Abstract
Embryonic stem (ES) cells are immortal and pluripotent cells derived from early mammalian embryos. Transcription factor Oct3/4 is essential for self-renewal of ES cells and early mouse development. However, only a few Oct3/4 target genes have been identified. In this study, we found that F-box-containing protein Fbx15 was expressed predominantly in mouse undifferentiated ES cells. Inactivation of Oct3/4 in ES cells led to rapid extinction of Fbx15 expression. Reporter gene analyses demonstrated that this ES cell-specific expression required an 18-bp enhancer element located approximately 500 nucleotides upstream from the transcription initiation site. The enhancer contained an octamer-like motif and an adjacent Sox-binding motif. Deletion or point mutation of either motif abolished the enhancer activity. The 18-bp fragment became active in NIH 3T3 cells when Oct3/4 and Sox2 were coexpressed. A gel mobility shift assay demonstrated cooperative binding of Oct3/4 and Sox2 to the enhancer sequence. In mice having a β-galactosidase gene knocked into the Fbx15 locus, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining was detected in ES cells, early embryos (two-cell to blastocyst stages), and testis tissue. Despite such specific expression of Fbx15, homozygous mutant mice showed no gross developmental defects and were fertile. Fbx15-null ES cells were normal in morphology, proliferation, and differentiation. These data demonstrate that Fbx15 is a novel target of Oct3/4 but is dispensable for ES cell self-renewal, development, and fertility.
Embryonic stem (ES) cells are derived from mammalian blastocysts and maintain pluripotency, an ability to differentiate into all types of somatic and germ cells (32). Another important property of ES cells is their robust and infinite growth equivalent to tumor cells despite their normal karyotype. ES cells were developed from mouse blastocysts in 1981 (8, 15) and have been extensively used to generate knockout mice. Human ES cells were established in 1998 (33) and are considered promising sources for cell transplantation therapy.
POU transcription factor Oct3/4 is expressed specifically in pluripotent cells, including ES cells, early embryos, and germ cells (27, 31). Targeted disruption of the Oct3/4 gene in the mouse results in early embryonic lethality (21). The inner cellular mass of Oct3/4-null blastocysts differentiates exclusively into trophoblasts. Furthermore, conditional deletion of Oct3/4 in ES cells leads to spontaneous differentiation into trophectoderm (25), demonstrating that Oct3/4 is essential for self-renewal of ES cells and mouse early development.
Only a few Oct3/4 target genes have been identified. These include FGF-4 (4) and Rex-1 (2), in which Oct3/4 binds to an octamer motif, ATT(T/A)GCAT, located in regulatory elements. In FGF-4, SRY-related transcription factor Sox2 binds to a motif adjacent to the octamer sequence and synergistically activates transcription (5). In Rex-1, hypothetical factor ROX1 functions in a similar manner (2). It is not clear whether synergetic interaction with other transcription factors is common among target genes. Even consensus nucleotide sequences of Oct3/4-binding sites have not been fully determined. For example, the Oct3/4-binding site in UTF1 is one nucleotide different from the octamer sequence (22). Furthermore, it remains largely unknown how Oct3/4 maintains self-renewal of ES cells. Identification of novel Oct3/4 target genes is crucial to answering these questions.
In this study, we utilized expression analyses, reporter gene analyses, and a gel mobility shift assay to demonstrate that Fbx15, which encodes an F-box-containing protein (35), is a novel target of Oct3/4. We also performed gene-targeting experiments to study physiological functions of Fbx15 in self-renewal of ES cells, mouse development, and fertility.
MATERIALS AND METHODS
Digital Differential Display.
We utilized the Digital Differential Display program (http://www.ncbi.nlm.nih.gov/UniGene/info_ddd.shtml) to analyze gene representation in cDNA libraries from ES cells and other tissues. Expressed sequence tag (EST) libraries derived from ES cells were no. 274 and 220 (a total of 27,705 entries). Libraries used to represent various somatic tissues were no. 467, 318, 198, 408, 239, 400, 453, 109, 16, 523, 161, 483, 258, 264, 419, 529, 327, 411, 417, 418, 399, 196, 271, 255, 495, 101, 98, 351, 416, 321, 251, 412, 379, 549, 329, 265, 449, 328, 516, 320, 436, 427, 297, 366, 390, 315, 228, 277, 292, 284, 285, and 30 (a total of 1,328,835 entries).
Cell culture.
The RF8 (16), JI (13), CGR8 (20), and MG1.19 (9) ES cell lines were cultured as previously described. Differentiation of ES cells was induced with retinoic acid as previously described (36). NIH 3T3 cells were cultured with Dulbecco's modified Eagle medium (Sigma) containing 10% fetal bovine serum (Sanko Junyaku, Tokyo, Japan) and maintained at 37°C with 5% CO2.
Northern blot analyses.
Total RNAs were prepared from RF8 and MG1.19 ES cells with Trizol (Invitrogen). Total RNAs from untreated and tetracycline-treated ZHBTc4 ES cells were prepared as previously described (23). Total RNAs from 12 adult mouse tissues were purchased from Sawady Technology (Tokyo, Japan) and Funakoshi (Tokyo, Japan). Northern blot analyses were performed with formalin agarose gels as previously described (36, 37). As a probe, we used a NotI/SalI fragment of an EST clone (accession number AA571680) containing Fbx15 cDNA.
Reverse transcription (RT)-PCR.
First-strand cDNA was synthesized from total RNA with ReverTra Ace-α (Toyobo). Fbx15, Oct3/4, and NAT1 were amplified with primers U1110 and L1431, Oct3-U474 and Oct3-L935, and NAT1-U10 and NAT1-L21, respectively. The sequences of the primers used in this study are available upon request.
Western blot analyses.
ES cell extracts were prepared with M-Per (Pierce), separated on sodium dodecyl sulfate (SDS)-14% polyacrylamide gels, and transferred to nitrocellulose membranes (Millipore). The primary antibodies used were anti-Skp1 (SC1568, 1/600 dilution; Santa Cruz), anti-hemagglutinin (anti-HA; SC1568, 1/600; Roche), anti-myc (SC789, 1/600; Santa Cruz), and anti-GST (SC459, 1/2,000; Santa Cruz). The secondary antibodies used were horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (SC2030, 1/5,000; Santa Cruz) and horseradish peroxidase-conjugated anti-rat immunoglobulin G (SC2006, 1/5,000; Santa Cruz).
5′ RACE.
Fbx15 transcription initiation sites in ES cells were determined with the 5′ rapid amplification of cDNA ends (5′ RACE) system, version 2.0 (Invitrogen). The primers used were L1102 for cDNA synthesis and L782 and L350 for PCR.
Determination of the nucleotide sequence of the 5′ flanking region.
A bacterial artificial chromosome (BAC) clone containing the Fbx15 gene was identified from mouse BAC library DNA pools (Research Genetics) by PCR-based screening with primers U889 and L1102. BAC DNA was directly sequenced with primers Fbx-AS1, Fbx-AS2, Fbx-AS3, and Fbx-AS4 as previously described (37).
Construction of luciferase reporter plasmids.
A DNA fragment (−526 to +145) was amplified from the BAC clone by PCR with primers mFbx-pro.U637 and mFbx-pro.L1337 and cloned into pCR2.1 (Invitrogen). An EcoRI/MunI fragment of this was cloned into the EcoRI site of pGV-BM2, which we made from pGV-B (Promega) by inserting an EcoRI/XbaI linker into the KpnI site. The resulting plasmid was designated p(−526/+114)LUC and used to construct reporter plasmids with longer fragments as follows. PCR products amplified from the BAC clone with the primers described below were cloned into the pCR2.1 vector. DNA inserts were then isolated by EcoRI digestion and cloned into the EcoRI site of p(−526/+114)LUC. The upper primers used were Fbx-pro.U95 and Fbx-pro.U 573 for DNA fragments starting at positions −1120 and −640. The common lower primer used was mFbx-pro.L688, whose 5′ end was at position −504.
Construction of reporter plasmids with Fbx15 enhancer and minimum thymidine kinase promoter.
A PCR product amplified from the BAC clone with primers Fbx-pro.U573 and mFbx-pro.L688 was cloned into pCR2.1. An EcoRI fragment of the resulting plasmid, which contained the Fbx15 enhancer (−640 to −527), was blunt end ligated into the BglII site of pTA-Luc (Clontech). We constructed reporter plasmids with deleted or mutated sequences by inserting various linkers into the MluI/BglII site of pTA-Luc. The linker sequences are available upon request.
Construction of entry vectors for the Gateway cloning system.
We utilized Gateway cloning technology (Invitrogen) to construct expression vectors for various genes. In this method, genes of interest were first cloned into pDONR201 to construct entry vectors, which were then recombined with destination vectors to construct expression vectors. Coding regions of Fbx15, Skp1, and Cul1 were amplified by PCR from EST clones (accession numbers: AA571680 for Fbx15, AA08144 for Skp1, and BE533604 for Cul1). The primers used were fbx-gw-S and fbx-gw-AS for Fbx15, fbxC-gw-S and fbxC-gw-AS for the C-terminal portion of Fbx15 (amino acids 297 to 473), Skp1-gw-S and Skp1-gw-AS for Skp1, and Cul1-gw-S and Cul1-gw-AS for Cul1. Sox2 cDNA was amplified by RT-PCR from mouse ES cell total RNA with primers Sox-2-gw-S and Sox-2-gw-AS. The PCR products were reamplified with primers attB1-AD and attB2-AD and cloned into pDONR201 to construct pDONR-Fbx15, pDONR-Fbx15C, pDONR-Skp1, pDONR-CulI, and pDONR-Sox2.
Construction of destination vectors for the Gateway cloning system.
An NdeI/EcoRI fragment of pCX-EGFP (enhanced green fluorescent protein) containing the CAG promoter (26) was inserted into the same site of pIRES-neo (Clontech) to construct pCAG-IRES-neo. the Gateway refA cassette was blunt end ligated into the EcoRI site to construct pCAG-gw-IRES-neo. The Gateway refC cassette was blunt end ligated into the EcoRI/XbaI site of pCS2+MT (17) to construct pMyc-gw. An NheI/BamHI fragment of pEGFP-C2 (Clontech) was inserted into the same site of pcDNA3.1 (Clontech) to construct pcDNA3.1-EGFP. The Gateway refC cassette was blunt end ligated into the XhoI site of pcDNA3.1-EGFP to construct pcDNA3.1-EGFP-gw. pDEST17 was purchased from Invitrogen.
Construction of expression vectors.
The expression vectors constructed by a recombination reaction between the entry vectors and destination vectors were pMyc-Fbx15, pMyc-Skp1, pCAG-EGFP-Skp1, pDEST17-Fbx15C, and pCAG-Sox2-IRES-neo. pCAG-Oct3/4-IRES-neo was constructed as follows. An EcoRI/BclI fragment of the plasmid containing Oct3/4 cDNA (27) was inserted into the EcoRI/BamHI site of pIRES-neo (Clontech) to construct pCMV-Oct3/4-IRES-Neo. An NdeI/EcoRI fragment of pCAG-IRES-Neo containing the CAG promoter was inserted into the same site of pCMV-Oct3/4-IRES-Neo to construct pCAG-Oct3/4-IRES-Neo.
Construction of episomal expression plasmids.
BamHI/NotI fragments of pMyc-Fbx15 and pMyc-Skp1 and an Eco47III/XbaI fragment of pCAG-EGFP-Skp1 were blunt end ligated into the XhoI site of pCAGIP (24) to construct pCAGIP-Myc-Fbx15, pCAGIP-Myc-Skp1, and pCAGIP-EGFP-Skp1. pCAGIP-HA-gw (K. Takahashi et al., unpublished data) was recombined with pDONR-Cul1 and pDONR-Fbx15 by LR reaction to construct pCAGIP-HA-Cul1 and pCAGIP-HA-Fbx15.
Luciferase reporter assay.
DNA transfection into ES and NIH 3T3 cells was performed with Lipofectamine 2000 (Invitrogen) in accordance with the manufacture's protocol. After 24 h, cell lysates were collected and luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) as described elsewhere (37).
Immunoprecipitation.
Twenty-microgram samples of expression vectors were introduced into MG1.19 ES cells grown in 10-cm-diameter dishes. After 24 h, cell lysates were corrected with 500 μl of M-PER (Pierce) supplemented with 10 μl of protease inhibitor cocktail (Nacalai Tesque). Myc-tagged proteins were precipitated with agarose-conjugated anti-myc antibody sc40AC (Santa Cruz), separated by SDS-polyacrylamide gel electrophoresis (PAGE), and analyzed by Western blotting.
Generation of anti-Fbx15 polyclonal antibody.
pDEST17-Fbx15C was transformed into BL21-SI (Invitrogen). Recombinant protein was induced in accordance with the manufacture's protocol. Purification of histidine-tagged Fbx15C was performed with Ni-nitrilotriacetic acid agarose (Qiagen) under denaturing conditions. In brief, cells were lysed with buffer A and incubated with Ni-nitrilotriacetic acid resin. After washing with buffers C and D, histidine-tagged protein was eluted with buffer E. Purified proteins were dialyzed in phosphate-buffered saline containing 6 M urea overnight at 4°C. New Zealand White rabbits were immunized with this recombinant protein to generate anti-Fbx15 serum.
Gel mobility shift assay.
pCAGIP-HA-Oct3/4 and pCAGIP-HA-Sox2 (M. Maruyama et al., unpublished data) were introduced into Cos7 cells with Fugene 6 (Roche). Cell lysates were collected with extraction buffer (20 mM HEPES [pH 7.8], 450 mM NaCl, 0.4 mM EDTA, 0.5 mM dithiothreitol, 25% glycerol, 0.5 mM phenylmethylsulfonyl fluoride) supplemented with protease inhibitor cocktail and then dialyzed to remove salt. F9 embryonic carcinoma cell extract was purchased from Funakoshi. The DNA mobility shift assay was performed as described by Dent and Latchman (7), except that 0.5× Tris-borate-EDTA buffer was used for electrophoresis, gels were prerun for 10 min, and poly(dG-dC) was used instead of poly(dI-dC).
Targeted disruption of the mouse Fbx15 gene.
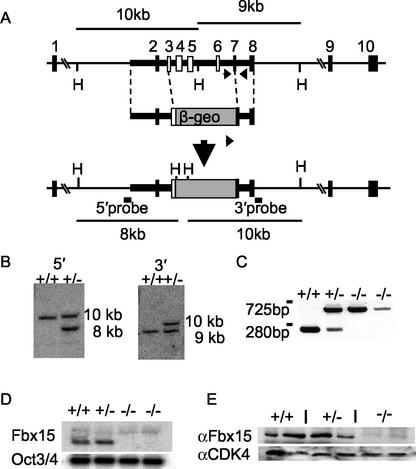
A targeting vector with promoter trap selection was designed to replace exons 3 to 7 of the mouse Fbx15 gene, which contain the F-box domain, with an IRES (internal ribosome entry site)-β-geo (fusion of β-galactosidase [β-gal]- and neomycin resistance-encoding genes) cassette (19). A 1.4-kbp fragment containing intron 1 to exon 3 and a 3.5-kbp fragment containing exons 7 and 8 were amplified from the BAC clone and used as the 5′ and 3′ homologous regions of the targeting vector. The 5′ arm was amplified by using a TOPO Walker Kit (Invitrogen) with primer L289 for extension and nested primer L255 for PCR. The 3′ arm was amplified by PCR with primers U939 and L1102. An IRES-β-geo cassette was ligated between the two DNA fragments. The resulting targeting vector was linearized by NotI digestion and introduced into RF8 ES cells by electroporation (16). Genomic DNAs from G418-resistant colonies were screened for homologous recombination by Southern blot analyses. For 5′ recombination, genomic DNA was digested with HindIII, separated on a 0.8% agarose gel, and transferred to a nylon membrane. Hybridization with a 600-bp probe from intron 1 produced a 10-kbp band from the wild-type locus and an 8-kbp band from the targeted locus. For 3′ recombination, the same membrane was hybridized with a 300-bp probe from intron 8, which produced a 9-kbp band from the wild-type locus and a 10-kbp band from the targeted locus.
Genotyping of mice and ES cells.
After identifying ES cell clones that were correctly targeted, we determined genotypes of mice and ES cells with a three-primer PCR. The first sense primer, −/−screeningS2, was designed from intron 6 to amplify the wild-type locus. The second sense primer, β-geo screening1, was designed from the β-geo cassette to amplify the targeted locus. A single antisense primer, −/−screeningAS2, was designed from intron 7 to amplify both the wild-type and targeted loci. PCR with these primers produces a 280-bp fragment from the wild-type locus and a 725-bp fragment from the targeted locus. PCR was performed with the Expand Long Template PCR system (Roche) in accordance with the manufacturer's protocol. The PCR program consisted of initial denaturation at 94°C for 2 min; 35 cycles of 94°C for 30 s, 53°C for 30 s, and 68°C for 1 min; and a final extension at 68°C for 7 min.
RESULTS
To isolate candidate genes that are expressed specifically in mouse ES cells under control of Oct3/4, we used digital differential display to compare gene representation between EST libraries derived from ES cells and those from various somatic tissues. We identified several genes that appeared only in ES cell-derived libraries. One of the candidate genes encoded Fbx15, a protein with an F-box domain (35). A BLAST search against whole-mouse dbEST databases (22 July 2000) with the full-length Fbx15 cDNA sequence as a query identified 16 cDNA clones corresponding to Fbx15 in libraries from ES cells and 39 clones in libraries from preimplantation embryos (16 2-cell embryo, 15 8-cell embryo, 2 16-cell embryo, and 6 blastocyst libraries [numbers of clones with scores of >200 bits]) but none in unfertilized egg, in vitro-fertilized egg, and other libraries.
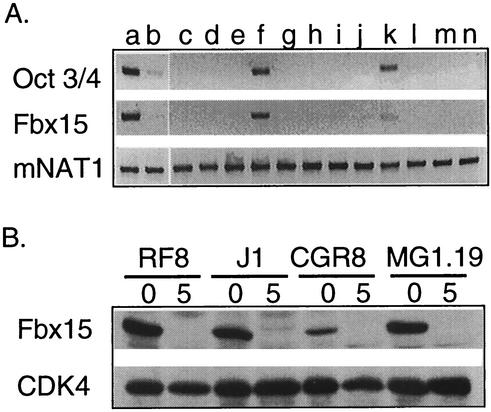
To determine whether Fbx15 is indeed expressed predominantly in ES cells, we studied its expression level by RT-PCR analyses. We found a high level of Fbx15 expression in MG1.19 ES cells (16) (Fig. 1A). Upon differentiation induced with retinoic acid, Fbx15 expression was repressed. Testis tissue also expressed Fbx15 but at a level lower than that at which undifferentiated ES cells did. There was faint expression in ovary tissue. No expression was detected in 10 other tissue types from adult mice. This Fbx15 expression pattern was nearly identical to that of Oct3/4 (Fig. 1A).
FIG. 1.
(A) RT-PCR analysis showing the expression profiles of mouse Oct3/4, Fbx15, and NAT1. Lanes: a, undifferentiated ES cells; b, differentiated ES cells; c, brain; d, heart; e, kidney; f, testis; g, spleen; h, muscle; i, lung; j, stomach; k, ovary; l, thymus; m, liver; n, skin. (B) Western blot analysis showing Fbx15 expression in the RF8, J1, CGR8, and MG1.19 ES cell lines. Lysates were collected from ES cells maintained undifferentiated (lanes 0) or treated with retinoic acid for 5 days (lanes 5). Western blotting was performed with anti-Fbx15 serum or anti-CDK4 antibody.
To confirm specific expression in ES cells at the protein level, we generated rabbit polyclonal antibody against the C-terminal portion (amino acids 297 to 473) of mouse Fbx15. Western blot analyses with the antiserum detected a single band of 55 kDa in four independent ES cell lines, RF8, J1, CGR8, and MG1.19 (Fig. 1B). Upon differentiation induced by retinoic acid, the Fbx15 protein disappeared in all four lines (Fig. 1B), confirming undifferentiated cell-specific expression at the protein level.
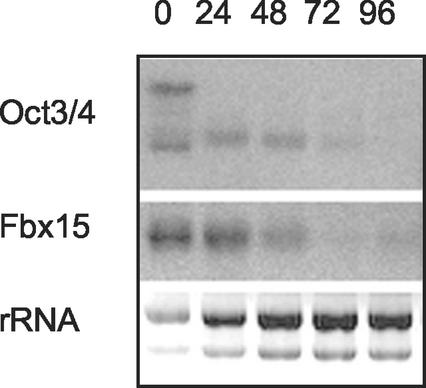
To study a putative role of Oct3/4 in the ES cell-specific expression of Fbx15, we utilized ZHBTc4 ES cells, in which both copies of the Oct3/4 gene were deleted while self-renewal was maintained by tetracycline-controlled expression of Oct3/4 (25). Because of a cryptic polyadenylation site, the transgene produced two transcripts that both encoded functional Oct3/4 protein (Fig. 2). Upon addition of tetracycline to the culture medium, the longer and shorter transcripts disappeared within 24 and 72 h, respectively. Expression of Fbx15 also disappeared within 72 h after tetracycline addition. This result suggests that Oct3/4 controls the expression of Fbx15 in ES cells.
FIG. 2.
Expression of Fbx15 in ZHBTc4 ES cells. Cells were treated with tetracycline, and the expression of Oct3/4 and Fbx15 was determined at the indicated time points by Northern blot analysis. Ethidium bromide staining of rRNAs was used as a loading control.
To further study the role of Oct3/4 in ES cell-specific expression of Fbx15, we analyzed the 5′ flanking region of the Fbx15 gene. We first determined the transcription initiation site by 5′ RACE. We obtained a single band from a PCR using gene-specific primers (data not shown), which was cloned into plasmid pCR2.1. We sequenced 10 clones and found that 7 of them started 17 nucleotides upstream from the 5′ end of the reported sequence. We consider this position a major transcription initiation site and refer to it as position +1 hereafter. Three other clones started at positions −26, −30, and −199, suggesting that multiple minor initiation sites also exist.TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCHJ.html) identified a putative binding site of the Sox family of transcription factors, AACAAT, between positions −534 and −528. It is noteworthy that an octamer-like sequence, TTTATCAT, was located immediately upstream of the Sox-binding site.
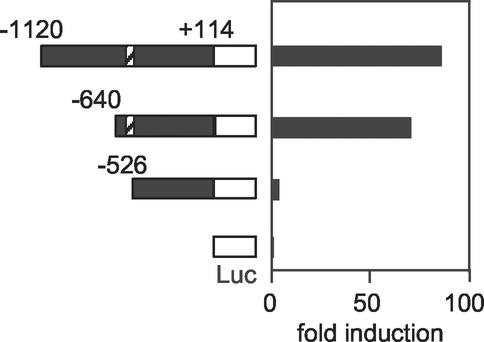
To study the roles of the octamer-like sequence and the Sox-binding motif, we constructed luciferase reporter vectors with DNA fragments that started at positions −1120, −640, and −526. These constructs were introduced into undifferentiated ES cells. The DNA fragment starting at position −640 showed marked enhancement in a luciferase activity compared to that starting at −526 (Fig. 3). Only a small enhancement was observed with the fragment starting at −1120. All constructs were inactive in differentiated ES cells and NIH 3T3 cells (data not shown). These data indicate that the DNA fragment (−640 to −527) containing the octamer-like motif and the Sox-binding site is essential for ES cell-specific expression of Fbx15.
FIG. 3.
Reporter gene analyses to define the Fbx15 enhancer. DNA fragments starting at positions −1120, −640, and −526 and ending at position +114 of the Fbx15 gene were placed in front of the firefly luciferase gene. These reporter constructs were introduced into undifferentiated ES cells. Shown is fold induction of normalized luciferase activity compared to that of the promoterless construct (pGV-BM2). Diagonally striped boxes represent the octamer-like and Sox-binding motifs.
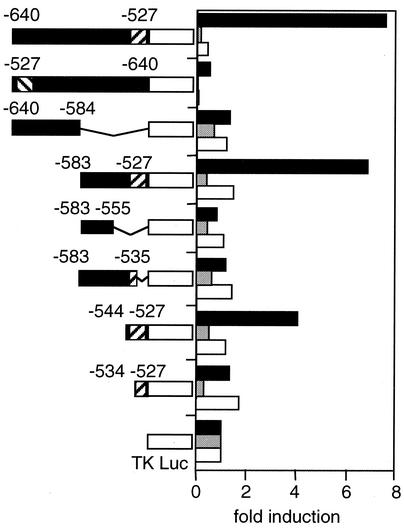
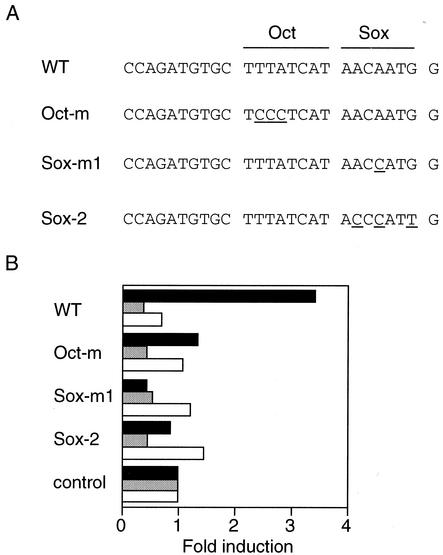
To further define the enhancer element, we located a DNA fragment corresponding to positions −640 to −527 and various deleted and/or mutated fragments into reporter constructs that contained luciferase cDNA driven by the minimum thymidine kinase promoter. The longest (114-bp) fragment was able to enhance luciferase activity in RF8 ES cells but not in differentiated ES cells or NIH 3T3 cells (Fig. 4). The same fragment inserted in the opposite orientation failed to enhance luciferase activity. Analyses by serial deletion demonstrated that the enhancer is located in an 18-bp fragment between positions −544 and −527 that contains the Sox-binding site and the octamer-like motif. Deletion of either of these two sites abolished the enhancer activity (Fig. 4). Furthermore, point mutation of the octamer-like sequence to TCCCTCAT or the Sox-binding site to AACCAT or ACCCAT (mutations are underlined) also abolished enhancer activity (Fig. 5). These data indicate that the octamer-like motif and the Sox-binding site are essential for the enhancer activity of Fbx15.
FIG. 4.
Enhancer analyses with the minimal thymidine kinase (TK) promoter. DNA fragments of various sizes were isolated from the Fbx15 gene and placed in front of the firefly luciferase gene driven by the minimal thymidine kinase promoter. These reporter constructs were introduced into undifferentiated ES cells (black column), differentiated ES cells (gray), and NIH 3T3 cells (white). Shown is fold induction of normalized luciferase activity compared to that of the enhancerless construct (pTA-Luc). Diagonally striped boxes represent the octamer-like and Sox-binding motifs.
FIG. 5.
Effects of point mutations in the octamer-like sequence or the Sox recognition motif. Reporter plasmids containing the Fbx15 enhancer sequence (WT) or mutated sequences (Oct-m, Sox-m1, and Sox-m2) were analyzed as described in the legend to Fig. 4.
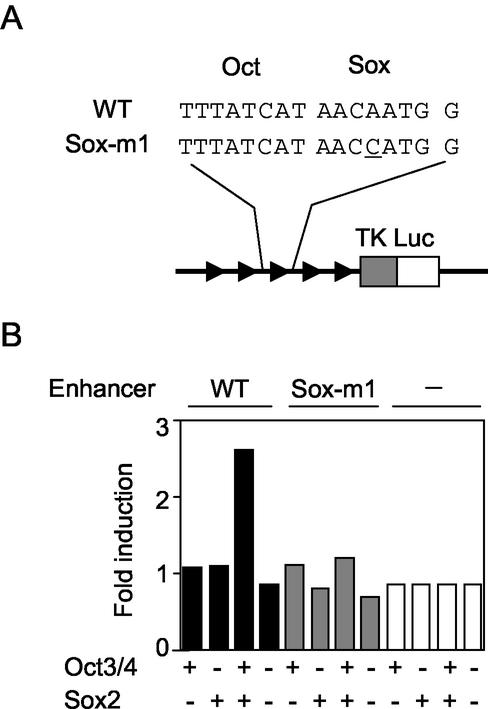
We next investigated whether Oct3/4 and Sox2 can activate the Fbx15 enhancer. We constructed a reporter gene in which the luciferase gene was driven by the minimum thymidine kinase promoter and five copies of the Fbx15 enhancer. When this construct was introduced into NIH 3T3 cells, luciferase activity was the same as the background level (Fig. 6). When it was transfected together with an Oct3/4 or Sox2 expression vector, no enhancement was observed. However, when it was cotransfected with both the Oct3/4 and Sox2 expression vectors, significant enhancement of luciferase activity was achieved. In contrast, Oct3/4 and Sox2 did not enhance a reporter gene that contained a single-nucleotide modification in the Sox-binding site. These data, taken together, demonstrate that Oct3/4 and Sox2 synergistically activated the Fbx15 enhancer.
FIG. 6.
Activation of the Fbx15 enhancer by Oct3/4 and Sox2 in NIH 3T3 cells. Reporter plasmids containing five copies of the Fbx15 enhancer sequence (WT) or a mutated sequence (Sox-m1) were introduced into NIH 3T3 cells with or without Oct3/4 and/or Sox2 expression vectors. Luciferase activities were analyzed as described in the legend to Fig. 4.
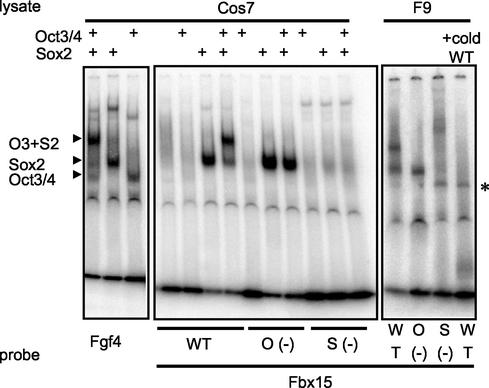
To further confirm the direct activation of the Fbx15 enhancer by Oct3/4 and Sox2, we performed a gel mobility shift assay. To determine the positions of bands corresponding to Oct3/4, Sox2, and their complex, we incubated 32P-labeled FGF-4 enhancer oligonucleotides with Cos7 cell extracts expressing Oct3/4, Sox2, or both (Fig. 7, left). When we incubated 32P-labeled Fbx15 enhancer oligonucleotides with a Cos7 extract expressing Oct3/4 alone, no shifted band was observed (Fig. 7 center). In contrast, when they were incubated with cell extracts expressing both Oct3/4 and Sox2, a shifted band corresponding to the complex was observed. This band was abolished by mutation either in the octamer-like sequence or in the Sox-binding site. When we incubated the 32P-labeled Fbx15 enhancer oligonucleotides with nuclear extracts of F9 embryonic carcinoma cells, we observed two shifted bands at the positions corresponding to the Oct3/4-Sox2 complex and the Sox2 monomer (Fig. 7, right). Both bands were eliminated by an excess amount of unlabeled Fbx15 enhancer oligonucleotides. Mutation either in the octamer-like sequence or in the Sox-binding site abolished the band corresponding to the Oct3/4-Sox2 complex. As expected, the band corresponding to the Sox2 monomer was abolished by the Sox mutation but not by the octamer mutation. These results demonstrate that Oct 3/4 directly binds to the Fbx15 enhancer with the help of Sox2.
FIG. 7.
Gel mobility shift assay. A 32P-labeled oligonucleotide (WT; ccagatgtgcTTTATCATAACAATGgaattcctaggggct) corresponding to the Fbx15 enhancer was incubated with F9 EC cell extract or Cos7 cell extract expressing Oct3/4, Sox2, or both. Oligonucleotides with mutations (underlined) in the octamer-like sequence [O (−); ccagatgtgcTCCCTCATAACAATGgaattcctaggggct] and the Sox-binding site [S (−); ccagatgtgcTTTATCATAACCATGgaattcctaggggct] were also tested. As a control, an FGF-4 enhancer oligonucleotide (tttaagtatcccATTAGCATccaAACAAAGagttttcta) was incubated with Cos7 cell extracts expressing Oct3/4, Sox2, or both. Shown on the left are the positions of bands corresponding to each transcription factor(s). The nature of the faint band indicated by the asterisk is not known. cold WT, unlabeled oligonucleotide corresponding to the Fbx15 enhancer.
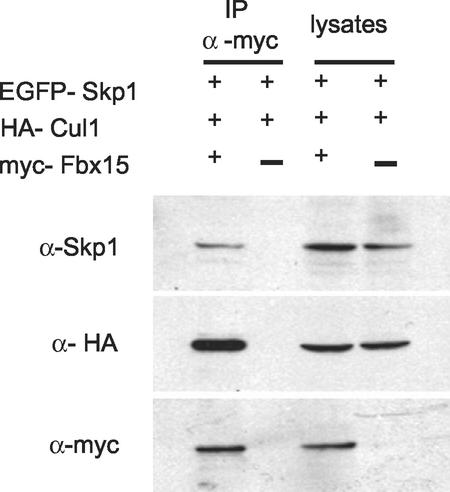
We next analyzed the functions of Fbx15. We first determined whether Fbx15 is a component of the SCF complex, as was the case with other F-box-containing proteins (1, 14). We constructed expression vectors of myc-tagged Fbx15, HA-tagged Cul1, and EGFP-tagged Skp1 and introduced them into MG1.19 ES cells. Expression of transgenes in cell lysates was confirmed by Western blotting with anti-Skp1, anti-HA, and anti-myc antibodies, respectively (Fig. 8). When myc-Fbx15 was immunoprecipitated with anti-myc antibody, both HA-cul1 and EGFP-Skp1 were copurified. In contrast, when myc-Fbx15 was omitted from the transfection, neither HA-Cul1 nor EGFP-Skp1 was precipitated with the anti-myc antibody. The association between Fbx15 and Skp1 was also demonstrated in the opposite direction. When myc-Skp1 and HA-Fbx15 were expressed and myc-Skp1 was precipitated, HA-Fbx15 was copurified (data not shown). These data indicate that Fbx15 forms the SCF complex.
FIG. 8.
Western blot analyses showing association of Fbx15 with Skp1 and Cul1. MG1.19 cells were transfected with myc-Fbx15, HA-Cul1, and EGFP-Skp1 expression vectors. Cell lysates and anti-myc antibody immunoprecipitates (IP) were separated by SDS-PAGE and immunoblotted with anti-Skp1 (top), anti-HA (middle), and anti-myc (bottom) antibodies. In a negative-control experiment, myc-Fbx15 was omitted from the transfection reaction mixture.
To elucidate the in vivo functions of Fbx15, we disrupted its mouse gene by homologous recombination. To this end, we constructed a targeting vector that replaced exons 3 to 7 containing the F box with a β-geo cassette (Fig. 9A). By introducing this vector into RF8 ES cells, we obtained 2 correctly targeted clones out of 200 screened. Both 5′ and 3′ junctions were confirmed by Southern blot analyses (Fig. 9B). These cells were injected into C57BL/6 blastocysts, and germ line transmission was obtained from one clone. Homozygous mutant mice were born from heterozygous intercrossing in accordance with Mendel's ratio (+/+, 22; +/−, 43; −/−, 18 [out of 83 examined]) and showed no gross abnormalities. Both male and female homozygous mice were fertile. These data showed that Fbx15 is not required for mouse development or fertility.
FIG. 9.
Targeted disruption of the mouse Fbx15 gene. (A) Structures of the Fbx15 genomic locus, a targeting vector, and the targeted locus generated by homologous recombination. The targeting vector contains the β-geo cassette in place of exons 3 to 7. The length of the diagnostic HindIII (H) restriction fragments and the locations of the 5′ and 3′ probes for Southern blot analyses are shown. Arrows indicate the primers for PCR analysis. The diagrams are not drawn to scale. (B) Southern blot analysis. Specific hybridization with the 5′ probe produces a 10-kb band from the wild-type locus and an 8-kb band from the targeted locus. Hybridization with the 3′ probe produces a 9-kb band from the wild-type locus and a 10-kb band from the targeted locus. +/+ and ± represent genotypes of Fbx15+/+ and Fbx15+/− cells, respectively. (C) PCR analysis. PCR with the three primers shown in panel A produces a 280-bp band form the wild-type locus and a 725-bp band from the targeted locus. (D) Northern blot analysis. Total RNA isolated from ES cells of each genotype was separated, blotted, and hybridized to cDNA probes of Fbx15 and Oct3/4. (E) Western blot analyses. Lysates isolated from ES cells of each genotype were separated by SDS-PAGE and immunoblotted with anti-Fbx15 serum or anti-CDK4 antibody.
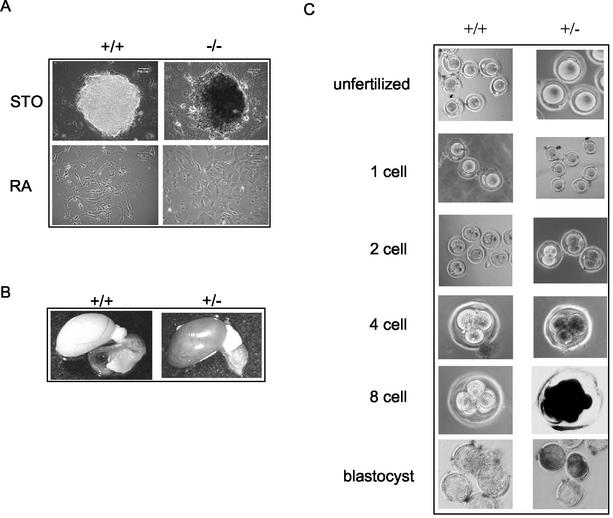
To study the functions of Fbx15 in ES cells, we established homozygous mutant ES cells by selecting heterozygous cells with a high concentration (6 mg/ml) of G418 (18). We obtained two homozygous clones (Fig. 9C). Northern (Fig. 9D) and Western (Fig. 9E) blot analyses demonstrated that Fbx15 transcripts and proteins were absent in both clones. Homozygous mutant cells were normal in morphology (Fig. 10A), Oct3/4 expression (Fig. 9D), and proliferation (data not shown). They were differentiated normally by retinoic acid (Fig. 10A) and formed normal embryoid bodies (data not shown). Furthermore, when injected subcutaneously into nude mice, they formed teratomas (5, 6) consisting of all three germ layers (data not shown).
FIG. 10.
Fbx15 expression represented by β-gal activity in mutant ES cells and mice. (A) Wild-type and Fbx15−/− ES cells were stained with X-Gal. Cells were either maintained undifferentiated on feeder cells or induced to differentiate with retinoic acid. (B) Whole-mount X-Gal staining of testes from wild-type and Fbx15+/− mice. (C) Whole-mount X-Gal staining of eggs from Fbx15+/+ and Fbx15+/− female mice mated with wild-type males.
We used mutant ES cells and mice to examine the expression profile of Fbx15 by means of the β-gal cassette knocked into the Fbx15 locus. Undifferentiated mutant ES cells showed strong β-gal activity (Fig. 10A). Upon differentiation, the activity was lost, indicating that β-gal expression mimicked the endogenous expression of Fbx15. In adult mice, 5-bromo-4-chloro-3-indolylphosphate (X-Gal) staining was only detected in testis tissue (Fig. 10B). No β-gal activity was detected in unfertilized eggs or fertilized one-cell stage eggs (Fig. 10C). X-Gal staining started to appear in two-cell stage embryos and reached a maximum in eight-cell embryos and blastocysts. These results confirmed that Fbx15 is specifically expressed in ES cells, early embryos, and testis tissue.
We also examined the function of Fbx15 in ES cells by a gain-of-function approach. We generated ES cells constitutively expressing Fbx15 cDNA from the CAG promoter. These cells expressed Fbx15 even after LIF removal or retinoic acid treatment (not shown). However, those cells differentiated normally when subjected to these treatments. They also formed normal embryoid bodies and teratomas (not shown). These findings indicate that Fbx15 alone is not capable of maintaining the undifferentiated state of ES cells.
DISCUSSION
Undifferentiated-cell-specific transcription factor Oct3/4 plays essential roles in self-renewal of ES cells and mouse early development. However, only a few Oct3/4 target genes have been identified. In this report, we presented three lines of evidence showing that Fbx15 is a novel target of Oct3/4. First, the expression profile of Fbx15 was nearly identical to that of Oct3/4 and was restricted in ES cells, early embryos, and testis tissue. In addition, conditional inactivation of Oct3/4 in ES cells led to rapid disappearance of Fbx15 transcripts. Second, analyses of the regulatory elements of Fbx15 demonstrated that an octamer-like motif and an adjacent Sox-binding motif are crucial for its expression in ES cells. Third, a gel mobility shift assay demonstrated direct and cooperative binding of Oct3/4 and Sox2 to the Fbx15 enhancer. These data suggested important roles of Fbx15 in ES cells and early development. Surprisingly, however, ES cells deficient in Fbx15 were normal in morphology, proliferation, and pluripotency. Homozygous mutant mice developed normally and were fertile. These data, taken together, demonstrate that mouse Fbx15 is a novel target of Oct3/4 but is dispensable for ES cell self-renewal, early development, and fertility.
The first evidence supporting the notion that Fbx15 is an Oct3/4 target is the high degree of similarity in expression profiles. Oct3/4 expression is restricted in pluripotent cells, including ES cells and germ cells (30). In unfertilized eggs and one-cell stage embryos, Oct3/4 is expressed at a low level (28). Its expression level becomes higher at the four-cell stage and reaches a maximum at the eight-cell and morula stages. Our Northern blot and RT-PCR analyses showed that Fbx15 expression was highest in ES cells, intermediate in testis tissue, and low in ovary tissue. No expression was detected in any of the other adult tissue types examined. X-Gal staining of β-gal knock-in mice demonstrated that Fbx15 expression was undetectable in unfertilized eggs and one-cell stage embryos. It was activated at the two-cell stage and was highest at the eight-cell stage. Thus, Fbx15 expression is restricted to cells and tissues where Oct3/4 is expressed at a high level, suggesting that Oct3/4 may control Fbx15 gene expression.
Additional data supporting the notion that Fbx15 is an Oct3/4 target came from ZHBTc4 ES cells, in which both copies of the Oct3/4 gene had been deleted by gene targeting (25). Self-renewal of these cells was maintained by transgene expression of Oct3/4 cDNA under control of the tetracycline-responsive element. Addition of tetracycline to culture medium resulted in rapid inactivation of the Oct3/4 transgene. Oct3/4 target genes, such as Rex-1, were rapidly repressed after transgene inactivation. Our study showed that Fbx15 expression disappeared within 72 h after tetracycline addition. Since inactivation of Oct3/4 led to spontaneous differentiation of ES cells, the decline in Fbx15 may be an indirect effect. However, the rapid reduction in Fbx15 argues that Oct3/4 controls Fbx15 expression.
Analyses of regulatory regions further support the notion that Fbx15 is a novel Oct3/4 target. Reporter gene analyses showed that the ES cell-specific enhancer (TTTATCATAACAAT) of the Fbx15 gene contained a sequence similar to the octamer motif (ATT[A/T]GCAT) (29) and an adjacent Sox-binding sequence (AACAAT) (11). Deletion or point mutation of either motif abolished the enhancer activity. The enhancer was inactive in NIH 3T3 cells but became active when Oct3/4 and Sox2 expression vectors were introduced. Furthermore, a gel mobility shift assay demonstrated direct binding of Oct3/4 and Sox2 to the enhancer. These data indicate that Oct3/4, together with Sox2, controls the Fbx15 enhancer.
The putative Oct3/4-binding site (TTTATCAT) in Fbx15 differs from the canonical octamer motif (ATT[A/T]GCAT) by two nucleotides (the mutations are underlined). Such variations have been documented in other cases. For example, although the octamer motif was first identified in and highly conserved among immunoglobulin genes, the human h100 gene has a motif, ATTTTCAT, that differs from the consensus by one nucleotide (29). The Oct3/4-binding site in UTF1 (ATCTGCAT) also differs from the consensus by one nucleotide (22). It is likely that these differences reduce affinity for Oct3/4. In consistent with this, our gel mobility shift assay suggested that Oct3/4 requires the cooperation of Sox2 for stable binding to the octamer-like sequence. This may explain why Fbx15 is undetectable in cells expressing low levels of Oct3/4, such as unfertilized eggs and one-cell stage embryos.
Fbx15 was identified as a novel F-box-containing protein. The F box was first defined in Skp2 and cyclin F as a Skp1-binding site (1). F-box-containing proteins bind Skp1 and Cul1 to form the SCF complexes that function as E3 ubiquitin ligases. In general, C-terminal portions of F-box proteins bind to substrate proteins, thus promoting their ubiquitin-dependent proteolysis. Our study showed that Fbx15 formed an SCF complex when Fbx15, Skp1, and Cul1 were overexpressed, suggesting its involvement in the ubiquitin-proteasome pathway. Fbx15 may play a role in determining the expression levels of regulatory proteins necessary for pluripotency. In B cells, the transcription coactivator OBF-1, which binds to Oct-1, Oct-2, and the octamer sequence, is regulated by ubiquitin proteasome-mediated degradation (34). A similar mechanism may operate in ES cells. However, whether endogenous Fbx15 forms the SCF complex has not been determined. Furthermore, Fbx15 lacks known domains for interaction with substrate proteins, such as leucine-rich and WD40 motifs. Recently, Fbx2 (38) and Fbx32 (3, 10), which both lack leucine-rich and WD40 motifs, were shown to function as ubiquitin ligases, suggesting the existence of novel substrate-interacting domains. Fbx15 may also bind to substrates via a novel interacting motif.
To understand the in vivo functions of Fbx15, we deleted its gene by homologous recombination in ES cells and mice. Despite its specific expression, Fbx15-null ES cells were normal in morphology, proliferation, and pluripotency. Mutant mice showed no gross abnormalities. Both male and female homozygous mice were fertile. These data suggest that Fbx15 does not play important roles. However, it is more likely that other genes play redundant roles, thus eliminating obvious phenotypes in Fbx15-null cells and mice. It is known that multiple ubiquitin ligases have the same substrates. For example, p27Kip1 is still degraded in Skp2−/− cells (12). An EST database search showed that ES cells express various F-box proteins, including Skp2, Fbl6, and Fbl12. Some of these may compensate Fbx15 functions.
In summary, we have demonstrated three lines of evidence supporting the idea that Fbx15 is a novel target of Oct3/4. We also generated ES cells and mice deficient in Fbx15. Lack of obvious phenotypes in these cells and mice suggested that other ubiquitin ligases compensate Fbx15. Identification of Fbx15 substrates and other ubiquitin ligases that use the same substrates and simultaneous gene knockout are required to understand the roles of the ubiquitin-proteasome pathway in self-renewal of ES cells, early mouse development, and fertility.
Acknowledgments
Y. Tokuzawa and E. Kaiho contributed equally to this work.
We are grateful to T. Ichisaka and Y. Samitsu for blastocyst microinjection. We thank C. Takigawa and J. Iida for technical assistance and R. Sasaki for plasmid construction.
This work was supported in part by grants from The Sumitomo Foundation, the Uehara Memorial Foundation, and the Japanese Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- 1.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodine, S. C., E. Latres, S. Baumhueter, V. K. Lai, L. Nunez, B. A. Clarke, W. T. Poueymirou, F. J. Panaro, E. Na, K. Dharmarajan, Z. Q. Pan, D. M. Valenzuela, T. M. DeChiara, T. N. Stitt, G. D. Yancopoulos, and D. J. Glass. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704-1708. [DOI] [PubMed] [Google Scholar]
- 4.Dailey, L., H. Yuan, and C. Basilico. 1994. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol. Cell. Biol. 14:7758-7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damjanov, I. 1978. Teratoma and teratocarcinoma in experimental animals. Natl. Cancer Inst. Monogr. 49:305-306. [PubMed] [Google Scholar]
- 6.Damjanov, I., and D. Solter. 1974. Experimental teratoma. Curr. Top. Pathol. 59:69-130. [DOI] [PubMed] [Google Scholar]
- 7.Dent, C. L., and D. S. Latchman. 1993. The DNA mobility shift assay, p. 1-26. In D. S. Latchman (ed.), Transcription factors: a practical approach. Oxford University Press, New York, N.Y.
- 8.Evans, M. J., and M. H. Kaufman. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154-156. [DOI] [PubMed] [Google Scholar]
- 9.Gassmann, M., G. Donoho, and P. Berg. 1995. Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 92:1292-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes, M. D., S. H. Lecker, R. T. Jagoe, A. Navon, and A. L. Goldberg. 2001. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 98:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haqq, C. M., C. Y. King, E. Ukiyama, S. Falsafi, T. N. Haqq, P. K. Donahoe, and M. A. Weiss. 1994. Molecular basis of mammalian sexual determination: activation of Mullerian inhibiting substance gene expression by SRY. Science 266:1494-1500. [DOI] [PubMed] [Google Scholar]
- 12.Hara, T., T. Kamura, K. Nakayama, K. Oshikawa, and S. Hatakeyama. 2001. Degradation of p27(Kip1) at the G0-G1 transition mediated by a Skp2-independent ubiquitination pathway. J. Biol. Chem. 276:48937-48943. [DOI] [PubMed] [Google Scholar]
- 13.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 14.Lyapina, S. A., C. C. Correll, E. T. Kipreos, and R. J. Deshaies. 1998. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. USA 95:7451-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, G. R. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78:7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meiner, V. L., S. Cases, H. M. Myers, E. R. Sande, S. Bellosta, M. Schambelan, R. E. Pitas, J. McGuire, J. Herz, and R. V. Farese, Jr. 1996. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc. Natl. Acad. Sci. USA 93:14041-14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsui, K., M. Nakanishi, S. Ohtsuka, T. H. Norwood, K. Okabayashi, C. Miyamoto, K. Tanaka, A. Yoshimura, and M. Ohtsubo. 1999. A novel human gene encoding HECT domain and RCC1-like repeats interacts with cyclins and is potentially regulated by the tumor suppressor proteins. Biochem. Biophys. Res. Commun. 266:115-122. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen, R. M., D. A. Conner, S. Chao, A. A. T. Geisterfer-Lowrance, and J. G. Seidman. 1992. Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol. 12:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mountford, P., B. Zevnik, A. Duwel, J. Nichols, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols, J., E. P. Evans, and A. G. Smith. 1990. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development 110:1341-1348. [DOI] [PubMed] [Google Scholar]
- 21.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 22.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa, H. 2001. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 26:137-148. [DOI] [PubMed] [Google Scholar]
- 24.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12:2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 26.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto, K., H. Okazawa, A. Okuda, M. Sakai, M. Muramatsu, and H. Hamada. 1990. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60:461-472. [DOI] [PubMed] [Google Scholar]
- 28.Palmieri, S. L., W. Peter, H. Hess, and H. R. Scholer. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166:259-267. [DOI] [PubMed] [Google Scholar]
- 29.Parslow, T. G., D. L. Blair, W. J. Murphy, and D. K. Granner. 1984. Structure of the 5′ ends of immunoglobulin genes: a novel conserved sequence. Proc. Natl. Acad. Sci. USA 81:2650-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pesce, M., and H. R. Scholer. 2001. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 19:271-278. [DOI] [PubMed] [Google Scholar]
- 31.Scholer, H. R., G. R. Dressler, R. Balling, H. Rohdewohld, and P. Gruss. 1990. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 9:2185-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, A. G. 2001. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 17:435-462. [DOI] [PubMed] [Google Scholar]
- 33.Thomson, J. A., J. Itskovitz-Eldor, S. S. Shapiro, M. A. Waknitz, J. J. Swiergiel, V. S. Marshall, and J. M. Jones. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145-1147. [DOI] [PubMed] [Google Scholar]
- 34.Tiedt, R., B. A. Bartholdy, G. Matthias, J. W. Newell, and P. Matthias. 2001. The RING finger protein Siah-1 regulates the level of the transcriptional coactivator OBF-1. EMBO J. 20:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winston, J. T., D. M. Koepp, C. Zhu, S. J. Elledge, and J. W. Harper. 1999. A family of mammalian F-box proteins. Curr. Biol. 9:1180-1182. [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka, S., X. Y. Zhang, M. Maeda, K. Miura, S. Wang, R. V. Farese, Jr., H. Iwao, and T. L. Innerarity. 2000. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 19:5533-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanaka, S., X. Y. Zhang, K. Miura, S. Kim, and H. Iwao. 1998. The human gene encoding the lectin-type oxidized LDL receptor (OLR1) is a novel member of the natural killer gene complex with a unique expression profile. Genomics 54:191-199. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, Y., T. Chiba, F. Tokunaga, H. Kawasaki, K. Iwai, T. Suzuki, Y. Ito, K. Matsuoka, M. Yoshida, K. Tanaka, and T. Tai. 2002. E3 ubiquitin ligase that recognizes sugar chains. Nature 418:438-442. [DOI] [PubMed] [Google Scholar]