused by hyperphosphorylation (7, 17, 31, 53, 63), rather than by sumoylation. It was shown in the earlier study (19) that sumoylation in vitro converted HSF1 into a DNA-binding form and that transcriptional competence, as measured by the luciferase reporter assay in HeLa cells, was completely abolished by K298R mutation. Our DNA-binding analysis of the K298R mutant, when transfected into hsf1−/− MEFs, does not support the conclusion of SUMO-1-regulated DNA binding in vivo, since, in response to heat shock, the sumoylation-deficient mutant was as capable of binding to HSE as was the wild type. Similarly, Hsp70 expression was equally induced when the wild-type HSF1 and the K298R mutant were expressed in MEFs lacking endogenous HSF1. According to the other study (19), HSF1 and SUMO-1 displayed complete colocalization in the nuclear bodies also during a prolonged exposure to heat shock. In our experiments, a prominent colocalization occurred only transiently, and HSF1 was still localized to the stress granules after dissociation of the SUMO-1-HSF1 complex. Moreover, the ability of S303A and K298R mutants to localize to the HSF1 stress granules, despite the lack of SUMO-1 conjugation and colocalization, is not in line with the hypothesis that sumoylation would regulate the localization of HSF1 to the stress granules. Notably, our immunofluorescence analyses were conducted in HeLa cells, since we were not able to detect HSF1 granules in hsf1−/− MEFs. In fact, the study by Hong and coworkers (19) is, to our knowledge, the only report where HSF1 granules have been detected in murine cells. In the light of our results, it is plausible that the sumoylated population of HSF1 performs stress-induced functions different from the regulation of heat shock gene expression. Alternatively, sumoylation might favor the HSF1 transactivation capacity, the changes being too subtle and dynamic to allow detection with the available techniques. In conclusion, it appears that HSF1 sumoylation is a more complex and dynamic process than originally anticipated, and statements about the role of SUMO conjugation in the multistep activation of HSF1 should therefore be treated with great caution.
The list of reports describing subnuclear compartmentalization of various proteins, including transcriptional regulators, splicing factors, and SUMO substrates, is growing fast. Although in many cases the functional significance is still missing, it is anticipated that the presence of certain regulatory proteins in the same restricted compartment in the nucleus is needed to enhance the interaction and cross talk between the various members of the same regulatory unit. SUMO-1 has been shown elsewhere to target many of its substrates into nuclear structures, known as PML bodies or PODs (for a review see reference 57). This does not seem to be the case for HSF1, since our analysis using the sumoylation-deficient HSF1 mutants provides evidence that, when conjugation is blocked, SUMO-1 and HSF1 localize to distinct structures. Because these HSF1 mutants localize into the nuclear stress granules similarly to the wild type, it is probable that SUMO-1 modification of HSF1 is not needed for the granule formation. This is in agreement with earlier studies by Cotto and coworkers (6) showing that HSF1 does not colocalize with the PML bodies. Nevertheless, the HSF1-SUMO-1 cogranules that we observed within a 5- to 15-min time window at the onset of heat shock response demonstrated the highly dynamic nature of the subnuclear structures. These results emphasize the great reorganization potential of nuclear bodies in response to specific stimuli, and therefore, the composition of nuclear structures that have been regarded as distinct entities may vary in a condition-dependent manner. Identification of novel members of the subnuclear compartments, although challenging, should provide more mechanistic insights into the biological role and the interplay of these nuclear structures.
Cooperative regulation of phosphorylation and sumoylation appears to be a common denominator for several SUMO targets. Desterro and coworkers (9) showed that SUMO-1 modification antagonizes ubiquitination and thus degradation of IκBα. The switch between the ubiquitination and the sumoylation mode is regulated by phosphorylation so that the unphosphorylated IκBα favors the SUMO-1 incorporation, whereas phosphorylation triggers the ubiquitination and degradation. Likewise, studies of PML and c-Jun have shown inhibition of sumoylation upon induction of phosphorylation (39, 40). In the case of c-Jun, the unphosphorylated state of serines 63 and 73 promotes sumoylation of lysine 229, demonstrating that mutual proximity of the modification sites in the primary structure is not necessarily required for the cooperative regulation (39). Intriguingly, our study of HSF1 provides an example opposite to those mentioned above, showing that phosphorylation can also positively regulate sumoylation. The spatial proximity of the sumoylation site and the regulatory phosphorylation site suggests that phosphorylation could change HSF1 conformation, thereby exposing the SUMO target lysine to the modification machinery. This hypothesis is supported by the finding that the S303/7D mutant mimicking phosphorylated HSF1 was a preferred substrate in the in vitro sumoylation assay that utilized only purified E1 and E2 enzymes together with SUMO-1. Furthermore, we did not detect any difference between the localization of the wild-type HSF1 and that of the S303A mutant, ruling out the possibility that phosphorylation would regulate HSF1 sumoylation through subcellular targeting. Ongoing studies of the in vivo components belonging to the HSF1-specific sumoylation machinery as well as the phosphorylation-mediated signaling cascade will hopefully further elucidate the interrelationship between these two posttranslational modifications and ultimately unravel the regulation of HSF1 SUMO-1 modification.
Acknowledgments
We thank Ivor J. Benjamin (Southwestern Medical Center) for the hsf1−/− MEFs and Anne Dejean, Ronald Hay, and Hetti Poukka for the plasmids. Lorenz Poellinger, Noora Kotaja, and John E. Eriksson are acknowledged for fruitful and encouraging discussions, and Helena Saarento is acknowledged for skillful technical assistance. We are grateful to Minna Poukkula and Pia Roos-Mattjus for critical comments on the manuscript.
This work was supported by the Academy of Finland, the Finnish Cancer Organizations, the Sigrid Jusélius Foundation, and the Borg Foundation (Åbo Akademi University). V.H. and M.H. were supported by the Turku Graduate School of Biomedical Sciences.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Neriah, Y. 2002. Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3:20-26. [DOI] [PubMed] [Google Scholar]
- 3.Bies, J., J. Markus, and L. Wolff. 2002. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 277:8999-9009. [DOI] [PubMed] [Google Scholar]
- 4.Chu, B., F. Soncin, B. D. Price, M. A. Stevenson, and S. K. Calderwood. 1996. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J. Biol. Chem. 271:30847-30857. [DOI] [PubMed] [Google Scholar]
- 5.Chu, B., R. Zhong, F. Soncin, M. A. Stevenson, and S. K. Calderwood. 1998. Transcriptional activity of heat shock factor 1 at 37°C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3 and protein kinases Cα and Cζ. J. Biol. Chem. 273:18640-18646. [DOI] [PubMed] [Google Scholar]
- 6.Cotto, J., S. Fox, and R. Morimoto. 1997. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci. 110:2925-2934. [DOI] [PubMed] [Google Scholar]
- 7.Cotto, J. J., M. Kline, and R. I. Morimoto. 1996. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J. Biol. Chem. 271:3355-3358. [DOI] [PubMed] [Google Scholar]
- 8.Dai, R., W. Frejtag, B. He, Y. Zhang, and N. F. Mivechi. 2000. c-Jun NH2-terminal kinase targeting and phosphorylation of heat shock factor-1 suppress its transcriptional activity. J. Biol. Chem. 275:18210-18218. [DOI] [PubMed] [Google Scholar]
- 9.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 10.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 11.Eloranta, J. J., and H. C. Hurst. 2002. Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumolated in vivo. J. Biol. Chem. 277:30798-30804. [DOI] [PubMed] [Google Scholar]
- 12.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, M., T. J. Schuetz, E. K. Sullivan, and R. E. Kingston. 1995. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol. Cell. Biol. 15:3354-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 16.He, B., Y. H. Meng, and N. F. Mivechi. 1998. Glycogen synthase kinase 3β and extracellular signal-regulated kinase inactivate heat shock transcription factor 1 by facilitating the disappearance of transcriptionally active granules after heat shock. Mol. Cell. Biol. 18:6624-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmberg, C. I., V. Hietakangas, A. Mikhailov, J. O. Rantanen, M. Kallio, A. Meinander, J. Hellman, N. Morrice, C. MacKintosh, R. I. Morimoto, J. E. Eriksson, and L. Sistonen. 2001. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmberg, C. I., S. A. Illman, M. Kallio, A. Mikhailov, and L. Sistonen. 2000. Formation of nuclear HSF1 granules varies depending on stress stimuli. Cell Stress Chaperones 5:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, Y., R. Rogers, M. J. Matunis, C. N. Mayhew, M. Goodson, O.-K. Park-Sarge, and K. D. Sarge. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276:40263-40267. [DOI] [PubMed] [Google Scholar]
- 20.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, P. K. 2001. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 15:3053-3058. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799-26802. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16:5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolly, C., and R. I. Morimoto. 1999. Stress and the cell nucleus: dynamics of gene expression and structural reorganization. Gene Expr. 7:261-270. [PMC free article] [PubMed] [Google Scholar]
- 26.Jolly, C., L. Konecny, D. L. Grady, Y. A. Kutskova, J. J. Cotto, R. I. Morimoto, and C. Vourc'h. 2002. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 156:775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolly, C., R. I. Morimoto, M. Robert-Nicoud, and C. Vourc'h. 1997. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell Sci. 110:2935-2941. [DOI] [PubMed] [Google Scholar]
- 28.Jolly, C., Y. Usson, and R. I. Morimoto. 1999. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl. Acad. Sci. USA 96:6769-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 30.Kirsh, O., J. S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, and A. Dejean. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kline, M. P., and R. I. Morimoto. 1997. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol. Cell. Biol. 17:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knauf, U., E. M. Newton, J. Kyriakis, and R. E. Kingston. 1996. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 10:2782-2793. [DOI] [PubMed] [Google Scholar]
- 33.Kotaja, N., U. Karvonen, O. A. Jänne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindquist, S., and E. A. Craig. 1988. The heat-shock proteins. Annu. Rev. Genet. 22:631-677. [DOI] [PubMed] [Google Scholar]
- 35.McMillan, D. R., X. Xiao, L. Shao, K. Graves, and I. J. Benjamin. 1998. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 273:7523-7528. [DOI] [PubMed] [Google Scholar]
- 36.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 37.Morimoto, R. I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12:3788-3796. [DOI] [PubMed] [Google Scholar]
- 38.Mosser, D. D., N. G. Theodorakis, and R. I. Morimoto. 1988. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 8:4736-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller, S., M. Berger, F. Lehembre, J. S. Seeler, Y. Haupt, and A. Dejean. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275:13321-13329. [DOI] [PubMed] [Google Scholar]
- 40.Müller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakai, A., M. Tanabe, Y. Kawazoe, J. Inazawa, R. I. Morimoto, and K. Nagata. 1997. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol. Cell. Biol. 17:469-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newton, E. M., U. Knauf, M. Green, and R. E. Kingston. 1996. The regulatory domain of human heat shock factor 1 is sufficient to sense heat stress. Mol. Cell. Biol. 16:839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 44.Pirkkala, L., T.-P. Alastalo, X. Zuo, I. J. Benjamin, and L. Sistonen. 2000. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol. Cell. Biol. 20:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirkkala, L., P. Nykänen, and L. Sistonen. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15:1118-1131. [DOI] [PubMed] [Google Scholar]
- 46.Poukka, H., U. Karvonen, O. A. Jänne, and J. J. Palvimo. 2000. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 97:14145-14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabindran, S. K., G. Giorgi, J. Clos, and C. Wu. 1991. Molecular cloning and expression of a human heat shock factor, HSF1. Proc. Natl. Acad. Sci. USA 88:6906-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654-12659. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 52.Sapetschnig, A., G. Rischitor, H. Braun, A. Doll, M. Schergaut, F. Melchior, and G. Suske. 2002. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarge, K. D., S. P. Murphy, and R. I. Morimoto. 1993. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13:1392-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarge, K. D., V. Zimarino, K. Holm, C. Wu, and R. I. Morimoto. 1991. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 5:1902-1911. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt, D., and S. Müller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuetz, T. J., G. J. Gallo, L. Sheldon, P. Tempst, and R. E. Kingston. 1991. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc. Natl. Acad. Sci. USA 88:6911-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seeler, J. S., and A. Dejean. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene 20:7243-7249. [DOI] [PubMed] [Google Scholar]
- 58.Shi, Y., P. E. Kroeger, and R. I. Morimoto. 1995. The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol. Cell. Biol. 15:4309-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian, L., M. D. Benson, and J. A. Iniguez-Lluhi. A synergy control motif within the attenuator domain of C/EBPα inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem., in press. [DOI] [PubMed]
- 60.Takahashi, Y., T. Kahyo, A. Toh-E, H. Yasuda, and Y. Kikuchi. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276:48973-48977. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, V. G., and D. Rangasamy. 2001. Intracellular targeting of proteins by sumoylation. Exp. Cell Res. 271:57-65. [DOI] [PubMed] [Google Scholar]
- 62.Wu, C. 1995. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11:441-469. [DOI] [PubMed] [Google Scholar]
- 63.Xia, W., and R. Voellmy. 1997. Hyperphosphorylation of heat shock transcription factor 1 is correlated with transcriptional competence and slow dissociation of active factor trimers. J. Biol. Chem. 272:4094-4102. [DOI] [PubMed] [Google Scholar]
- 64.Xiao, X., X. Zuo, A. A. Davis, D. R. McMillan, B. B. Curry, J. A. Richardson, and I. J. Benjamin. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong, S., S. Müller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]
- 66.Zuo, J., D. Rungger, and R. Voellmy. 1995. Multiple layers of regulation of human heat shock transcription factor 1. Mol. Cell. Biol. 15:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]