Abstract
Expression of the prosurvival Bcl-2 homologue Bfl-1/A1 is induced by NF-κB-activating stimuli, while B and T cells from c-rel knockout mice show an absolute defect in bfl-1/a1 gene activation. Here, we demonstrate NF-κB-dependent assembly of an enhanceosome-like complex on the promoter region of bfl-1. Binding of NF-κB subunit c-Rel to DNA nucleated the concerted binding of transcription factors AP-1 and C/EBPβ to the 5′-regulatory region of bfl-1. Optimal stability of the complex was dependent on proper orientation and phasing of the NF-κB site. Chromatin immunoprecipitation analyses demonstrated that T-cell activation triggers in vivo binding of endogenous c-Rel, c-Jun, C/EBPβ, and HMG-IC to the bfl-1 regulatory region, coincident with selective recruitment of coactivators TAFII250 and p300, SWI/SNF chromatin remodeling factor component BRG-1, and basal transcription factors TATA-binding protein (TBP) and TFIIB, as well as hyperacetylation of histones H3 and H4. These results highlight a critical role for NF-κB in bfl-1 transcription and point to the need for a complex and precise regulatory network to control bfl-1 expression. To our knowledge, this is the first demonstration of enhanceosome-mediated regulation of a cell death inhibitor.
Activation of the Rel/NF-κB transcription factors was implicated in inhibiting cell death triggered by a variety of stimuli, ranging from radiation to genotoxic agents (9, 10, 38, 61, 62, 71). This survival response is dependent on the nuclear localization of NF-κB, because expression of a physiological inhibitor (IκBαM) suppressed its protective activity. Consistent with this observation, a number of Rel/NF-κB target genes have been described as effectors of its antiapoptotic effects (53, 64; reviewed in reference 7). Among them, our group and others previously identified the prosurvival Bcl-2 family member Bfl-1/A1 as an important transcriptional target of NF-κB (23, 32, 63, 72).
bfl-1 gene expression is generally confined to immune cells and tissues, in a pattern similar to that of NF-κB itself, and is strongly induced by cytokine stimulation of leukemic, endothelial, and hemopoietic cells (27, 34, 42, 60). Constitutively elevated levels of bfl-1 transcripts are seen in mature neutrophils and are selectively induced in long-lived peripheral B cells (60). These observations suggest an important role for Bfl-1 in the survival and selection of distinct subsets of cells in the immune system. Consistent with this hypothesis, Bfl-1 can suppress apoptosis triggered by the proinflammatory cytokine tumor necrosis factor α (TNF-α), tumor suppressor p53, B-cell receptor aggregation, proapoptotic factors Bax and Bad, and chemotherapeutic agents (13, 15, 17, 23, 25, 26, 31, 44, 49, 60, 63). Importantly, B and T cells from c-rel−/− mice exhibit an absolute defect in the ability to express bfl-1/a1 in response to cell activation (23). These results emphasize the notion that bfl-1 gene expression is strictly controlled and that NF-κB must play a critical role in this process.
The transcription of eukaryotic genes is highly regulated and relies upon the binding of the correct constellation of factors to particular sites within a defined gene locus. For a select number of tightly regulated genes, transcription also depends upon specific DNA-protein and protein-protein interactions in a highly ordered complex referred to as an enhanceosome. This precisely orchestrated complex allows for a limited subset of genes, such as those regulated in a tissue-specific manner, to appropriately respond to a defined group of transcription factors and the multistimulatory environment they inhabit. In this context, synergistic transcriptional activation derives from the cooperative binding of transcription factors and architectural factors to recruit coactivators and basal transcription factors through a novel activating surface (29, 39, 40). The enhanceosome also provides a platform for the binding of covalent histone modifiers to alter chromatin in a location-specific manner (35). The best-characterized enhanceosome-regulated genes are those encoding beta interferon (IFN-β) and T-cell receptor α (TCRα) (1, 21, 29, 35, 40, 59, 67, 68).
In prior studies, we showed that various NF-κB-inducing stimuli led to upregulation of bfl-1 gene expression in different cells (72). For instance, bfl-1 transcripts were sharply elevated in human Jurkat T cells activated with phorbol 12-myristate 13-acetate (PMA) plus ionomycin. This activation was dependent on NF-κB, because bfl-1 mRNA induction was substantially reduced in cells expressing IκBαM. Here, we have characterized how NF-κB regulates bfl-1 gene expression. We show that the sole NF-κB DNA-binding site in the 5′ regulatory region of bfl-1 promotes the cooperative binding of C/EBPβ and AP-1. In this context, the c-Rel and p50 subunits of NF-κB nucleated an enhanceosome-like complex containing NF-κB, AP-1, and C/EBPβ together with chromatin architectural factor HMG-I and transcriptional coactivators. Chromatin immunoprecipitation (ChIP) analyses demonstrated that T-cell activation triggers in vivo recruitment of these factors to the endogenous bfl-1 locus, coincident with histone acetylation. These results indicate that bfl-1 transcription is regulated by an NF-κB-dependent enhanceosome-like complex, highlighting the need for a complex and precise regulatory network to control its expression.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The −1374/+83 bfl-1:luc reporter construct was generated by subcloning the −1374/+83 region of bfl-1 from bfl-1:CAT (72) into the promoterless pGL3 · Basic reporter (Promega). The −1374/+83 bfl-1 region was cloned into pALTER-1 (Promega) and subjected to site-directed mutagenesis to inactivate the binding sites for NF-κB (−833 GTTTATTTACC), AP-1 (−864 GTCCTA), or C/EBPβ (−927 TCGCT; Altered Sites Mutagenesis System; Promega) (mutated residues are underlined). Site-directed mutagenesis was used to alter the orientation of the NF-κB site in the −1374/+83 bfl-1 region (rκB; 5′-GGTAAATCCCC-3′), or its phasing by repositioning it +6 or +10 bp toward the transcription start site of bfl-1 (QuickChange Site-Directed Mutagenesis kit; Stratagene). −933/−773:−100/+83bfl-1:luc was generated by amplifying the −933/−773 region of bfl-1 with primers 5′-CGACGCGTCTCCCGGGTTCAAGCAAT-3′ and 5′-TCGTGCGTAGGCTGTGCGGGCGGATTGCCT-3′ and amplifying the −100/+83 region with primers 5′-AGGCAATCCGCCCGCACAGCCTACGCACGA-3′ and 5′-GAAGATCTGCTGCCTGGTGGAGAGCA-3′. Products from these reactions were combined with overlap-anneal PCR and cloned in the MluI and BglII sites of pGL3-Basic. pcDNA3:C/EBPβ (46), pCB6+:c-jun, pCB6+:c-fos (5), and CMVβ:p300 (18) were gifts from D. E. Zhang (The Scripps Research Institute), T. Curran (St. Jude Children's Research Hospital), and D. Livingston (Dana Farber Cancer Institute and Harvard Medical School), respectively. pCMV-c-Rel was described previously (66). Where indicated, pcDNA3.1HisC (Invitrogen) was used as a control.
Cell transfection, luciferase, and CAT assays.
The HtTA-1-derived cell line HtTA-CCR43 expressed c-Rel under tetracycline-regulated control (8). Jurkat E6.1 T-lymphocytic leukemia cells were obtained from the American Type Culture Collection. Endogenous NF-κB activity was induced by cell treatment with PMA (50 ng/ml in dimethyl sulfoxide [DMSO]) plus ionomycin (1 μM in DMSO) overnight or with DMSO alone as a control. Jurkat T cells (2 × 106) were transiently transfected with a total of 4 μg of DNA with DMRIE-C reagent (4 μl; Invitrogen). Where indicated, cells were equally divided into two wells at 24 h posttransfection and treated with PMA plus ionomycin or with DMSO as a control. Cells were harvested at 48 h posttransfection and washed once with phosphate-buffered saline (PBS), and extracts were prepared with 1× passive lysis buffer (Promega). Extracts were normalized for protein content (11) and assayed for luciferase activity by using the Promega luciferase assay reagent on a TD-20/20 luminometer (Turner Designs). Cos-7 cells (1.4 × 105) were transiently transfected with a total of 6 μg of DNA by using calcium phosphate. Cells were harvested 48 h later. Extracts were normalized for protein content, and chloramphenicol acetyltransferase (CAT) activity was assayed on 50 μg of total protein for 2 h.
Gel retardation assays.
Nuclear extracts from human Jurkat T cells treated with PMA plus ionomycin for 2 or 18 h, or with DMSO as a control, were prepared as described previously (52). DNA probes consisted of 160-bp wild-type or mutant −933/−773 fragments corresponding to the 5′ regulatory region of bfl-1 cloned into pBLCAT5 (American Type Culture Collection). Fragments were isolated from pBLCAT5 by restriction with XbaI and SalI, 32P labeled with Klenow enzyme, and used as probes in gel shift assays. Nuclear extracts (3 μg) were incubated with DNA probes (4 × 104 cpm) in a mixture containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 7.5 mM MgCl2, 1 mM EDTA, 5% glycerol, 5% sucrose, 0.1% NP-40, 0.5 μg of poly(dI-dC), 5 mM dithiothreitol (DTT), and 0.5 mg of bovine serum albumin (BSA) per ml, and analyzed on 5% native polyacrylamide gels. Where indicated, DNA-protein complexes were supershifted with anti-hc-Rel or anti-p50 antibodies (1136 and 1263, respectively; a gift from N. Rice, ABL-National Cancer Institute, Frederick, Md.), anti-p65, anti-c-Jun, anti-C/EBPβ (sc-109, -822, and -150; Santa Cruz Biotechnology), anti-p300 (14991A; Pharmingen), or anti-HMGI-C (a gift from K. Chada, Robert Wood Johnson Medical School, Piscataway, N.J.) antibodies. Normal rabbit serum (NRS; Calbiochem) was used as a control.
In competition gel retardation assays, 32P-labeled probes were incubated with nuclear extracts as described above. After a 30-min incubation, an aliquot of each binding reaction mixture was loaded onto a nondenaturing gel, and a 250-fold molar excess of unlabeled double-stranded oligonucleotide probe containing the C/EBPβ binding site from the bfl-1 regulatory region (5′-CCCGGGTTCAAGCAATTCCCTCTCCTT-3′) was added to the binding reaction mixtures. Aliquots were taken at time intervals and loaded onto a native 5% polyacrylamide gel, which was allowed to run between loadings. Bound complexes were quantified with ImageQuant software (Molecular Dynamics).
Northern blot analysis.
Total RNA (30 μg) was extracted with RNAzol B (TEL-TEST), fractionated on a 1% agarose-formaldehyde gel, and transferred onto a Hybond NX membrane (Amersham). The membrane was baked for 10 min at 80°C under vacuum and UV cross-linked with a Stratalinker (Stratagene). Probes were generated by random priming with Klenow polymerase in the presence of [32P]dCTP and [32P]dGTP (19). The membrane was hybridized in Perfect Hyb Plus hybridization buffer (Sigma) at 68°C overnight. The membrane was washed twice in a mixture containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.05% sodium dodecyl sulfate (SDS) at room temperature and twice in 0.1× SSC-0.1% SDS at 50°C, followed by autoradiography.
DNA-protein immunoprecipitation assays.
Nuclear extracts (5 μg) from HtTA-CCR43 cells uninduced or induced to produce c-Rel upon removal of tetracycline for 96 h were incubated with 32P- and bromodeoxyuridine (BrdU)-labeled wild-type or mκB mutant −933/−773 bfl-1 enhancer probes (50 fmol) in a mixture of 20 mM Tris-HCl (pH 7.8), 50 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 15% glycerol, 2 mM DTT, 0.01% NP-40, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 100 μg of BSA per ml, and 1 μg of poly(dI-dC) per 17 μl. Binding reactions were subjected to UV cross-linking and immunoprecipitated with anti-c-Jun, anti-C/EBPβ, anti-HMGI-C, or anti-c-Rel antibodies (22) or with normal rabbit immunoglobulin G (IgG) as a control (Calbiochem). Complexes were resolved on SDS-5% polyacrylamide gels.
ChIP assays and real-time PCR analysis.
ChIP assays were performed as described (45, 47) with the following modifications. Jurkat T cells (5 × 107) were treated with PMA plus ionomycin for 2 or 18 h or with DMSO alone as a control. After cross-linking with formaldehyde, chromatin was extracted, sonicated, and purified on CsCl gradients. Fractions containing chromatin were pooled and immunoprecipitated with antibodies specific for c-Rel (1136; N. Rice, ABL-National Cancer Institute), p65/RelA, c-Jun, C/EBPβ, p/CAF, or TAFII250 (sc-109, -45, -150, -8999, and -735; Santa Cruz Biotechnology), HMGI-C (K. Chada, Robert Wood Johnson Medical School), p300 (14991A; Pharmingen), CBP (D. M. Livingston, Dana-Farber Cancer Institute, Boston, Mass.), BRG-1 (W. Wang, National Institute on Aging, National Institutes of Health), hTBP or hTFIIB (D. Reinberg, Robert Wood Johnson Medical School), or acetylated histone H3 or H4 (06-599 and 06-866, respectively; Upstate Biotechnology). After washing, cross-linking was reversed, and the precipitated chromatin was extracted with phenol-chloroform. After ethanol precipitation, the DNA was resuspended in water (25 μl) and PCR amplified.
In standard PCRs, immunoprecipitated DNA (1 μl) was amplified for 25 cycles with Amplitaq (Applied Biosystems). In real-time PCR analyses, immunoprecipitated DNA (1 μl) was amplified in 50-μl reaction mixtures containing SYBR Green master mix (25 μl; Applied Biosystems) and oligonucleotide primers specific for the 5′ regulatory region of bfl-1 (300 μM each; forward, 5′-TGGAGACAGAGTCTCGCTCTGTT-3′; reverse, 5′-GAACCCGGGAGACAGAAGTTG-3′), the proximal promoter region of bfl-1 (forward, 5′-GGATATTATAAAGTGATGCAAACAGAAATT-3′; reverse, 5′-TGAGGCAATGTGCTGAGAATG-3′), or the 5′ regulatory regions of the IκBα gene (forward, 5′-GGCTCATCGCAGGGAGTTT-3′; reverse, 5′-GAACTGGCTTCGTCCTCTGCTA-3′) and the BLR1 chemokine receptor gene (forward, 5′-TGAAATGCTTGACTAGCACTGATG-3′; reverse, 5′-CGCGACCTGCCTCACAAC-3′) as controls. Incorporation of SYBR Green dye into PCR products was monitored in real time with an ABI PRISM 7700 sequence detection system (PE Applied Biosystems), allowing determination of the threshold cycle (CT) at which exponential amplification of PCR products begins. Each reaction was performed in triplicate. The CT values for PCR products corresponding to individual loci were used to calculate the amount of chromatin immunoprecipitated in each reaction. Relative enrichment over the no-antibody control was first determined for each sample. Relative factor recruitment in PMA- plus ionomycin-treated cells was then normalized to the DMSO control for each antibody. The specificity of DNA amplification by primer pairs was verified by standard PCR (40 cycles) under the cycling conditions used for real-time analysis and evaluated by agarose gel electrophoresis and ethidium bromide staining. Absence of primer-dimer formation was confirmed in amplification reactions without template.
RESULTS
T-cell activation induces binding of NF-κB, AP-1, C/EBPβ, HMGI-C, and p300 to the 5′ bfl-1 regulatory region.
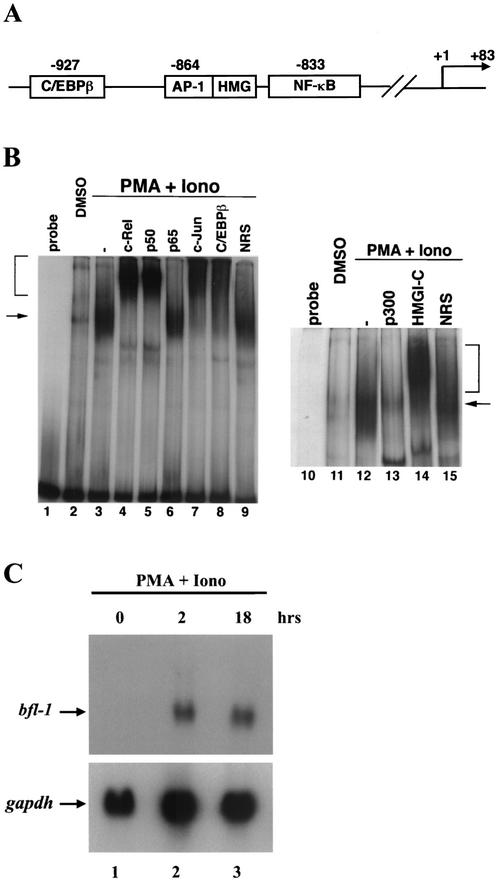
Detailed analysis of a 1.4-kbp genomic fragment comprising the 5′ regulatory region of bfl-1 revealed consensus binding sites for transcription factors C/EBPβ (−927) and AP-1 (−864) located within 100 bp of the sole NF-κB DNA binding motif (−833) that we previously implicated in regulating bfl-1 transcription (72) (Fig. 1A). An AT-rich sequence corresponding to the recognition motif for chromatin modeling factor HMG-I (AATTTTTT) overlapped the putative AP-1 binding site. The binding of endogenous NF-κB, AP-1, and C/EBPβ proteins to the 5′ bfl-1 regulatory region was investigated with gel retardation assays. Incubation of nuclear extracts from human Jurkat T cells stimulated with PMA plus ionomycin with a 160-bp probe (−933/−773) derived from the 5′ bfl-1 regulatory region induced formation of a large-molecular-weight complex compared to results with extracts from control cells treated with DMSO (Fig. 1B, compare lanes 3 and 12 to lanes 2 and 11). Parallel Northern blot analysis showed that this was correlated with a sharp induction of endogenous bfl-1 transcripts (Fig. 1C, compare lanes 2 and 3 to lane 1). Supershift analyses revealed the presence of endogenous c-Rel, p50, c-Jun, and C/EBPβ in this complex (Fig. 1B, lanes 4, 5, 7, and 8, respectively). An antibody specific for chromatin architectural factor HMGI-C supershifted the complex, while an antibody for coactivator p300 interrupted its formation (lanes 13 and 14). HMGI-C is related to HMG-I(Y), which is found in the IFN-β enhanceosome, and enhances NF-κB function in a manner similar to HMG-I(Y) (37, 58, 67). In contrast, antibodies to RelA/p65 and HMG-I(Y) or NRS had no such effect (lanes 6 and 9) (data not shown). These results demonstrate the binding of endogenous NF-κB, c-Jun, C/EBPβ, HMGI-C, and p300 proteins to the 5′ regulatory region of bfl-1 and indicate that the primary NF-κB dimer regulating bfl-1 in activated Jurkat T cells is a c-Rel/p50 heterodimer.
FIG. 1.
T-cell activation causes binding of c-Rel, p50/NF-κB1, c-Jun, C/EBPβ, HMGI-C and p300 to the 5′ regulatory region of bfl-1. (A) Schematic representation of the 5′ regulatory region of the human bfl-1 gene. Binding sites for NF-κB, AP-1, C/EBPβ, and HMGI are indicated. The sequence of the 1.4-kbp 5′-regulatory region of bfl-1 is available in GenBank (accession no. AF479683). (B) Gel retardation analysis of factor binding to the 5′ regulatory region of bfl-1. Jurkat T cells were treated with PMA plus ionomycin (Iono) for 18 h or with DMSO as a control. Nuclear extracts were incubated with a 160-bp probe derived from the −933/−773 region of bfl-1. Where indicated, antibodies to c-Rel, p50, p65, c-Jun, C/EBPβ, p300, and HMGI-C or NRS was added to the binding reaction mixtures. Arrows indicate the DNA-protein complex, and brackets indicate supershifted complexes. (C) Northern blot analysis of endogenous bfl-1 transcripts in Jurkat T cells stimulated with PMA plus ionomycin for 2 or 18 h (lanes 2 and 3) or with DMSO as a control (lane 1). RNA was successively hybridized to probes specific for genes coding for Bfl-1 or GAPDH.
NF-κB binding to DNA promotes the binding of AP-1, C/EBPβ, HMGI-C, and p300 to the −933/−773 bfl-1 probe.
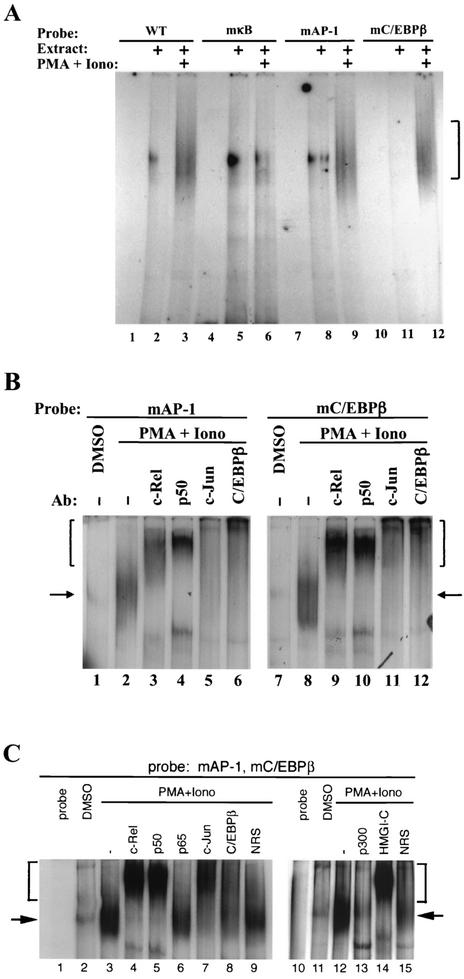
The binding of HMGI-C to the 5′ regulatory region of bfl-1 and its close proximity to binding sites for NF-κB, AP-1, and C/EBPβ are somewhat reminiscent of the features of its homologue, HMG-I(Y), and transcription factors ATF2/c-Jun, IRF1, and NF-κB in the enhanceosome-regulated IFN-β-positive regulatory domains (36, 39, 43, 59). We thus explored whether individual mutation of the NF-κB, AP-1, and C/EBPβ sites in the −933/−773 bfl-1 probe would affect the binding of the other factors in the complex. The loss of transcription factor binding to the individual mutant sites was confirmed in gel retardation assays (data not shown). In the context of the −933/−773 bfl-1 regulatory probe, mutation of the NF-κB site alone suppressed induction of the high-molecular-weight DNA-protein complex upon activation of Jurkat T cells with PMA plus ionomycin compared to the DMSO control (mκB; Fig. 2A, compare lanes 5 and 6 to lanes 2 and 3). Surprisingly, mutation of either the unique AP-1 (mAP-1) or C/EBPβ binding sites (mC/EBPβ) in the −933/−773 probe still allowed formation of a DNA-protein complex with a mobility similar to that formed on the wild-type probe (lanes 9 and 12). This raised the possibility that the binding of a c-Rel/p50 heterodimer to the bfl-1 regulatory region may help to nucleate the binding of AP-1 and C/EBPβ to the probe. Alternatively, the compositions of the DNA-protein complexes on the wild-type and mutant bfl-1 probes may differ despite their apparently similar mobilities.
FIG. 2.
The binding of NF-κB to DNA promotes the binding of AP-1, C/EBPβ, HMGI-C, and p300 to the −933/−773 bfl-1 probe. (A) Nuclear extracts from Jurkat T cells treated with PMA plus ionomycin (Iono), or with DMSO as control, were incubated with 160-bp probes derived from the −933/−773 region of bfl-1. The probes were either wild type (WT) or mutated at the NF-κB, AP-1, or C/EBPβ binding sites as indicated. The bracket highlights the large DNA-protein complex. (B) Supershift analysis of DNA-protein complexes formed on mutant AP-1 or C/EBPβ probes with antibodies (Ab) specific to each factor. Arrows point to the DNA-protein complex, and brackets indicate supershifted complexes. (C) Supershift analysis of the complex bound to a −933/−773 probe in which the AP-1 and C/EBPβ DNA sites were mutated simultaneously.
To discriminate between these two possibilities, the composition of complexes formed on the mutant AP-1 and C/EBPβ probes was analyzed with supershift assays. Interestingly, c-Rel, p50, c-Jun, and C/EBPβ were all found to be present in complexes that formed on both the mAP-1 and mC/EBPβ −933/−773 bfl-1 probes, despite the mutation of their respective DNA recognition sites (Fig. 2B, lanes 4 to 7 and 11 to 14). Similar results were obtained with a −933/−773 bfl-1 probe in which the C/EBPβ and AP-1 sites were mutated simultaneously (mAP-1,C/EBPβ). A high-molecular-weight DNA-protein complex still formed on the mutant probe with the NF-κB DNA site as the sole remaining recognition motif (Fig. 2C, lanes 3 and 12). As with the wild-type probe, supershift analysis revealed the presence of c-Rel, p50, c-Jun, C/EBPβ, HMGI-C, and p300 in this complex (lanes 4, 5, 7, 8, 13, and 14, respectively). These data indicated that the multiprotein complexes that formed on the mutant bfl-1 probes contained the same transcription factors as the complex bound to the wild-type probe. In conjunction with the data on the mκB probe (Fig. 2A), these results suggest that efficient complex formation on the 5′ regulatory region of bfl-1 is dependent on the binding of a c-Rel/p50 heterodimer to the NF-κB site. Although we do not rule out the possibility that the high concentration of factors and probe available for in vitro binding may allow protein-protein interactions to suffice in gel retardation assays, it is also possible that the DNA recognition motifs for AP-1 and C/EBPβ play a secondary role in vitro. In this scenario, the well-documented protein-protein interactions between NF-κB, AP-1, and C/EBPβ are likely to play an important role in stabilizing the complex (12, 16, 51, 54-56, 70).
NF-κB nucleates the binding of a multiprotein complex containing NF-κB, AP-1, C/EBPβ, and HMGI-C to the bfl-1 regulatory region.
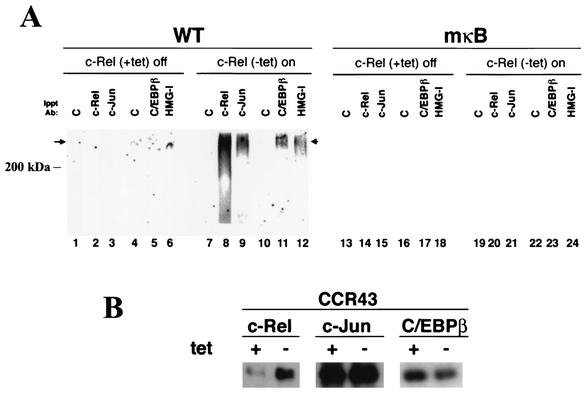
Since an intact NF-κB DNA recognition motif was necessary to promote efficient binding of c-Jun and C/EBPβ to the −933/−773 bfl-1 probe, we went on to test the hypothesis that NF-κB participates in recruiting these factors. To this end, we used nuclear extracts from HtTA-CCR43 cells, which conditionally expressed c-Rel under tetracycline-regulated control. Extracts from uninduced cells (+tet) or cells induced to express c-Rel (−tet) were tested for endogenous transcription factor binding to the 5′ regulatory region of bfl-1 in DNA-protein immunoprecipitation assays. After incubation with 32P- and BrdU-labeled wild-type or mκB mutant −933/−773 bfl-1 probes, DNA-protein complexes were subjected to UV cross-linking and immunoprecipitated with antibodies specific for c-Rel, c-Jun, C/EBPβ, or HMGI-C. DNA-bound protein complexes were resolved by SDS-polyacrylamide gel electrophoresis and revealed by autoradiography.
In the absence of c-Rel induction, no DNA-protein complex was immunoprecipitated with any of the antibodies specific for c-Rel, c-Jun, C/EBPβ, or HMGI-C (Fig. 3A, lanes 2, 3, 5, and 6, respectively), despite the presence of endogenous c-Jun and C/EBPβ proteins in the cells as detected by Western blotting (+tet; Fig. 3B). In sharp contrast, induction of c-Rel expression upon removal of tetracycline was sufficient to provoke immunoprecipitation of very-large-molecular-mass DNA-protein complexes (>200 kDa) on the wild-type bfl-1 probe with anti-c-Rel, -c-Jun, -C/EBPβ, or -HMGI-C antibodies (Fig. 3A, lanes 8, 9, 11, and 12, respectively). This assay was specific, because no complex was immunoprecipitated with a normal rabbit IgG control (lanes 7 and 10). Moreover, mutation of the NF-κB DNA site in the −933/−773 bfl-1 probe abolished immunoprecipitation of DNA-protein complexes with all antibodies, further suggesting that the binding of c-Rel to DNA is necessary to allow efficient binding of endogenous c-Jun, C/EBPβ, and HMGI-C to this region (mκB; Fig. 3A, lanes 20, 21, 23, and 24). Altogether, these data indicate that the binding of NF-κB to the 5′ regulatory region of bfl-1 nucleates the cooperative binding of AP-1, C/EBPβ, and HMGI-C. In conjunction with the experiments described above suggesting that NF-κB may help to anchor this complex through protein-protein interactions, we postulate that bfl-1 gene expression is regulated by an enhanceosome-like complex that is nucleated by and dependent on NF-κB DNA binding.
FIG. 3.
NF-κB nucleates formation of a DNA-protein complex containing c-Rel/p50, c-Jun, C/EBPβ, and HMGI-C on the −933/−773 bfl-1 probe. (A) Nuclear extracts from uninduced (+tet, lanes 1 to 6 and 13 to 18) or induced (−tet, lanes 7 to 12 and 19 to 24) HtTA-CCR43 cells were incubated with a BrdU- and 32P-labeled wild-type −933/−773 bfl-1 probe (lanes 1 to 12) or a mutant probe in which the NF-κB site was inactivated (lanes 13 to 24). After UV cross-linking, DNA-protein complexes were immunoprecipitated (Ippt) with antibodies (Ab) specific for c-Rel, c-Jun, C/EBPβ, and HMGI-C or control normal rabbit IgG, followed by autoradiography. (B) Western blot analysis of endogenous c-Rel, c-Jun, and C/EBPβ proteins in nuclear extracts from HtTA-CCR43 cells uninduced (+tet) or induced to express c-Rel (−tet).
The orientation and phasing of the NF-κB site affect the stability of the complex.
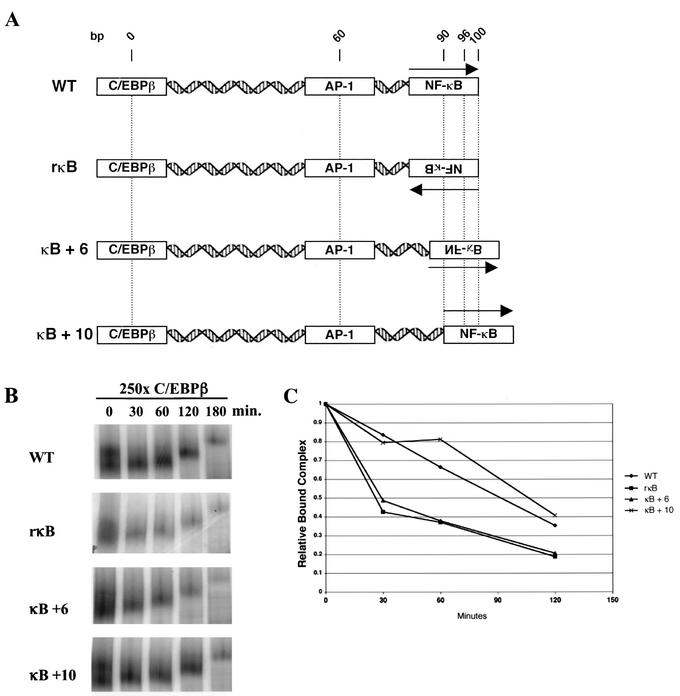
The well-documented interactions between NF-κB, AP-1, and C/EBPβ (12, 16, 51, 54-56, 70) led us to assess possible effects of alterations in the orientation and phasing of the NF-κB binding site on the formation or stability of the complex. In the −933/−773 bfl-1 probe, the center of the C/EBPβ site is located six helical turns away from the center of the AP-1 binding site. The center of the AP-1 binding site is three helical turns away from the center of the NF-κB site. To address the orientation requirement, the NF-κB site was placed on the opposite strand of the DNA, reversing its orientation with respect to the AP-1 and C/EBPβ sites in the −933/−773 probe (rκB). The need for proper phasing was investigated by repositioning the NF-κB site 6 or 10 bp away from the AP-1 and C/EBPβ sites (κB+6 and κB+10, respectively). This effectively moved the NF-κB site one-half or a full helical turn away from the rest of the bfl-1 complex (Fig. 4A).
FIG. 4.
Phasing and orientation of the NF-κB dimer affect the stability of the complex on the −933/−773 bfl-1 probe. (A) Schematic diagram of −933/−773 bfl-1 positional mutants. Arrows indicate the orientation of the NF-κB dimer with respect to the rest of the 5′ bfl-1 regulatory region. (B) Nuclear extracts from Jurkat T cells stimulated with PMA plus ionomycin were incubated with wild-type (WT) or mutant −933/−773 bfl-1 probes (rκB, κB+6, and κB+10). Aliquots of the binding reaction mixtures were loaded onto a running nondenaturing gel at time zero and at intervals following addition of a 250-fold molar excess (250x) of an unlabeled competitor oligonucleotide. (C) Bound complexes were quantitated with ImageQuant software (Molecular Dynamics). Relative bound complex represents the ratio of DNA-bound complex remaining at indicated time points over the amount of bound complex at time zero. The data are representative of three independent experiments.
Competition gel retardation assays revealed significant differences in the stability of complexes forming on the wild-type and κB+10 probes compared to those on the rκB and κB+6 probes, in which the NF-κB binding site is located on the opposite face of the helix. In these assays, DNA-binding reactions on the wild-type, rκB, κB+6, and κB+10 probes were challenged with a 250-fold molar excess of an unlabeled C/EBPβ competitor oligonucleotide. Aliquots of the binding reaction mixtures were loaded onto a running native gel at intervals following addition of the competitor (Fig. 4B). As plotted in Fig. 4C, the amount of remaining complex bound to the rκB and κB+6 probes following addition of competitor was approximately 50% of that of the remaining complexes bound to the wild-type and κB+10 probes. This indicated that improper phasing and orientation of the NF-κB site with respect to the AP-1 and C/EBPβ motifs adversely affect the stability of the complex (rκB and κB+6), in contrast to the wild-type and κB+10 probes, in which proper phasing was restored. Similar results were obtained with gel retardation assays performed in the presence of the anionic detergent Sarkosyl (data not shown). Altogether, these data support the premise that proper orientation and phasing of the NF-κB dimer relative to the AP-1 and C/EBPβ dimers are required for optimal stability of the complex.
Transcription coactivator p300 synergizes with NF-κB, AP-1, and C/EBPβ to activate the bfl-1 promoter.
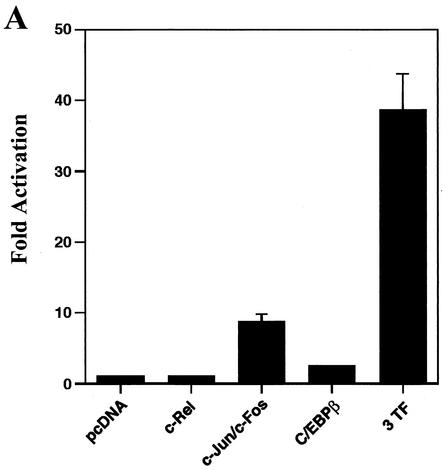
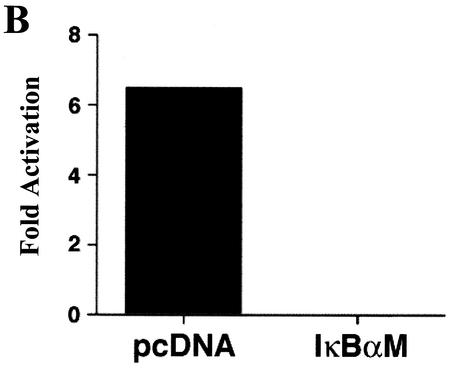
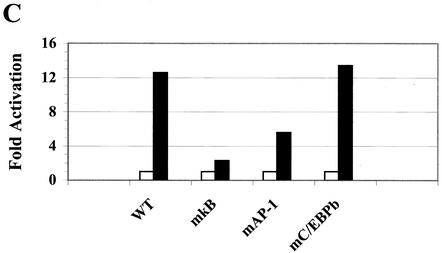
Consistent with their concerted binding to the −933/−773 bfl-1 probe, NF-κB, AP-1, and C/EBPβ synergistically activated expression from the bfl-1 promoter (Fig. 5A). Jurkat T cells cotransfected with a −1374/+83bfl-1:luc reporter and small amounts of expression vectors for c-Rel or C/EBPβ alone showed virtually no activation under these conditions, whereas the use of c-Fos and c-Jun (AP-1) led to a modest increase in luciferase activity. However, cotransfection of all three transcription factors together led to a sharp 39-fold synergistic activation of the promoter. Moreover fusion of the −933/−773 fragment from the 5′ bfl-1 regulatory region that encompasses only the C/EBPβ, AP-1, NF-κB, and HMG-I sites to the proximal promoter region of bfl-1 was strongly activated in response to PMA plus ionomycin, and this was suppressed by a dominant IκBαM inhibitor of NF-κB (−933/−773:−100/+83bfl-1:luc; Fig. 5B). Similar results were observed following CD3/CD28 stimulation (data not shown). These results underscore an important role for the −933/−773 region in the transcriptional regulation of bfl-1 and demonstrate that this region can function at a distance from or in close proximity to the bfl-1 promoter region to drive gene expression. Consistent with the in vitro DNA-binding studies on mutant bfl-1 probes (Fig. 2), mutation of the NF-κB recognition motif abolished reporter gene activation, whereas mutation of the C/EBPβ site had no significant effect (Fig. 5C). Interestingly, mutation of the AP-1 site led to a 50% decrease in activation, suggesting that it is not completely dispensable in vivo. Altogether, these results further emphasize an important role for NF-κB in allowing factors to synergize on the bfl-1 promoter.
FIG. 5.
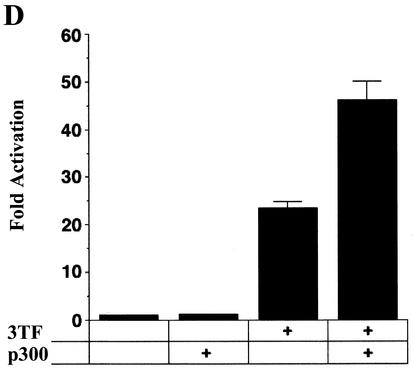
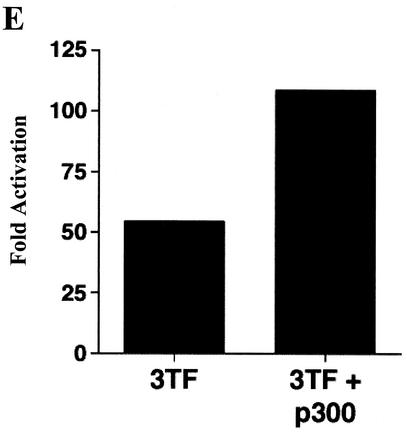
NF-κB, AP-1, and C/EBPβ synergistically activate transcription from the bfl-1 regulatory region with coactivator p300. (A) Jurkat T cells were transiently cotransfected with a −1374/+83bfl-1:luc reporter construct (1.33 μg) and expression vectors for c-Rel, c-Jun, c-Fos, and C/EBPβ (667 ng each), alone or together (3 TF). The total amount of transfected DNA was kept constant by addition of empty pcDNA3.1HisC vector. The average of three independent experiments is shown. (B) Transcriptional activity of the −933/−773 bfl-1 regulatory region. Jurkat T cells were transfected with 360 ng of −933/−773:−100/+83 bfl-1:luc or control −100/+83 bfl-1:luc reporter and 3.6 μg of pcDNA3 control or pcDNA3:IκBαM. Cells were treated with DMSO or stimulated with PMA plus ionomycin for 18 h prior to harvest. The results show the average fold activation of −933/−773:−100/+83 bfl-1:luc by treatment with PMA plus ionomycin from which background activity of the −100/+83 bfl-1:luc control was subtracted. (C) Cells were transfected with wild-type or mκB, mAP-1, or mC/EBPβ mutant −993/−773 bfl-1:CAT reporters (1.5 μg) and 1.5 μg each of CMV-c-Rel, CMV-c-Jun, and CMV-C/EBPβ DNA or 4.5 μg of an empty pcDNA3.1HisC vector as a control. CAT activity was assayed at 48 h posttransfection. (D) Jurkat T cells were cotransfected with a total of 4 μg of DNA containing −1374/+81 bfl-1:luc (730 ng) together with expression vectors for c-Rel, c-Jun, c-Fos, and C/EBPβ (360 ng each; TF mix), alone or together with p300 (1.8 μg). The average of three independent experiments is shown. (E) Assays were conducted essentially as in panel D, but with the −933/−773:−100/+83 bfl-1:luc reporter construct. The average of three independent experiments is shown.
NF-κB, AP-1, and C/EBPβ have all been shown to interact physically and functionally with transcriptional coactivators p300 and CBP (3, 6, 20, 41, 50). In light of the critical role of coactivators in regulating the IFN-β enhanceosome (40), the recruitment of p300 to the −933/−773 probe raised the possibility of a functional role in the activation of bfl-1. To address this issue, an expression vector for p300 was cotransfected into Jurkat T cells along with expression vectors for c-Rel, c-Jun, c-Fos, C/EBPβ, and the −1374/+83bfl-1:luc and −933/−773:−100/+83bfl-1:luc reporter constructs. As expected, transfection of p300 alone failed to increase transcription of the −1374/+83bfl-1:luc reporter (Fig. 5D). However it doubled activation observed with the combination of NF-κB, AP-1, and C/EBPβ on either the −1374/+83bfl-1:luc or the −933/−773:−100/+83bfl-1:luc reporter (Fig. 5D and E). Combined with the gel shift data presented above, this demonstrates that p300 can synergistically activate transcription from the bfl-1 promoter along with NF-κB, AP-1, and C/EBPβ.
T-cell activation induces complex assembly on the 5′ regulatory region of bfl-1 in chromatin, leading to coactivator recruitment and histone hyperacetylation.
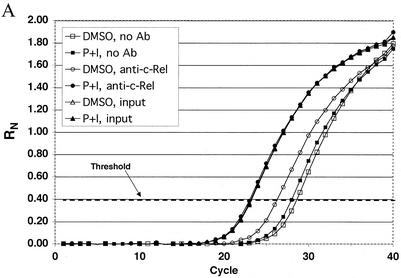
To demonstrate their participation in a physiologically relevant context, in vivo assembly of endogenous factors on the 5′ regulatory region of bfl-1 was investigated by ChIP. Human Jurkat T cells were activated with PMA plus ionomycin for 2 or 18 h, or with DMSO as a control, and treated with formaldehyde to cross-link endogenous protein-DNA and protein-protein complexes. After sonication, chromatin was immunoprecipitated with factor-specific antibodies followed by quantitative analysis by real-time PCR with primers specific for the 5′ regulatory region of bfl-1. A representative real-time PCR profile is shown in Fig. 6A. DNA enrichment in antibody-containing samples relative to the no-antibody controls was calculated by using the difference in the average threshold cycle (CT) from three PCR amplifications for each immunoprecipitation.
FIG. 6.
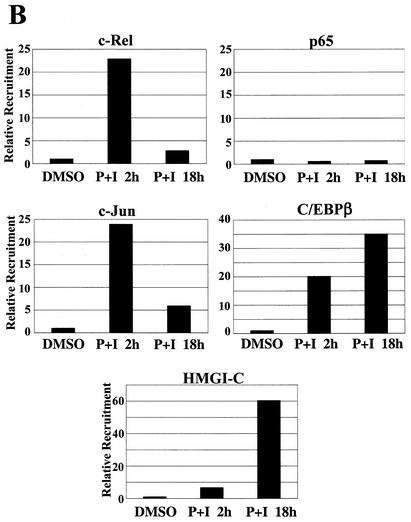
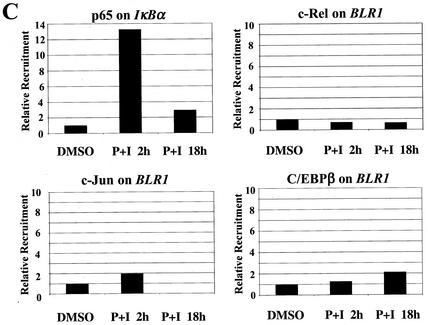
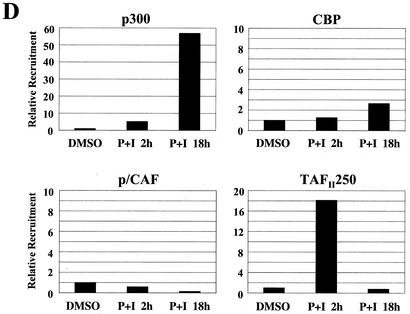
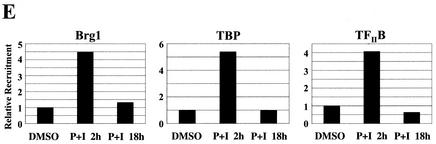
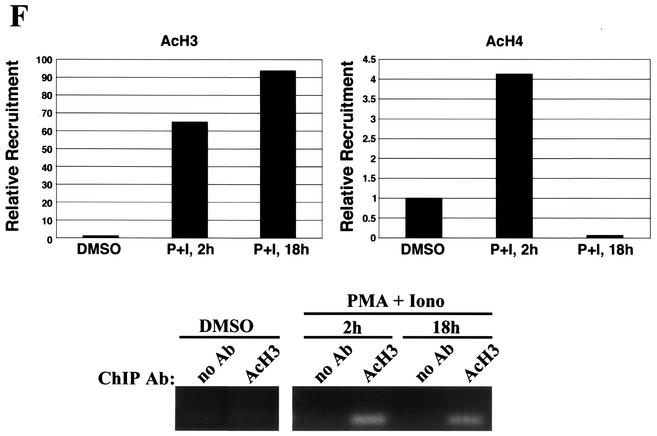
ChIP analysis of in vivo complex formation on the 5′ regulatory region of bfl-1 upon T-cell activation. (A) Representative real-time PCR analysis of ChIP assays with an anti-c-Rel antibody (Ab). Accumulation of product in cells treated with PMA plus ionomycin or with DMSO as a control normalized to reporter fluorescence (RN; y axis) is plotted as a function of cycle number (x axis). The threshold (dashed line) was calculated by multiplying the standard deviation of the RN of the baseline (cycles 3 to 15) by 10. The average of three independent reactions is shown. (B) ChIP analysis of transcription factors c-Rel, p65/RelA, c-Jun, and C/EBPβ and of architectural factor HMGI-C binding to the bfl-1 locus in human Jurkat T cells stimulated with PMA plus ionomycin (P+I) for 2 or 18 h or with a DMSO control. Factor binding was evaluated by determining the difference in the average number of PCR cycles at which amplification of the bfl-1 regulatory region reached the threshold between chromatin samples immunoprecipitated with transcription factor-specific antibodies and the no-antibody control (2−ΔCT). Standard deviations were calculated by using the formula s12 + s22 (ABI Prism 7700 Sequence Detection System, user bulletin 2, 1997. Applied Biosystems, Foster City, Calif.). (C) Control ChIP analysis showing p65/RelA recruitment to the 5′ regulatory region of the NF-κB target gene encoding IκBα in Jurkat T cells activated with PMA plus ionomycin and of c-Rel, c-Jun, and C/EBPβ recruitment to the upstream regulatory region of the B-cell-specific chemokine receptor gene BLR1 as a negative control. (D) ChIP analysis of coactivator p300, TAFII250, CBP, and p/CAF recruitment in Jurkat T cells treated with DMSO or following activation with PMA plus ionomycin. (E) ChIP analysis of SWI/SNF chromatin remodeling factor component BRG-1 and basal transcription factors TBP and TFIIB recruitment to the proximal bfl-1 promoter region in human Jurkat T cells stimulated with PMA plus ionomycin or with a DMSO control. (F) ChIP analysis of histone H3 and H4 acetylation on the bfl-1 regulatory region before and after Jurkat T-cell activation with PMA plus ionomycin (top panel). ChIP samples from unstimulated or PMA-plus-ionomycin-treated Jurkat T cells were immunoprecipitated with an antibody for hyperacetylated histone H3 and subjected to 25 rounds of standard nonquantitative PCR. The products were resolved on an agarose gel and revealed by ethidium bromide staining (bottom panel).
T-cell activation led to a strong increase in endogenous c-Rel binding to the 5′ regulatory region of bfl-1 at the 2-h time point compared to that in the DMSO control (23-fold; Fig. 6B), consistent with the sharp induction in endogenous bfl-1 transcripts in Jurkat T cells treated with PMA plus ionomycin for 2 h (Fig. 1C) (72). In agreement with the in vitro gel retardation analyses, no binding of p65/RelA to the 5′ regulatory region of bfl-1 was detected under these conditions (Fig. 6B). In contrast, p65 was efficiently recruited to the 5′ regulatory region of the NF-κB target gene encoding IκBα in control ChIP assays (Fig. 6C). Akin to c-Rel, binding of endogenous c-Jun and C/EBPβ to the bfl-1 regulatory region was also sharply induced upon T-cell activation compared to that in DMSO-treated cells (∼20-fold; Fig. 6B). While detection of c-Rel and c-Jun was reduced at the 18-h time point, that of C/EBPβ remained strong. Since efficient binding of c-Rel to DNA was detected at 2 and 18 h poststimulation in in vitro gel retardation assays, coincident with bfl-1 transcript expression (Fig. 1B and C) (data not shown), it is possible that in the context of chromatin, some epitopes in c-Rel may become masked at later time points, thereby precluding its efficient detection by ChIP at 18 h (Fig. 6B). The fact that a different anti-c-Rel antibody could detect sevenfold more c-Rel bound at the bfl-1 locus in cells activated for 18 h compared to the level in the DMSO control is consistent with this hypothesis (data not shown). Concomitant with the recruitment of c-Rel, c-Jun, and C/EBPβ, recruitment of architectural factor HMGI-C was observed at 2 h and was further increased at 18 h poststimulation (Fig. 6B). The specificity of these analyses was confirmed in control ChIP assays examining factor recruitment to the 5′ regulatory region of the gene for chemokine receptor BLR1, a B-cell-specific NF-κB target that is not expressed in Jurkat T cells (2, 65). As anticipated, little or no recruitment of c-Rel, c-Jun, or C/EBPβ to the BLR1 locus was observed following T-cell stimulation (Fig. 6C). Combined, these results demonstrate efficient in vivo recruitment of endogenous c-Rel, c-Jun, C/EBPβ, and HMGI-C proteins to the 5′ regulatory region of bfl-1 in response to T-cell activation.
In vivo assembly of c-Rel, c-Jun, C/EBPβ, and HMGI-C at the bfl-1 locus coincided with preferential recruitment of transcriptional coactivators TAFII250 and p300, but not the closely related coactivator CBP or associated factor P/CAF (Fig. 6D). Moreover, coactivator binding to the complex was correlated with transient recruitment of the BRG-1 ATPase component of the SWI/SNF remodeling complex and general transcription factors TBP and TFIIB at the proximal promoter region of bfl-1 (Fig. 6E). Importantly, the presence of acetyltransferases p300 and TAFII250 in the complex was correlated with hyperacetylation of histones H3 and H4 (Fig. 6F). As with the IFN-β enhanceosome, acetylation of histone H4 was transient and was dramatically reduced at the 18-h time point when TAFII250 was no longer present. In contrast, association of coactivator p300 with the bfl-1 locus was sharply elevated at 18 h poststimulation and was rather correlated with hyperacetylation of histone H3. Altogether, these studies in the context of intact chromatin demonstrate that T-cell activation provokes the in vivo binding of NF-κB, AP-1, C/EBPβ, and HMGI to the bfl-1 regulatory region and recruitment of histone acetyltransferases, chromatin remodeling complex SWI/SNF, and basal transcription factors. In conjunction with our gel retardation, DNA coimmunoprecipitation, and transactivation assays, these findings support a model in which NF-κB promotes assembly of an enhanceosome-like complex to acetylate histones and induce bfl-1 gene expression upon lymphoid cell activation (Fig. 7).
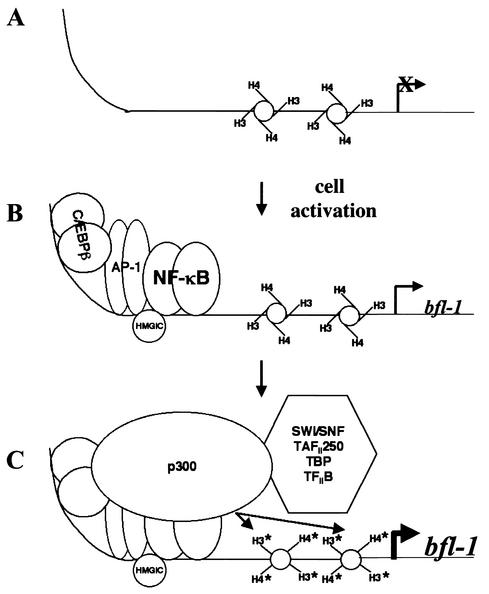
FIG. 7.
Model for induction of bfl-1 gene transcription by an NF-κB-dependent enhanceosome-like complex. (A) In unstimulated cells, histones located in the 5′ regulatory region of bfl-1 are hypoacetylated and the bfl-1 gene is not expressed. (B) Upon cell activation, NF-κB enters the nucleus and promotes assembly of transcription factors AP-1, C/EBPβ, and architectural factor HMGI on the 5′ regulatory region of bfl-1. (C) This enhanceosome-like complex recruits coactivators TAFII250 and p300 and the SWI/SNF chromatin-remodeling complex, leading to acetylation of histone tails and recruitment of basal transcription factors to drive bfl-1 gene expression.
DISCUSSION
The transcriptional regulation of eukaryotic genes is an intricate process. Nucleosomes, ATP-dependent chromatin remodeling factors, covalent chromatin modifiers, and sequence-specific DNA-binding proteins play key roles in determining whether a particular transcript is expressed or not (reviewed in reference 33). This complexity is reflected in the enhanceosome mechanism, described in detail for the IFN-β gene (40, 58, 59, 67, 68). Our description here of the assembly of an NF-κB-dependent enhanceosome-like complex on the bfl-1 promoter is the first to suggest an enhanceosome mechanism to control transcription of a cell death inhibitor. The NF-κB-mediated regulation of bfl-1 can be defined as enhanceosome-like based on (i) the cooperativity in transcription factor binding observed in gel retardation, protein-DNA immunoprecipitation, and ChIP assays; (ii) extensive protein-protein interactions highlighted by the ability of NF-κB to recruit and anchor AP-1, C/EBPβ, HMGI-C, and p300 on the DNA; and (iii) synergy in transactivation between these transcription factors and with transcriptional coactivators.
The IFN-β enhanceosome complex is dependent on the orientation of individual transcription factor binding sites (29, 59). Here, we have examined the requirements for formation of the bfl-1 enhanceosome-like complex and the subsequent events that occur in vivo. While the IFN-β enhanceosome is proximal to the transcription start site, the bfl-1 enhanceosome is located distally. However, repositioning the −933/−773 regulatory region in proximity to the core bfl-1 promoter retained 40 to 50% activity in response to PMA plus ionomycin compared to the wild type −1374/+83 bfl-1 reporter. Supershift and gel retardation assays revealed a dependence on proper orientation and phasing of the NF-κB site in the bfl-1 regulatory region for optimal stability of the complex. Relocation of this site to the opposite face of the helix, by reversing its orientation or moving it by half of a helical turn, destabilized the complex. The fact that complexes could assemble on mutant rκB and κB+6 probes was not entirely unexpected, since gel retardation assays indicated that an intact NF-κB site was sufficient to allow efficient complex formation on −933/−773 probes in vitro. These findings are reminiscent of the transcriptionally active enhanceosome-like complex that assembles on the nur77 promoter, which depends on intact DNA binding of MEF2D but does not require the DNA-binding activity of the synergistically acting transcription factor NFATp. In this context, NFATp acted as a coactivator for MEF2D (69). Although DNA contact with AP-1 and C/EBPβ was not absolutely required for in vitro formation of the complex on the −933/−773 bfl-1 probe when NF-κB was present, our transactivation assays with the mAP-1 mutant reporter suggest that in a natural context, cooperative protein-protein and protein-DNA interactions involving the NF-κB and AP-1 binding sites are likely to be involved in efficient formation of the complex and optimal activation of bfl-1 gene expression in vivo.
ChIP analysis of endogenous factor recruitment to the bfl-1 regulatory region in intact chromatin demonstrated induction of c-Rel, c-Jun, and C/EBPβ binding to the bfl-1 locus upon T-cell activation, in agreement with our in vitro data. These studies also revealed selective recruitment of coactivators p300 and TAFII250 and a sharp induction in histone acetylation at the bfl-1 locus. While further studies are needed to establish the precise timing of factor recruitment to the bfl-1 regulatory region, it is intriguing to note the transient binding of TAFII250, BRG-1 (SWI/SNF), TBP, and TFIIB. Whereas binding of these factors was sharply elevated at the 2-h time point, their detection had declined to basal levels by 18 h. Although it is not possible at this time to distinguish between departure of the factors and epitope masking at the later time point, these data may suggest interesting differences between coactivators TAFII250 and p300 in regulating factor recruitment, histone modification, and bfl-1 gene expression in response to cell activation. Together, our data highlight interplay between the factors that regulate bfl-1 through specific protein-protein and protein-DNA interactions and support a model in which NF-κB promotes assembly of an enhanceosome-like complex to acetylate histones and induce chromatin remodeling to promote bfl-1 expression upon lymphoid cell activation (Fig. 7).
Our finding that bfl-1 gene expression may involve an NF-κB-dependent enhanceosome-like complex is physiologically relevant, because it is consistent with and provides an explanation for the failure of B and T cells from c-Rel knockout mice to express endogenous bfl-1/a1 upon cell activation (23). In this regard, similar binding sites for AP-1, C/EBPβ, and HMG-I are found in close proximity to the unique NF-κB recognition motif in the 5′ regulatory region of the mouse a1 gene. Our findings are also consistent with prior studies from our group and others showing that stable expression of a dominant-negative form of IκBα repressed endogenous bfl-1/a1 transcription (15, 31, 44, 63, 72). Our in vitro and in vivo analyses indicating that the major NF-κB dimer that controls bfl-1 gene expression in activated Jurkat T cells is a c-Rel/p50 heterodimer agree with prior studies indicating basal expression of endogenous bfl-1/a1 in mouse WEHI-231 B cells, in which endogenous nuclear NF-κB complexes consist of c-Rel/p50 heterodimers (72). However, it is possible that RelA might contribute to bfl-1 gene expression in other cell types or in response to particular stimuli. Although we do not rule out the possibility that other transcription factors binding to sites that lie outside of the −933/−773 bfl-1 regulatory region may also participate in regulating bfl-1, our studies revealed a critical role for NF-κB in this process.
The coordinated involvement of AP-1, C/EBPβ, and NF-κB in bfl-1 gene expression implies an important role for Bfl-1 in rescuing cells from apoptosis in the immune system. Indeed, pairwise combinations of these factors act synergistically with one another in regulating expression of many genes involved in the acute phase of inflammation. Their combination is also responsible for activation of the inflammatory cytokines interleukin 1 (IL-1), IL-6, and TNF-α (24, 30, 57, 70). Architectural factor HMGI-C was implicated in enhancing NF-κB-mediated transcription and is rearranged in mesodermally derived tumors, such as B- and T-cell leukemias (4, 37). The strong induction of bfl-1 transcripts by proinflammatory cytokines in endothelial, leukemic, and hemopoietic cells is consistent with a role in protecting them from apoptotic extinction (14, 27, 34, 42). The observation that Bfl-1/A1 gene expression is also sharply induced in mature B cells that have entered the long-lived pool highlights its possible contribution to cell survival during negative selection (60). In this context, Bfl-1 might be viewed as a key determinant of cell fate for immature peripheral B cells.
Elevated bfl-1 gene expression was reported in several human cancers (28, 48). Moreover, cell treatment with the chemotherapeutic drug etoposide elevated bfl-1 transcripts in HT1080 fibrosarcoma cells, and this elevation was in turn responsible for their resistance to the killing effects of this drug (49, 63). Our findings that bfl-1 gene regulation is a multicomponent affair suggest that the engagement of signaling pathways leading to activation of NF-κB, AP-1, and C/EBPβ may need to be monitored to avoid desensitizing oncogenic cells to chemotherapeutic treatment. We have presented here the first evidence of an enhanceosome-like mechanism regulating expression of an antiapoptotic gene. It is not unreasonable to propose that other cell death regulators may be controlled in this manner, given the potentially devastating consequences associated with their inappropriate expression. Since NF-κB regulates a number of antiapoptotic genes and is involved in several different enhanceosome complexes, NF-κB-dependent enhanceosome control of other anti- or proapoptotic genes may come to light. Future studies will help to elucidate how their transcriptional regulation is controlled in normal cells and in cancer cells.
Acknowledgments
This work was supported by Public Health Service grants (CA83937 and CA54999) from the National Cancer Institute and by a grant from The Charlotte Geyer Foundation to C.G. We acknowledge partial support from an NIH predoctoral training grant in biotechnology (GM08339 [L.E.]) and an NIH predoctoral training grant in biochemistry and molecular biology (GM08360 [L.E., L.L., and M.S.]).
We are grateful to K. Chada, T. Curran, D. Livingston, D. Reinberg, N. Rice, W. Wang, and D.E. Zhang for the kind gift of reagents. We are indebted to S. Lomvardas and D. Thanos for the ChIP protocol and to E. White for sharing her real-time PCR instrument. We thank C. Abate-Shen, A. B. Rabson, and J. Suh for modified gel retardation assay procedures and M. Hampsey, A. B. Rabson, R. Steward, and members of the Gélinas laboratory for fruitful discussions and suggestions during the course of this work. We also thank M. Hampsey, A. Rabson, and B. Rayet for helpful comments on the manuscript.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, N. Diamant, and R. Dikstein. 2002. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 22:6354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-229. [DOI] [PubMed] [Google Scholar]
- 4.Ashar, H. R., M. S. Fejzo, A. Tkachenko, X. Zhou, J. A. Fletcher, S. Weremowicz, C. C. Morton, and K. Chada. 1995. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 82:57-65. [DOI] [PubMed] [Google Scholar]
- 5.Baker, S. J., T. K. Kerppola, D. Luk, M. T. Vandenberg, D. R. Marshak, T. Curran, and C. Abate. 1992. Jun is phosphorylated by several protein kinases at the same sites that are modified in serum-stimulated fibroblasts. Mol. Cell. Biol. 12:4694-4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister, A. J., and T. Kouzarides. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14:4758-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkett, M., and T. D. Gilmore. 1999. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18:6910-6924. [DOI] [PubMed] [Google Scholar]
- 8.Bash, J., W.-X. Zong, and C. Gélinas. 1997. c-Rel arrests the proliferation of HeLa cells and affects critical regulators of the G1/S-phase transition. Mol. Cell. Biol. 17:6526-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baud, V., and M. Karin. 2001. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11:372-377. [DOI] [PubMed] [Google Scholar]
- 10.Beg, A. A., and D. Baltimore. 1996. An essential role for NF-κB in preventing TNF-α-induced cell death. Science 274:782-784. [DOI] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Catron, K. M., J. R. Brickwood, C. Shang, Y. Li, M. F. Shannon, and T. P. Parks. 1998. Cooperative binding and synergistic activation by RelA and C/EBPβ on the intercellular adhesion molecule-1 promoter. Cell Growth Differ. 9:949-959. [PubMed] [Google Scholar]
- 13.Cheng, Q., H. H. Lee, Y. Li, T. P. Parks, and G. Cheng. 2000. Upregulation of Bcl-x and Bfl-1 as a potential mechanism of chemoresistance, which can be overcome by NF-κB inhibition. Oncogene 19:4936-4940. [DOI] [PubMed] [Google Scholar]
- 14.Choi, S. S., I.-C. Park, J. W. Yun, Y. C. Sung, S.-I. Hong, and H.-S. Shin. 1995. A novel Bcl-2 related gene, Bfl-1, is overexpressed in stomach cancer and preferentially expressed in bone marrow. Oncogene 11:1693-1698. [PubMed] [Google Scholar]
- 15.Craxton, A., P. I. Chuang, G. Shu, J. M. Harlan, and E. A. Clark. 2000. The CD40-inducible Bcl-2 family member A1 protects B cells from antigen receptor-mediated apoptosis. Cell Immunol. 200:56-62. [DOI] [PubMed] [Google Scholar]
- 16.Diehl, J. A., and M. Hannink. 1994. Identification of a C/EBP-Rel complex in avian lymphoid cells. Mol. Cell. Biol. 14:6635-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Sa-Eipper, C., T. Subramanian, and G. Chinnadurai. 1996. Bfl-1, a Bcl-2 homologue, suppresses p53-induced apoptosis and exhibits potent cooperative transforming activity. Cancer Res. 56:3879-3882. [PubMed] [Google Scholar]
- 18.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 20.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giese, K., C. Kingsley, J. R. Kirshner, and R. Grosschedl. 1995. Assembly and function of a TCR α enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995-1008. [DOI] [PubMed] [Google Scholar]
- 22.Gilmore, T. D., and H. M. Temin. 1986. Different localization of the product of the v-rel oncogene in chicken fibroblasts and spleen cells correlates with transformation by Rev-T. Cell 44:791-800. [DOI] [PubMed] [Google Scholar]
- 23.Grumont, R. J., I. J. Rourke, and S. Gerondakis. 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 13:400-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guha, M., W. Bai, J. L. Nadler, and R. Natarajan. 2000. Molecular mechanisms of tumor necrosis factor α gene expression in monocytic cells via hyperglycemia-induced oxidant stress-dependent and -independent pathways. J. Biol. Chem. 275:17728-17739. [DOI] [PubMed] [Google Scholar]
- 25.Holmgreen, S. P., D. C. Huang, J. M. Adams, and S. Cory. 1999. Survival activity of Bcl-2 homologs Bcl-w and A1 only partially correlates with their ability to bind pro-apoptotic family members. Cell Death Differ. 6:525-532. [DOI] [PubMed] [Google Scholar]
- 26.Karsan, A., E. Yee, and J. M. Harlan. 1996. Endothelial cell death induced by tumor necrosis factor-alpha is inhibited by the Bcl-2 family member, A1. J. Biol. Chem. 271:27201-27204. [DOI] [PubMed] [Google Scholar]
- 27.Karsan, A., E. Yee, K. Kaushansky, and J. M. Harlan. 1996. Cloning of a human Bcl-2 homologue: inflammatory cytokines induce human A1 in cultured endothelial cells. Blood 87:3089-3096. [PubMed] [Google Scholar]
- 28.Kenny, J. J., T. J. Knobloch, M. Augustus, K. C. Carter, C. A. Rosen, and J. C. Lang. 1997. GRS, a novel member of the Bcl-2 gene family, is highly expressed in multiple cancer cell lines and in normal leukocytes. Oncogene 14:997-1001. [DOI] [PubMed] [Google Scholar]
- 29.Kim, T. K., and T. Maniatis. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol. Cell 1:119-129. [DOI] [PubMed] [Google Scholar]
- 30.Koj, A. 1996. Initiation of acute phase response and synthesis of cytokines. Biochim. Biophys. Acta 1317:84-94. [DOI] [PubMed] [Google Scholar]
- 31.Lee, H. H., H. Dadgostar, Q. Cheng, J. Shu, and G. Cheng. 1999. NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. USA 96:9136-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, R. M., G. Gillet, J. Burnside, S. J. Thomas, and P. Neiman. 1999. Role of Nr13 in regulation of programmed cell death in the bursa of Fabricius. Genes Dev. 13:718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 34.Lin, E. Y., A. Orlofsky, M. S. Berger, and M. B. Prystowsky. 1993. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J. Immunol. 151:1979-1988. [PubMed] [Google Scholar]
- 35.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 36.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-β enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 37.Mantovani, F., S. Covaceuszach, A. Rustighi, R. Sgarra, C. Heath, G. H. Goodwin, and G. Manfioletti. 1998. NF-κB mediated transcriptional activation is enhanced by the architectural factor HMGI-C. Nucleic Acids Res. 26:1433-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayo, M. W., and A. S. Baldwin. 2000. The transcription factor NF-κB: control of oncogenesis and cancer therapy resistance. Biochim. Biophys. Acta 1470:M55-M62. [DOI] [PubMed] [Google Scholar]
- 39.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 40.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 41.Mink, S., B. Haenig, and K.-H. Klempnauer. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell. Biol. 17:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreb, J. S., and M. Schweder. 1997. Human A1, a Bcl-2-related gene, is induced in leukemic cells by cytokines as well as differentiating factors. Leukemia 11:998-1004. [DOI] [PubMed] [Google Scholar]
- 43.Munshi, N., Y. Yie, M. Merika, K. Senger, S. Lomvardas, T. Agalioti, and D. Thanos. 1999. The IFN-β enhancer: a paradigm for understanding activation and repression of inducible gene expression. Cold Spring Harbor Symp. Quant. Biol. 64:149-159. [DOI] [PubMed] [Google Scholar]
- 44.Noble, K. E., R. G. Wickremasinghe, C. DeCornet, P. Panayiotidis, and K. L. Yong. 1999. Monocytes stimulate expression of the Bcl-2 family member, A1, in endothelial cells and confer protection against apoptosis. J. Immunol. 162:1376-1383. [PubMed] [Google Scholar]
- 45.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 46.Pan, Z., C. J. Hetherington, and D. E. Zhang. 1999. CCAAT/enhancer-binding protein activates the CD14 promoter and mediates transforming growth factor beta signaling in monocyte development. J. Biol. Chem. 274:23242-23248. [DOI] [PubMed] [Google Scholar]
- 47.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 48.Park, I. C., S. H. Lee, D. Y. Whang, W. S. Hong, S. S. Choi, H. S. Shin, T. B. Choe, and S. I. Hong. 1997. Expression of a novel Bcl-2 related gene, Bfl-1, in various human cancers and cancer cell lines. Anticancer Res. 17:4619-4622. [PubMed] [Google Scholar]
- 49.Perera, L. P., and T. A. Waldmann. 1998. Activation of human monocytes induces differential resistance to apoptosis with rapid down regulation of caspase-8/FLICE. Proc. Natl. Acad. Sci. USA 95:14308-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 51.Ruocco, M. R., X. Chen, C. Ambrosino, E. Dragonetti, W. Liu, M. Mallardo, G. De Falco, C. Palmieri, G. Franzoso, I. Quinto, S. Venuta, and G. Scala. 1996. Regulation of HIV-1 long terminal repeats by interaction of C/EBP(NF-IL6) and NF-κB/Rel transcription factors. J. Biol. Chem. 271:22479-22486. [DOI] [PubMed] [Google Scholar]
- 52.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ′mini-extracts,' prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stehlik, C., R. de Martin, I. Kumabashiri, J. A. Schmid, B. R. Binder, and J. Lipp. 1998. Nuclear factor (NF)-κB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor α-induced apoptosis. J. Exp. Med. 188:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein, B., and A. S. Baldwin, Jr. 1993. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol. Cell. Biol. 13:7191-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stein, B., A. S. Baldwin, Jr., D. W. Ballard, W. C. Greene, P. Angel, and P. Herrlich. 1993. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 12:3879-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein, B., P. C. Cogswell, and A. S. Baldwin, Jr. 1993. Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol. Cell. Biol. 13:3964-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stylianou, E., and J. Saklatvala. 1998. Interleukin-1. Int. J. Biochem. Cell Biol. 30:1075-1079. [DOI] [PubMed] [Google Scholar]
- 58.Thanos, D., and T. Maniatis. 1992. The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 71:777-789. [DOI] [PubMed] [Google Scholar]
- 59.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFNβ gene expression requires the assembly of enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 60.Tomayko, M. M., and M. P. Cancro. 1998. Long-lived B cells are distinguished by elevated expression of A1. J. Immunol. 160:107-111. [PubMed] [Google Scholar]
- 61.Van Antwerp, D. J., S. J. Martin, T. Kafri, D. R. Green, and I. M. Verma. 1996. Suppression of TNF-α-induced apoptosis by NF-κB. Science 274:787-789. [DOI] [PubMed] [Google Scholar]
- 62.Van Antwerp, D. J., S. J. Martin, I. M. Verma, and D. R. Green. 1998. Inhibition of TNF-induced apoptosis by NF-κB. Cell Biol. 8:107-111. [DOI] [PubMed] [Google Scholar]
- 63.Wang, C.-Y., D. C. Guttridge, M. W. Mayo, and A. S. Baldwin, Jr. 1999. NF-κB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol. Cell. Biol. 19:5923-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, C. Y., M. W. Mayo, R. G. Korneluk, D. V. Goeddel, and A. S. Baldwin, Jr. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680-1683. [DOI] [PubMed] [Google Scholar]
- 65.Wolf, I., V. Pevzner, E. Kaiser, G. Bernhardt, E. Claudio, U. Siebenlist, R. Forster, and M. Lipp. 1998. Downstream activation of a TATA-less promoter by Oct-2, Bob1, and NF-κB directs expression of the homing receptor BLR1 to mature B cells. J. Biol. Chem. 273:28831-28836. [DOI] [PubMed] [Google Scholar]
- 66.Xu, X., C. Prorock, H. Ishikawa, E. Maldonado, Y. Ito, and C. Gélinas. 1993. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol. Cell. Biol. 13:6733-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMG I(Y) in the assembly and function of the IFN-β enhanceosome. EMBO J. 18:3074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-β enhanceosome activates transcription. Proc. Natl. Acad. Sci. USA 96:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Youn, H. D., T. A. Chatila, and J. O. Liu. 2000. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 19:4323-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zagariya, A., S. Mungre, R. Lovis, M. Birrer, S. Ness, B. Thimmapaya, and R. Pope. 1998. Tumor necrosis factor alpha gene regulation: enhancement of C/EBPβ- induced activation by c-Jun. Mol. Cell. Biol. 18:2815-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zong, W. X., J. Bash, and C. Gélinas. 1998. Rel blocks both anti-Fas- and TNFα-induced apoptosis and an intact Rel transactivation domain is essential for this effect. Cell Death Differ. 5:963-972. [DOI] [PubMed] [Google Scholar]
- 72.Zong, W. X., L. C. Edelstein, C. Chen, J. Bash, and C. Gélinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev. 13:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]