Abstract
HMGI-Y is an architectural transcription factor that regulates gene expression in vivo by controlling the formation of stereospecific multiprotein complexes on the AT-rich regions of certain gene promoters. Recently, we demonstrated that HMGI-Y is required for proper transcription of the insulin receptor (IR) gene. Here we provide evidence that transcriptional activation of the human IR promoter requires the assembly of a transcriptionally active multiprotein-DNA complex which includes, in addition to HMGI-Y, the ubiquitously expressed transcription factor Sp1 and the CCAAT-enhancer binding protein β (C/EBPβ). Functional integrity of this nucleoprotein complex is required for full transactivation of the IR gene by Sp1 and C/EBPβ in cells readily expressing IRs. We show that HMGI-Y physically interacts with Sp1 and C/EBPβ and facilitates the binding of both factors to the IR promoter in vitro. Furthermore, HMGI-Y is needed for transcriptional synergism between these factors in vivo. Repression of HMGI-Y function adversely affects both Sp1- and C/EBPβ-induced transactivation of the IR promoter. Together, these findings demonstrate that HMGI-Y plays significant molecular roles in the transcriptional activities of these factors in the context of the IR gene and provide concordant support for the hypothesis that, in affected individuals, a putative defect in these nuclear proteins may cause decreased IR expression with subsequent impairment of insulin signaling and action.
Insulin produces a wide range of biological effects by binding to its receptor located on the plasma membrane (14, 34). Insulin receptors (IRs) are ubiquitous in vertebrate organisms, and this ubiquity reflects the fundamental and general significance of insulin in regulation of cellular metabolism and growth. Although the IR is ubiquitously expressed, data suggest that its expression can be regulated by a wide variety of factors under different environmental conditions. For example, glucocorticoids enhance transcription of the IR gene, whereas insulin down-regulates its own receptor (20). Also, IR mRNA levels increase during myocyte and adipocyte differentiation (1). Additionally, developmental regulation of IR gene expression has been documented (8).
Expression of the IR in the classical insulin target tissues, muscle, liver, and fat, is high. However, very little is known about the regulatory mechanisms controlling the IR at the level of gene expression. The IR is of major importance in certain states of insulin resistance in humans, in which decreased expression of the receptor may lead to defective transmembrane signaling (35, 37). Moreover, even though it is an open question whether the IR plays a critical role in aging and longevity in mammals as well as in Caenorhabditis elegans, IR signaling in the central nervous system plays an important role in regulation of energy disposal, fuel metabolism, and reproduction (4). Thus, regulation of IR gene expression seems to be important from both biological and clinical aspects.
As a step toward understanding the molecular basis of regulation of IR gene expression, the promoter region of the human IR gene has previously been identified and characterized (1, 20). This region extends over 1,800 bases 5′ upstream from the IR gene ATG codon, is extremely GC rich, and contains a series of GGGCGG repeats that are putative binding sites for the mammalian transcription factor Sp1 (27). The IR gene promoter lacks a TATA box or consensus initiator sequences and includes multiple transcription initiation sites primarily within the first 300-bp GC-rich region (20). Like other housekeeping promoters, the IR gene promoter confers a basal level of transcriptional activity common to all cells, whereas significantly higher transcriptional activity is induced in the muscle, liver, fat, and brain, suggesting the existence of tissue-specific transcriptional regulation for this promoter.
Previously, we identified two unique AT-rich sequences within the IR gene promoter, C2 and E3, which had a significant ability to drive transcription when introduced into mammalian cells (1). We have shown recently that both these sequences are positively regulated by the architectural transcription factor HMGI-Y, a distinct member of the high-mobility group (HMG) protein family (3). HMGI-Y binds to AT-rich regions in the minor groove of DNA via highly conserved DNA-binding peptide motifs called AT hooks and contributes importantly to the transcriptional activation of numerous mammalian genes in vivo by modifying DNA conformation and by recruiting transcription factors to the transcription start site (5, 31). Promoters of genes that are activated in a tissue-specific manner are often regulated by a combination of tissue-specific and ubiquitous transcription factors, where the ubiquitous element facilitates or enhances the action of one or more tissue-specific transcription factors (26). In this paper we demonstrate for the first time that HMGI-Y acts on the IR promoter as an element necessary for the formation of a transcriptionally active multiprotein-DNA complex involving, in addition to the HMGI-Y protein, the ubiquitously expressed transcription factor Sp1 and the C/EBPβ. We show that HMGI-Y physically interacts with Sp1 and C/EBPβ in vitro and in vivo and greatly enhances transactivation of the IR promoter by both these transcription factors in HepG2 human hepatoma cells, a cell line readily expressing IR. Additionally, overexpression of HMGI-Y in Epstein-Barr virus (EBV)-transformed lymphoblasts from a patient with type 2 diabetes associated with defects in HMGI-Y nuclear protein and reduced expression of IR significantly increases IR gene transcription and efficiently restores cell surface expression of the receptor and insulin-binding capacity.
MATERIALS AND METHODS
Cells and protein extracts.
HepG2 human hepatoma cells, 3T3-L1 mouse fibroblasts, and MCF-7 human breast cancer cells (>500 passages) (American Type Culture Collection, Manassas, Va.) were maintained in Dulbecco's modified Eagle medium (Gibco BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS). 3T3-L1 fibroblasts were differentiated into 3T3-L1 adipocytes as described elsewhere (12). Lymphoblast cell lines transformed with EBV were maintained in RPMI 1640 medium supplemented with 10% FBS. Nuclear extracts from these cell lines were prepared, with only minor modifications, as described previously (3), and the final protein concentration in the extract was determined by the modified Bradford method (Bio-Rad Laboratories, Hercules, Calif.).
Oligonucleotides and EMSA.
Wild-type and mutant C2 (300-bp) and E3 (300-bp) sequences of the human IR promoter region were generated by PCR amplification using the previously described recombinant plasmids pCAT-C2 and pCAT-E3 (1) and their mutagenized clones as templates (see below) and the following primers: C2 for, 5′-TCGAGTCACCAAAATAAACAT-3′ (for wild-type and mutated oligonucleotides); C2 rev, 5′-TGCAGGGGAGGGAGGTGCCGC-3′ (for wild-type oligonucleotides); pCAT-C2 rev, 5′-ATTGGGGATATATCAACGGTGGTATATCC-3′ (for mutated oligonucleotides); pCAT-E3 for, 5′-TACGCCAAGCTTGCATGCCTGCAG-3′ (for mutated oligonucleotides); E3 for, 5′-AGATCTCTGGCCATTGCACTCCAG-3′ (for wild type oligonucleotides); and E3 rev, 5′-TTCAATAAACAGTTTGCTAGGAGC-3′ (for wild type and mutated oligonucleotides). 32P-labeled C2 and E3 were used in electrophoretic mobility shift assays (EMSAs) as previously described (1). Double-stranded oligonucleotides containing wild-type or mutated binding sites for C/EBPβ (Santa Cruz Biotechnology, Santa Cruz, Calif.) were used in competition studies. Double-stranded 45-mer oligonucleotides containing relevant binding elements for HMGI-Y (5′-GAGAAAAACTCCATCTaaaaaaaaaaaaaaaaaaaaaAAAAAACA-3′) or Sp1 (5′-GGGAGGCggGGAGgCGgGCGGGGCggGGCGGGACCGgGCgGCACC-3′) were synthesized chemically (Life Technologies, Gaithersburg, Md.) from E3 and C2, respectively, and used in decoy experiments (only the coding strand is shown; mutagenized bases are identified by lowercase letters; HMGI-Y and Sp1 binding sites are underlined).
GST pull-down assay.
35S-labeled proteins (hemagglutinin [HA]-tagged HMGI, Sp1, and C/EBPβ) were synthesized in vitro by using the TNT-T7 quick-coupled transcription/translation system (Promega, Madison, Wis.). Glutathione S-transferase (GST) fusion protein expression vectors including those for HMGI and derivatives, a kind gift from D. Thanos (University of Columbia, New York, N.Y.), and Sp1 and derivatives, a kind gift from H. Rotheneder (University of Vienna, Vienna, Austria), and the pGEX-2TK control vector (Amersham Pharmacia Biotech, Piscataway, N.J.) were transformed into the BL21 strain of Escherichia coli (Stratagene, La Jolla, Calif.), expanded in suspension culture, and induced for 2 h with 0.5 mM isopropyl-d-thiogalactopyranoside. Bacteria were pelleted, sonicated in ice-cold phosphate-buffered saline lysis buffer containing 1% NP-40, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg of pepstatin A, leupeptin, and aprotinin/ml, and 0.4 mg of lysozyme/ml, and centrifuged. The resultant supernatant was then added to 300 μl of glutathione-agarose beads, mixed on a rotating wheel at 4°C for 2 h, and centrifuged. Bound GST-fused proteins in the pellet were washed five times with lysis buffer and resuspended in 300 μl of binding buffer (50 mM NaCl, 20 mM Tris-HCl [pH 8.0], 0.05% NP-40, 0.25% bovine serum albumin [BSA], 1 mM PMSF, 1 mM DTT). Bound protein was quantitated with the Coomassie protein assay reagent (Pierce, Rockford, Ill.), and 0.5 μg of each GST-protein bound to glutathione-agarose beads was incubated with 7 μl of in vitro-translated 35S-labeled protein in 150 μl of binding buffer at 4°C for 2 h. Reactions were terminated by centrifugation, the precipitate was washed three times with protein binding buffer and subjected to a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE), and proteins were visualized by autoradiography.
Immunoprecipitations and immunoblotting.
For immunoprecipitations, aliquots of HepG2 or 3T3-L1 cell nuclear extract or HMGI-Y and pure Sp1 together were incubated for 3 h with rotation at 4°C with 10 μl of antibody-coupled protein A beads. Beads were recovered by gentle centrifugation and washed three times with 500 μl of NETN wash buffer (0.1% NP-40, 150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl [pH 8.0]) for 5 min. Protein was removed from the beads by boiling in sample buffer for 5 min and analyzed by SDS-PAGE and immunoblotting as described previously (3). For immunoprecipitation studies with the Sp1 antibody, primary antibody-agarose conjugate (PEP 2; 10 μg; Santa Cruz Biotechnology) was added to nuclear extract from 3T3-L1 cells and protein was analyzed as described above. Western blot analyses of HMGI-Y, Sp1, and C/EBPβ in nuclear extracts from HepG2, MCF-7, and EBV-transformed lymphoblasts from healthy and diabetic individuals were performed as described previously (3). Antibodies used for these studies were as follows: anti-HMGI-Y (3), anti-Sp1 (PEP 2; 1:250), and anti-C/EBPβ (C-19; 1:1,000) (Santa Cruz Biotechnology). C/EBPβ (Δ198) antigen (Santa Cruz Biotechnology) was used as a positive control.
For covalent coupling of antibodies to protein A-Sepharose (Amersham Pharmacia), an anti-HMGI-Y or anti-C/EBPβ polyclonal antibody was mixed with beads and bound for 1 h with rotation at room temperature. After extensive washing with 200 mM sodium borate (pH 8.0), solid dimethyl pimelimidate (Sigma, St. Louis, Mo.) was added to a final concentration of 20 mM and the components were mixed on roller for 30 min at room temperature. To stop the reaction, the beads were washed twice in 200 mM ethanolamine, pH 8.0, and incubated on roller for 2 h at room temperature in 200 mM ethanolamine. Antibody-coupled protein A beads were washed twice in phosphate-buffered saline and used in immunoprecipitation studies.
Immunodepletion and in vitro transcription.
Protein A beads (100 μl) were suspended in 400 μl of transcription buffer (30 mM HEPES [pH 7.9], 3 mM MgCl2, 140 mM KCl) and mixed gently with 100 μl of either anti-Sp1 or anti-HMGI-Y polyclonal antibodies or control (unrelated rabbit serum immunoglobulin G) antibodies by end-over-end rotation for 2 h at 4°C. HeLa nuclear extracts (400 μl; Promega) containing 0.1 mg of BSA/ml were gently mixed with 100 μl of antibody-coupled beads and incubated for 30 min at 4°C. This step was repeated three times, and the extracts were then transferred to UFCMC ultrafiltration units (Millipore, Bedford, Mass.) and centrifuged at 2,000 × g for about 2 h at 4°C to achieve an approximately threefold concentration. Immunodepleted HeLa extracts were employed in in vitro transcription studies using the HeLa cell extract in vitro transcription kit (Promega) in the presence of the linearized pCAT-IR plasmid (1) as the DNA template, with or without HMGI-Y (3) and/or pure Sp1, a kind gift from R. Tjian (University of California, Berkeley). Hybridization of RNA transcripts with a 32P-labeled chloramphenicol acetyltransferase (CAT) primer and reverse transcription were carried out as reported previously (39). For competition experiments, double-stranded oligonucleotides containing wild-type or mutated HMGI-Y and/or Sp1 or C/EBPβ binding sites were added to untreated HeLa extracts and the extracts were incubated for 15 min at room temperature before exposure to the IR-CAT template.
Plasmids and mutagenesis.
Eukaryotic expression plasmids used in this study were as follows: pcDNA1-HMGI-Y (sense and antisense), a kind gift from T. Maniatis (Harvard University, Cambridge, Mass.), pEVR2/Sp1, a kind gift from G. Suske (Philipps University Marburg, Marburg, Germany), and pSG5-C/EBPβ, a kind gift from T. Penning (University of Pennsylvania, Philadelphia). Plasmids pSG5-C/EBPβ and pcDNA3-Sp1, a kind gift from K.-S. Chang (University of Texas, Houston), and the HA-I expression vector for HA-tagged HMGI, a kind gift from G. Manfioletti (University of Trieste, Trieste, Italy) were used in in vitro transcription and translation.
Site-directed mutagenesis of DNA binding sites for HMGI-Y and/or Sp1 in pCAT-C2 and pCAT-E3 was performed by the overlap extension method (11) using wild-type C2 and E3 sequences as initial templates in PCRs and the following primers (Life Technologies Inc.): C2-HMGI-Y for, 5′-GCCCACTATGAACccAATAGCAAccTGGTAGAGAAAGG-3′, and C2-HMGI-Y rev, 5′-CCTTTCTCTACCAggTTGCTATTggGTTCATAGTGGGC-3′; C2-Sp1 for, 5′-CCCGGCACAGGGAGGCttGGAGaCGtGCGGGGCG-3′, and C2-Sp1 rev, 5′-CGCCCCGCaCGtCTCCaaGCCTCCCTGTGCCGGG-3′ (first round); C2-Sp1 for, 5′-GACGTGCGGGGCttGGCGGGACCGaGCtGCACCTCCCTCC-3′, and C2-Sp1 rev, 5′-GGAGGGAGGTGCaGCtCGGTCCCGCCaaGCCCCGCACGTC-3′ (second round); E3-HMGI-Y for, 5′-AACTCCATCTcttctagAAAAAAAAAAAAAAAAAAAACAG-3′, and E3-HMGI-Y rev, 5′-CTGTTTTTTTTTTTTTTTTTTTTctagaagAGATGGAGTT-3′ (first round); E3-HMGI-Y for, 5′-ATCTCTTCTAGAAgcgctagctAAAAAAAAACAGAGAG-3′ and E3-HMGI-Y rev, 5′-CTCTCTGTTTTTTTTTagctagcgcTTCTAGAAGAGAT-3′ (second round); E3-Sp1 for, 5′-AAGGAAACGGAACTGtGatGAtGATTTGCAAAAATATG-3′, and E3-Sp1 rev, 5′-CATATTTTTGCAAATCaTCtaCaCTGTTCCGTTTCCTT-3′. Mutagenized bases are in lowercase letters. Mutations were confirmed by sequence analysis.
Transfection studies.
Recombinant vectors containing wild-type or mutant versions of the C2 or E3 promoter element and effector vectors for HMGI-Y (HMGI isoform protein), Sp1, or C/EBPβ were transiently transfected into HepG2 and MCF-7 cells by the calcium phosphate precipitation method, and CAT activity was assayed 48 h later as previously described (1). EBV-transformed lymphoblasts (4 × 106 cells per plate) were transfected by the DEAE-dextran method (3, 30) in 60-mm-diameter dishes and cells were harvested 72 h after transfection. As an internal control of transfection efficiency, β-galactosidase activity was measured in addition to protein expression levels (3). For antisense experiments, reporter vectors were cotransfected into cells with the expression plasmid pcDNA1 containing the HMGI-Y cDNA in the sense or antisense orientation in the absence or presence of Sp1 antisense (5′-CTGAATATTAGGCATCACTCCAGG-3′) or sense (5′-CCTGGAGTGATGCCTAATATTCAG-3′) commercially synthesized phosphorothioated deoxynucleotides (Life Technologies) (40). 125I-insulin binding to HepG2 cells and EBV-transformed lymphoblasts treated with HMGI-Y and/or Sp1 antisense deoxynucleotides was measured 72 h after transfection, as described previously (3). To assess the effect of distamycin A (Sigma) and/or bisanthracycline WP631 (Vinci-Biochem Alexis, Vinci, Italy) on the IR promoter, transfected HepG2 cells were incubated with medium containing DNA-binding drugs and analyzed for CAT activity 48 h after transfection. For decoy experiments, MCF-7 cells and EBV-transformed lymphoblasts were cotransfected with CAT reporter vectors plus 20 μg of double-stranded oligonucleotides containing wild-type or mutated C/EBPβ binding site sequences and CAT activity was measured as described above.
RESULTS
Nuclear protein-DNA interactions within C2 and E3 regions of the IR promoter.
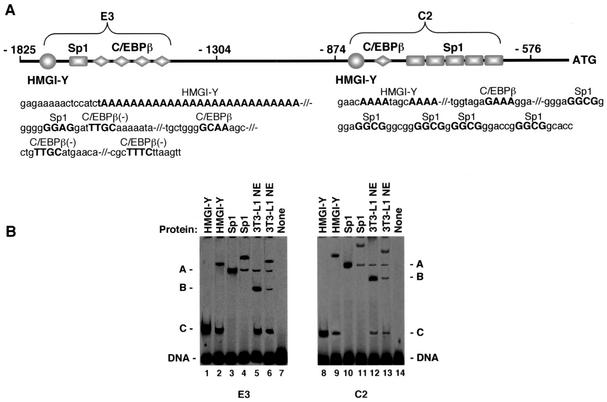
In an attempt to identify the biochemical mechanisms that underlie the stimulatory role of HMGI-Y in IR gene expression, we performed experiments designed to investigate whether HMGI-Y was necessary for recruitment and binding of transcription factors to the IR promoter. Figure 1A shows a schematic representation of the promoter region of the human IR gene, along with the two elements of interest, C2 and E3. Consensus transcription factor-binding sites within these two elements were explored by using the TRANSFAC database searched with MatInspector (29), and the TFMATRIX transcription factor-binding site profile database was searched with TFSEARCH (10). Putative binding sites for Sp1 and C/EBPβ, both flanked by AT-rich HMGI-Y binding sites, were identified in each region and were characterized by EMSA using radiolabeled fragment C2 or E3. As shown in Fig. 1B, the binding of either HMGI-Y (pure recombinant HMGI isoform protein [3]) or pure Sp1 to these probes produced a protein-DNA complex that was recognized and supershifted to a slower-migrating form by the anti-HMGI-Y or anti-Sp1 antibody, respectively. Nuclear extracts from 3T3-L1 cells were used as a source of C/EBPβ nuclear protein. The binding of C/EBPβ to C2 and E3 probes yielded a protein-DNA complex that was recognized and supershifted by the anti-C/EBPβ antibody. The two additional complexes, other than that including C/EBPβ, in 3T3-L1 nuclear extracts included Sp1 and HMGI-Y, as detected by antibody supershift analyses (not shown). To show individual binding of HMGI-Y, Sp1, and C/EBPβ, the formation of a large complex involving the simultaneous binding of all three proteins on the same probe was prevented by using relatively small amounts of nuclear extract in the presence of competitor DNA.
FIG. 1.
(A) Schematic representation of the regulatory region of the human IR gene with HMGI-Y, Sp1, and C/EBPβ binding sites (uppercase) within C2 and E3 promoter elements. Contiguous binding sites are separated by shills. -, site on the minus strand. (B) EMSA of radiolabeled fragments C2 and E3 (0.2 ng each) with 2.5 ng of either HMGI-Y or pure Sp1, in the presence of 2.0 μg of BSA or 0.5 μg of 3T3-L1 nuclear extract (NE) and 0.2 μg of poly(dI-dC) as the competitor DNA. In supershift assays the protein was preincubated with 1 μg of the polyclonal antibody to HMGI-Y (lanes 2 and 9), Sp1 (lanes 4 and 11), or C/EBPβ (lanes 6 and 13) before addition of the probe. A, B, and C indicate binding of Sp1, C/EBPβ, and HMGI-Y, respectively.
HMGI-Y enhances the binding of Sp1 to the IR promoter.
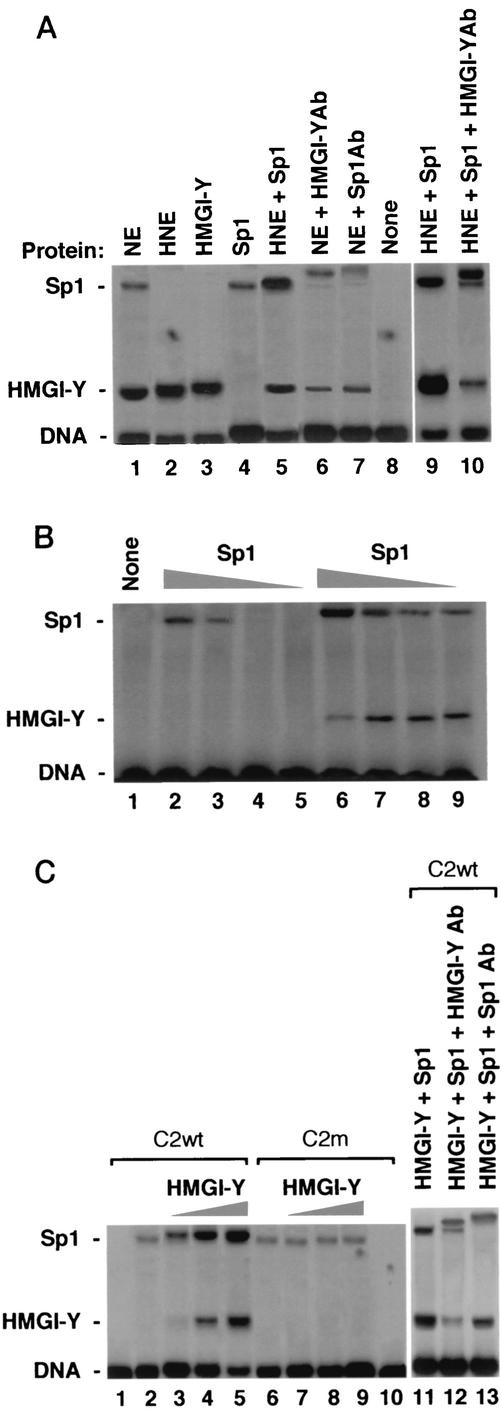
To analyze whether HMGI-Y exerted any influence on the binding of Sp1 to the IR promoter, we initially performed EMSAs by incubating radiolabeled fragment C2 or E3 with either untreated or heat-treated nuclear extract (HNE) from HepG2 cells, a cell line naturally expressing both HMGI-Y and Sp1 nuclear proteins and widely used in IR expression studies (3). Whereas heat treatment of nuclear extract eliminates the DNA binding of endogenous Sp1, HMG proteins are heat stable (38). As shown in Fig. 2A, the binding of pure Sp1 to probe C2 increased when HNE was added to the reaction mixture. Evidence that HMGI-Y was present in the HNE and enhanced Sp1 binding to DNA was provided by the addition of HMGI-Y antibody to the reaction mixture. In untreated nuclear extract, the presence of HMGI-Y in the more rapidly migrating complex was verified by its supershifting in the presence of the anti-HMGI-Y antibody, whereas the presence of Sp1 in the slowly migrating complex was verified by its supershifting in the presence of the anti-Sp1 antibody. The reduction of either Sp1 or HMGI-Y binding in the presence of the HMGI-Y or Sp1 antibody, respectively, suggests that both factors are simultaneously binding the same labeled probe. No effect on DNA-protein binding was observed in the presence of the control unrelated antibody (data not shown). In titration experiments, the binding of Sp1 to probe C2 was observed with as little as 0.25 ng of pure Sp1 in the presence of pure HMGI-Y, whereas in the absence of HMGI-Y an Sp1-DNA complex was first detected with 1 ng of Sp1 protein (Fig. 2B), indicating that HMGI-Y substantially stimulates Sp1 binding to its DNA binding site.
FIG. 2.
Binding of Sp1 to the IR promoter is enhanced by HMGI-Y and is prevented by mutational interference with HMGI-Y binding sites. (A) EMSA with probe C2 and 0.5 μg of either untreated nuclear extract (NE) or HNE from HepG2 cells and pure HMGI-Y or Sp1 protein (2 ng each). In supershift assays, NE was preincubated with 0.5 μl of an antibody (Ab) against either HMGI-Y or Sp1. Supershift analysis with HNE was performed with the HMGI-Y antibody in the presence of pure Sp1. (B) Probe C2 was incubated with decreasing amounts of pure Sp1 (lanes 2 and 6, 2 ng; lanes 3 and 7, 1 ng; lanes 4 and 8, 0.5 ng; lanes 5 and 9, 0.25 ng) in the absence (lanes 2 to 5) or presence (lanes 6 to 9) of pure HMGI-Y (2 ng). Lane 1, probe alone. (C) EMSA was performed with 2.5 ng of pure Sp1 plus increasing amounts of pure HMGI-Y (lanes 2 and 6, 0 ng; lanes 3 and 7, 1 ng; lanes 4 and 8, 2 ng; lanes 5, 9, and 11, 4 ng), in the presence of wild-type (wt) or mutant (m) probe C2. Lanes 1 and 10, probe alone. Supershift analysis with pure HMGI-Y and Sp1 proteins was performed with an antibody against either HMGI-Y or Sp1 (lanes 12 and 13).
To analyze whether the binding of HMGI-Y to DNA is a prerequisite for the stimulation of Sp1 binding, we performed titration experiments with wild-type probe C2 and mutant versions, in which mutations within the A stretches were generated. Such mutant probes should still bind Sp1 but should prohibit the binding of HMGI-Y. As shown in Fig. 2C, the binding of Sp1 to wild-type probe C2 was enhanced and slightly retarded in the presence of pure HMGI-Y, confirming that HMGI-Y and Sp1 were simultaneously binding adjacent sites on the same probe. HMGI-Y-DNA complexes were totally undetectable with probe C2m, and the binding of Sp1 remained virtually unaffected by HMGI-Y. Similar results were obtained with a mutant version of probe E3, which was generated by eliminating 21 bases of the A27 stretch within this sequence (data not shown). Under no conditions did we observe an additional complex of low mobility in the presence of HMGI-Y, suggesting that a ternary complex of C2 or E3, HMGI-Y, and Sp1, if it existed, was either transient or unstable under our assay conditions. The existence of HMGI-Y in a cocomplex with Sp1 was confirmed in supershift analyses using anti-HMGI-Y and anti-Sp1 antibodies (Fig. 2C). Taken together, these results clearly indicate that the binding of HMGI-Y to the IR promoter is required for the enhancement of Sp1 binding and suggest a molecular mechanism for HMGI-Y stimulation of the IR gene. The absence of any effect on Sp1 binding when contact between HMGI-Y and DNA was prevented is consistent with the notion that the binding of HMGI-Y may alter DNA structure so as to make it more favorable for Sp1 recognition.
HMGI-Y interacts with Sp1 in the absence of DNA.
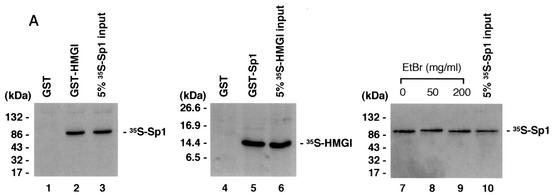
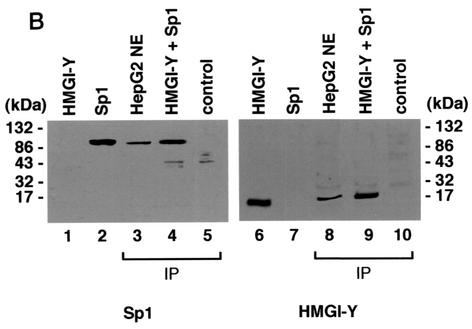
Direct physical association between HMGI-Y and multiple transcription factors has been reported (31, 36). To analyze the ability of HMGI-Y and Sp1 to interact with each other in vitro, in the absence of DNA, we first performed a GST pull-down assay, in which in vitro-translated 35S-labeled Sp1 was analyzed for its ability to be specifically retained by a GST-HMGI affinity resin. As shown in Fig. 3A, Sp1 was retained by GST-HMGI but not GST alone, suggesting that Sp1 interacts physically with HMGI-Y in vitro. In a reciprocal experiment, in vitro-synthesized 35S-labeled HMGI bound specifically to a resin carrying GST-Sp1 but not to a resin carrying GST alone. A bona fide interaction between HMGI and Sp1 was observed in the presence of high concentrations of ethidium bromide, which has been shown to disrupt DNA-dependent protein-protein contact (15). Interaction between HMGI-Y and Sp1 was further investigated in coimmunoprecipitation studies with either cell nuclear extract or HMGI-Y and pure Sp1 and an antibody against HMGI-Y immobilized on protein A beads. As shown in Fig. 3B, immunoprecipitation of HMGI-Y from HepG2 nuclear extracts followed by Western blot analysis for Sp1 revealed a major specific band, which migrated in a position corresponding to the size of Sp1. An identical result was obtained with HMGI-Y and pure Sp1, indicating that the binding of HMGI-Y to Sp1 is direct and not mediated by additional nuclear factors. When the same transfer was reprobed with the anti-HMGI-Y antibody, a unique specific band, which migrated in a position corresponding to the size of HMGI-Y was detected. Taken together, these data unequivocally indicate that HMGI-Y and Sp1 physically interact either in vitro or in vivo in the context of the intact cell and suggest that physical and functional cooperation between HMGI-Y and Sp1 on the IR promoter might occur through direct contact as well.
FIG. 3.
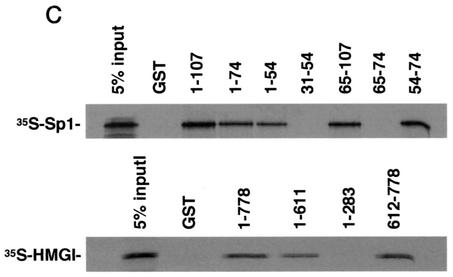
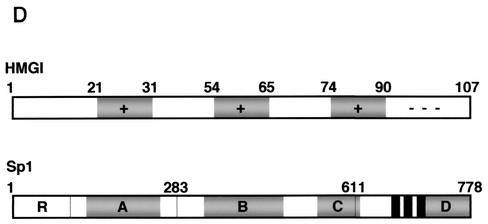
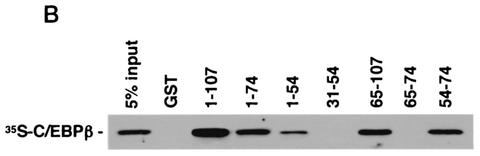
Physical association between HMGI-Y and Sp1. (A) SDS-PAGE of 35S-Sp1 bound to GST or GST-HMGI resin (lanes 1 to 3), 35S-HMGI bound to GST or GST-Sp1 resin (lanes 4 to 6), and 35S-Sp1 bound to GST-HMGI resin in the presence of ethidium bromide (EtBr) (lanes 7 to 10). In lanes 3, 6, and 10, the labeled protein was added directly onto the gel without binding to and elution from GST-protein resins. (B) Immunoprecipitation (IP) of HMGI-Y and Sp1 by using the anti-HMGI-Y antibody followed by immunoblotting with the anti-Sp1 antibody (lanes 1 to 5) or the anti-HMGI-Y antibody (lanes 6 to 10) after reprobing the same transfer. Lanes: 1, 10 ng of pure HMGI-Y; 2, 10 ng of pure Sp1; 3, HepG2 nuclear extract (NE; 500 μg); 4, HMGI-Y and Sp1 (20 ng each). In lanes 1 and 2, protein was directly applied to the gel without binding to and elution from protein A beads. To prove specificity, pure Sp1 (20 ng) was used for immunoprecipitation by the anti-HMGI-Y antibody (lane 5, control). (C) Localization of the interacting domains within HMGI-Y and Sp1. 35S-Sp1 or 35S-HMGI was incubated with resins containing normalized amounts of wild-type and various mutant versions of either GST-HMGI or GST-Sp1, respectively. Specifically bound proteins were resolved by SDS-PAGE and visualized by autoradiography. (D) Domain structures of HMGI and Sp1. +, basic repeats; −, acidic C-terminal tail; R, repression domain; A, B, C, and D, activation domains; black bars, zinc finger motifs. Numbers correspond to the positions of amino acids with relevance to the text.
To elucidate which regions of HMGI-Y and Sp1 were required for physical interactions, we carried out GST pull-down assays using GST-linked deletion mutant versions of both proteins. As shown in Fig. 3C, HMGI mutants 1-74 and 1-54, which lack the C-terminal tail plus the third and the second and third basic repeats, respectively, displayed a progressively lower binding activity for Sp1 than the wild-type (1-107) protein. Protein mutants carrying either the second or the third basic repeat (clones 54-74 and 65-107) were similar to the wild-type protein in their ability to retain Sp1, whereas HMGI mutants lacking any basic repeat (clones 31-54 and 65-74) were unable to bind Sp1. These data indicate that, in the context of HMGI-Y, the minimal region required for specific interaction with Sp1 corresponds to the middle basic repeat flanked by the spacer region between the middle and the third basic repeats (amino acids 54 to 74), revealing, in this respect, a behavior similar to that shown by the interaction between HMGI-Y and NF-κB (39). Similar experiments were performed with GST-Sp1 mutants and in vitro-synthesized 35S-labeled HMGI. As shown in Fig. 3C, Sp1 mutant 612-778, containing the zinc finger motifs plus activation domain D, was able to bind HMGI with high affinity. Sp1 DNA binding-deficient mutant 1-611, which retains the transcription activation domains A, B, and C, interacted also with HMGI, although to a lesser extent than the full-length (1-778) GST-Sp1. Conversely, the amino-terminal mutant 1-283, containing Sp1 repression domain R plus glutamine-rich domain A, was totally unable to retain any labeled HMGI. Thus, these findings indicate that Sp1 may interact with HMGI-Y through multiple functional domains that can function as potent activators of transcription in the presence of HMGI-Y.
Cooperative interaction between HMGI-Y and Sp1 is required for IR gene transcription and IR protein expression.
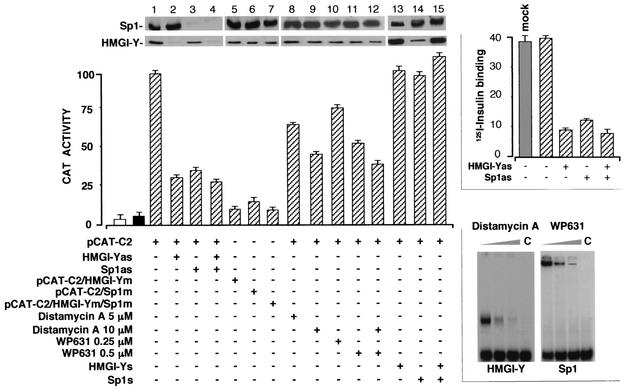
In the light of the above-mentioned data indicating that HMGI-Y physically interacts with Sp1 and facilitates its binding to the IR promoter, it was important to ask whether any perturbation of this interaction could have a negative effect on IR promoter function in vivo. For this purpose, the ability of C2 or E3 IR promoter elements to drive the expression of the CAT gene was measured in HepG2 cells, in which endogenous levels of HMGI-Y and Sp1 were specifically lowered by using an antisense approach (3, 36). As shown in Fig. 4, the HMGI-Y antisense expression plasmid inhibited the activity of the C2-containing vector (pCAT-C2), as judged by the decrease in CAT activity in transfected cells, and significantly reduced the binding of Sp1 to C2 and E3 probes (data not shown). Similar results were obtained when Sp1 antisense cDNA was used, either alone or in combination with HMGI-Y antisense cDNA. The absence of a further reduction in IR promoter activity with the knockout of both HMGI-Y and Sp1 suggests that the two factors functionally cooperate in delivering an activation signal to the basic transcription machinery. CAT activity correlated with HMGI-Y and Sp1 protein expression levels, as measured by Western blot analyses, and was not inhibited in cells transfected with an expression vector containing HMGI-Y cDNA in the sense orientation or Sp1 sense oligonucleotides (Fig. 4). In concert with these findings, perturbation of the HMGI-Y-Sp1 protein complex in vivo had inhibitory effects on the cell surface expression of the IR, as demonstrated by the reduction of 125I-insulin binding to HepG2 cells exposed to HMGI-Y and/or Sp1 antisense cDNA (Fig. 4).
FIG. 4.
Inhibition of IR promoter activity by antisense RNA and mutational and chemical interference with HMGI-Y and Sp1 DNA binding. HepG2 cells were transfected with CAT reporter plasmids (2 μg) containing wild-type or mutant (m) versions of the C2 IR promoter element in the presence or absence of an effector plasmid (5 μg) containing the HMGI-Y cDNA in either the antisense (as) or sense (s) orientation or Sp1 antisense or sense oligonucleotides (2 μM). For chemical interference experiments, transfected cells were incubated with increasing amounts of distamycin A and/or WP631 and CAT activity was measured 36 h later. CAT activity is expressed as percentages of the reporter activity in the presence of the pCAT-C2 vector alone. Open bar, mock (no DNA); solid bar, pCAT-basic (vector without an insert). Results are the means ± standard errors (SE) for three separate transfections. Equal amounts of cell protein extract were used for immunoblot analyses with antibodies against HMGI-Y and Sp1. Upper right, specific 125I-labeled insulin binding to HepG2 cells subjected to HMGI-Y and/or Sp1 antisense oligonucleotide treatment. Binding is expressed as the percentage of total insulin bound per 107 cells. Binding activity in the absence of antisense oligonucleotide treatment refers to untransfected control cells. Mock, no DNA. Results are means ± SE for three separate experiments. Lower right, EMSA analysis of the effect of distamycin A and WP631 on HMGI-Y and Sp1 binding to probe C2, respectively. Increasing amounts of distamycin A (0, 1, and 10 μM) or WP631 (0, 2.5, and 5 μM) were preincubated with probe C2 (15 min at room temperature) before 2 ng of either HMGI-Y or pure Sp1 was added. C, probe alone.
To extend these studies, we generated CAT constructs with C2 and E3 regulatory elements mutated at either or both of the HMGI-Y and Sp1 binding sites. As shown in Fig. 4, mutation of the HMGI-Y or Sp1 site in the C2 element (pCAT-C2/HMGI-Ym or pCAT-C2/Sp1m, respectively) reduced CAT activity in HepG2 cells to 10 to 15% of that with the native (pCAT-C2) promoter. Mutation of both binding sites together (pCAT-C2/HMGI-Ym/Sp1m) resulted in a similar ∼90% reduction in CAT activity, and this, in addition to the fact that mutation of the HMGI-Y site profoundly diminished IR promoter activity, even when the Sp1 binding site was intact, provides further indication of the existence of important functional interactions between HMGI-Y and Sp1. Similar results were obtained with the pCAT-E3 reporter plasmid (data not shown). In concert with these findings, treatment of transfected HepG2 cells with distamycin A and/or bisanthracycline WP631, which have been found to selectively block DNA binding by HMGI-Y and Sp1, respectively (38, 23), inhibited IR promoter-induced CAT activity in a dose-dependent manner, and this inhibition correlated with the decrease in DNA-binding activity by the C2 probe when both chemical agents were used in EMSAs with either HMGI-Y or pure Sp1 (Fig. 4).
HMGI-Y physically interacts with C/EBPβ and enhances its binding to the IR promoter.
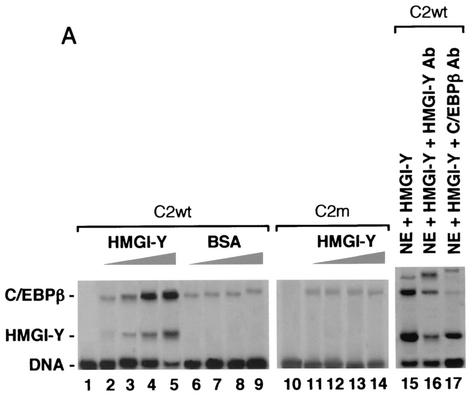
Levels of C/EBPβ are normally reduced in hepatoma cell lines compared with levels in normal liver (7). Therefore, DNA-protein and protein-protein interaction studies with HMGI-Y and C/EBPβ were performed with nuclear extracts from 3T3-L1 cells, whose differentiation into adipocytes is followed by an early increase in C/EBPβ protein levels (21). To see whether HMGI-Y had any influence on the binding of C/EBPβ to the IR promoter, we carried out EMSAs by using a fixed amount of nuclear extracts from early-differentiated 3T3-L1 adipocytes in the presence of increasing quantities of pure HMGI-Y. Under these conditions, the binding of C/EBPβ to probe C2 was progressively augmented (Fig. 5A) and a slightly increased retardation of the C/EBPβ complex was visible, meaning that HMGI-Y and C/EBPβ were simultaneously binding adjacent sites on the same probe, as confirmed in supershift analyses using either the anti-HMGI-Y or anti-C/EBPβ antibody. The binding of C/EBPβ did not change in the presence of increasing amounts of BSA and did not vary with probe C2m, containing a mutation at the HMGI-Y binding site, suggesting that the binding of C/EBPβ to the IR promoter may depend on the interaction of HMGI-Y with its DNA binding element, revealing, in this respect, a behavior virtually identical to that shown by the interaction between HMGI-Y and Sp1. Similar data were obtained with wild-type and mutant versions of probe E3 (data not shown).
FIG. 5.
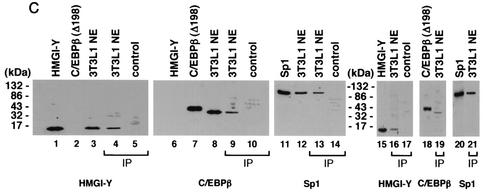
HMGI-Y physically interacts with C/EBPβ and enhances its binding to the IR promoter. (A) EMSAs were performed with 0.5 μg of 3T3-L1 nuclear extract (NE) and increasing amounts (lanes 2, 6, and 11, 0.25 ng; lanes 3, 7, and 12, 0.5 ng; lanes 4, 8, and 13, 1.0 ng; lanes 5, 9, 14, and 15, 2.0 ng) of pure HMGI-Y or BSA, in the presence of wild-type (wt) or mutant (m) probe C2. Lanes 1 and 10, probe alone. Supershift analysis with 3T3-L1 NE plus pure HMGI-Y was performed with the antibody (Ab) against either HMGI-Y or C/EBPβ (lanes 16 and 17). (B) Localization of HMGI-Y domains interacting with C/EBPβ. 35S-C/EBPβ was incubated with resins containing normalized amounts of wild-type GST-HMGI and various mutant derivatives of GST-HMGI. Specifically bound proteins were resolved by SDS-PAGE and visualized by autoradiography. (C) Immunoprecipitation (IP) of HMGI-Y, C/EBPβ, and Sp1 by using the anti-C/EBPβ antibody (lanes 4, 5, 9, 10, 13, and 14) or the anti-Sp1 antibody (lanes 16, 17, 19, and 21) followed by immunoblotting with the anti-HMGI-Y antibody (lanes 1 to 5 and 15 to 17), the anti-C/EBPβ antibody (lanes 6 to 10 [after reprobing the same transfer], 18, and 19), or the anti-Sp1 antibody (lanes 11 to 14, 20, and 21). Lanes 1 and 15, 10 ng of pure HMGI-Y; lanes 2 and 18, 20 ng of C/EBPβ (Δ198) antigen; lanes 3 and 12, 3T3-L1 NE (30 μg); lanes 4, 13, 16, 19, and 21, 3T3-L1 NE (500 μg); lanes 11 and 20, 10 ng of pure Sp1. In lanes 1, 2, 3, 11, 12, 15, 18, and 20 the protein was directly applied to the gel without binding to and elution from protein A beads. To prove the specificity of the immunoprecipitation, the anti-C/EBPβ antibody was tested with either HMGI-Y (lane 5, control) or pure Sp1 (lane 14, control) alone. The specificity of the immunoprecipitation with the anti-Sp1 antibody was tested with pure HMGI-Y alone (lane 17, control).
To gain support for the assumption that HMGI-Y and C/EBPβ interact directly at the IR promoter, we first tried to determine whether the two factors bind to each other in solution. To this end, a pull-down assay using GST-fused HMGI and in vitro-synthesized 35S-labeled C/EBPβ was performed. As can be seen in Fig. 5B, C/EBPβ was efficiently retained by wild-type (1-107) GST-HMGI but not GST alone. Pull-down assays using GST-fused HMGI deletion mutants were performed to map the region of HMGI-Y required for contact with C/EBPβ. As shown in Fig. 5B, mutant 1-74 bearing the deletion of the third basic repeat was able to bind C/EBPβ with high affinity. The HMGI-C/EBPβ interaction was considerably reduced after further truncation of the protein up to the loss of the middle basic repeat, a result closely resembling a recent one for HEK-293 cells overexpressing both HMGI-Y and C/EBPβ (24). HMGI derivatives carrying either the second or the third basic repeat (clones 65-107 and 54-74) were both able to retain C/EBPβ, whereas HMGI mutants lacking any AT hook (clones 31-54 and 65-74) did not display any tendency to bind to C/EBPβ. Therefore, these data indicate that C/EBPβ interacts physically with HMGI-Y in vitro and that the minimal region of HMGI required for this interaction is the region between amino acids 54 and 74, a result identical to the one described above for the interaction of HMGI-Y with Sp1.
To verify that the interaction between HMGI-Y and C/EBPβ observed in vitro resembles the interaction in vivo, we performed coimmunoprecipitation studies using an antibody against C/EBPβ immobilized on protein A beads. As shown in Fig. 5C, immunoprecipitation of C/EBPβ from nuclear extracts from early-differentiated 3T3-L1 cells followed by Western blot analysis for HMGI-Y revealed a unique specific band, which migrated in a position corresponding to the size of HMGI-Y. When the same transfer was reprobed with an anti-C/EBPβ antibody, a major band which migrated in a position corresponding to the size of C/EBPβ was observed. Thus, these findings indicate that these two factors physically interact in vivo. Moreover, when immunoprecipitation of C/EBPβ from 3T3-L1 nuclear extracts was followed by immunoblot analysis for Sp1, it was found that the Sp1 protein can be detected as well (Fig. 5C). Further evidence of the interaction between all three transcription factors was obtained by performing coimmunoprecipitation experiments with nuclear extracts from 3T3-L1 cells and an antibody against Sp1 immobilized on protein A beads. As expected, immunoprecipitation of Sp1, followed by immunoblot analyses with specific antibodies, allowed the identification of three proteins including, in addition to Sp1, HMGI-Y and C/EBPβ (Fig. 5C). Therefore, we conclude that a physical complex consisting of HMGI-Y, C/EBPβ, and Sp1 exists in vivo, in the context of the intact cell.
Activation of IR gene transcription by C/EBPβ requires HMGI-Y and is enhanced by Sp1.
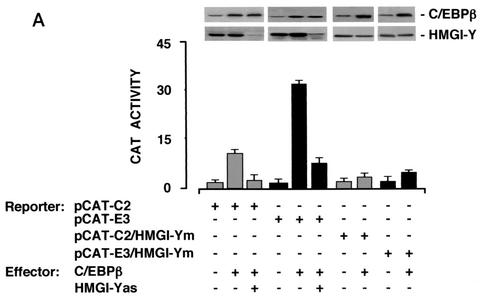
We next performed experiments to see whether HMGI-Y and C/EBPβ cooperate to activate the IR promoter at the transcriptional level. To test this possibility, HepG2 cells were cotransfected transiently with CAT reporter plasmids containing the C2 (pCAT-C2) or E3 (pCAT-E3) IR promoter sequence and the C/EBPβ effector vector, in the absence or presence of the HMGI-Y antisense expression plasmid. As shown in Fig. 6A, overexpression of C/EBPβ in HepG2 cells, significantly increased the CAT activity of both C2 and E3 promoter elements. However, CAT activity was significantly reduced in the presence of the HMGI-Y antisense expression plasmid, indicating that transactivation of the IR promoter by C/EBPβ requires HMGI-Y. Similar results were obtained with CAT reporter constructs pCAT-C2/HMGI-Ym and pCAT-E3/HMGI-Ym, containing mutated HMGI-Y binding elements. With either of these constructs, transactivation of the IR promoter by C/EBPβ was effectively prevented, even in the presence of endogenous HMGI-Y and overexpressed C/EBPβ.
FIG. 6.
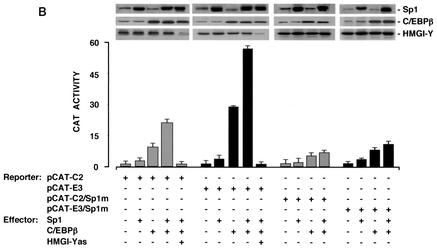
Functional significance of C/EBPβ for IR gene transcription. (A) The indicated CAT reporter vectors (2 μg each) were transfected into HepG2 cells with an effector plasmid for C/EBPβ (5 μg), in either the absence or presence of the HMGI-Y antisense (as) expression vector (5 μg). (B) HepG2 cells were transfected as in panel A along with C/EBPβ and/or Sp1 expression plasmids (5 μg each) in the absence or presence of the HMGI-Y (10 μg) antisense expression vector. In either case, data represent the means ± standard errors for three separate experiments; values are expressed as the factors by which induced activity increased above the level of CAT activity obtained in transfections with the wild-type or mutant reporter vector alone, which is assigned an arbitrary value of 1. Western blots of HMGI-Y, Sp1, and C/EBPβ in each condition are shown in the autoradiograms.
To investigate whether a functional cooperativity between C/EBPβ and Sp1 occurred at the IR promoter, transfections were carried out with either pCAT-C2 or pCAT-E3 in HepG2 cells, in which forced expression of both C/EBPβ and Sp1 was induced simultaneously. As shown in Fig. 6B, Sp1 and C/EBPβ together activated the IR promoter to levels of 20- and 55-fold above that observed with pCAT-C2 or pCAT-E3 alone, respectively. These increases were higher than the sum of the levels observed for transactivations with the two proteins individually, implying that Sp1 and C/EBPβ act synergistically to drive IR gene transcription. When Sp1 and C/EBPβ expression plasmids were cotransfected along with pCAT-C2/Sp1m or pCAT-E3/Sp1m, containing mutated Sp1 binding sites, the C/EBPβ-induced CAT activity was significantly impaired, indicating that transcriptional stimulation of the IR promoter by C/EBPβ is dependent on the integrity of the adjacent Sp1 binding site. Induction of CAT activity by both Sp1 and C/EBPβ together was virtually abolished in the presence of the HMGI-Y antisense expression plasmid, confirming the central importance of HMGI-Y in this scenario.
HMGI-Y, Sp1, and C/EBPβ are required for full activation of the IR promoter in in vivo and in vitro transcription studies.
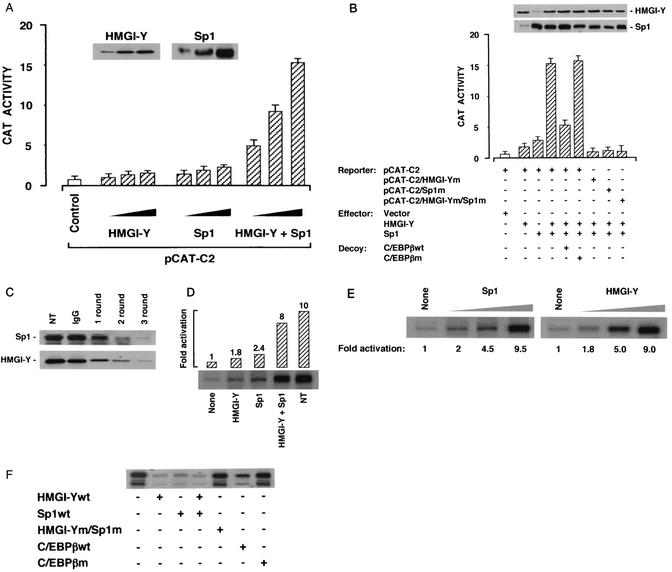
Finally, we asked whether the interplay among HMGI-Y, Sp1, and C/EBPβ could also be demonstrated to have functional sequelae in late-passage MCF-7 human breast cancer cells, a cell line ideally suited for studying the effects of these proteins on transcription since it does not express appreciable levels of either HMGI-Y or Sp1 (18, 32), while C/EBPβ is constitutively expressed (6). As shown in Fig. 7A and B, simultaneous overexpression of HMGI-Y and Sp1 in these cells led to a significant increment in CAT activity that exceeded that seen with either factor alone and that was at least in part dependent on endogenous C/EBPβ, as revealed by a C/EBPβ decoy strategy (28). As shown in Fig. 7B, cotransfection of a cis element decoy against the C/EBPβ binding site resulted in a 75% reduction in CAT activity, whereas transfection of the mutated C/EBPβ decoy element, which fails to bind the C/EBPβ protein in vitro, was ineffective. Induction of CAT activity required intact binding sites for HMGI-Y and Sp1, as demonstrated by diminished activity of constructs mutated at either or both of these sites. Similar results were obtained with wild-type and mutant pCAT-E3 reporter plasmids (data not shown).
FIG. 7.
Functional significance of HMGI-Y, Sp1, and C/EBPβ for IR gene transcription in MCF-7 cells and in HeLa nuclear extract (NE) immunodepleted of HMGI-Y and endogenous Sp1 gene activators. (A) MCF-7 cells were transiently transfected with the pCAT-C2 reporter vector (5 μg) plus increasing amounts (1, 5, or 10 μg) of HMGI-Y and Sp1 expression plasmids, either alone or in combination. (B) CAT activity in MCF-7 cells is reduced by decoy oligonucleotides with the C/EBPβ binding sequence and is eliminated by transfection of a pCAT-C2 reporter construct carrying mutated binding elements for HMGI-Y and/or Sp1. In either case, data represent the means ± standard errors for three separate experiments; values are expressed as factors by which induced activity increased above the level of CAT activity obtained in transfections with pCAT-C2 control vector alone, which is assigned an arbitrary value of 1. Immunoblots of HMGI-Y and Sp1 in each condition are shown in the autoradiograms. (C) Immunoblot analyses of HeLa NE with polyclonal antibodies against HMGI-Y and Sp1. Extracts were not treated (NT) or subjected to one, two, or three rounds of depletion with immobilized anti-HMGI-Y and anti-Sp1 antibodies or a control (unrelated rabbit serum immunoglobulin G [IgG]) antibody. (D) Transcription of the IR gene in immunodepleted extracts during 1 h of incubation at 30°C with the IR-CAT DNA as the template DNA in the absence or presence of the indicated proteins (100 ng each). Transcripts were identified by primer extension with a CAT primer and resolved on a 8% denaturing polyacrylamide gel. An activation value of 1 denotes the basal level of transcription in the absence of activators. A representative of three separate assays is shown. (E) Transcription of the IR gene was performed as in panel D, except that increasing amounts (0, 50, and 100 ng) of either Sp1 or HMGI-Y were used in the presence of a fixed amount (100 ng) of HMGI-Y or Sp1, respectively. (F) IR gene transcription was measured in untreated HeLa NE, in the absence or presence of oligonucleotides (10 ng each) bearing either a wild-type (wt) or mutant (m) binding sequence for HMGI-Y, Sp1, or C/EBPβ.
This observation correlated closely with the results from in vitro transcription studies using HeLa nuclear extracts from which HMGI-Y and endogenous Sp1 had been progressively removed by immunodepletion (Fig. 7C). Once again, the addition of a small amount of either HMGI-Y or pure Sp1 to these extracts stimulated transcription only weakly; however, simultaneous addition of both activators led to a significant increment in transcription (Fig. 7D). These results were confirmed in activator titration experiments with increasing amounts of either factor (Fig. 7E) and were supported further by in vitro transcription studies using untreated HeLa nuclear extracts in the presence of synthetic oligonucleotides with either the HMGI-Y, Sp1, or C/EBPβ binding sequence (Fig. 7F).
Studies with EBV-transformed lymphoblasts from diabetic patients and nondiabetic control subjects.
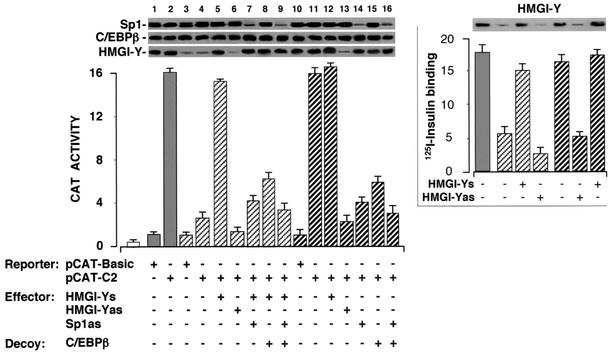
The above observations were supported further by the evaluation of transcriptional regulation of the IR gene in EBV-transformed lymphoblasts from one patient with the usual features of type 2 diabetes, in which defects in nuclear proteins regulating the IR gene were previously reported (2). In EBV-transformed lymphoblasts from this patient, IR expression was decreased despite the fact that the IR gene was normal (2). As shown in Fig. 8, the expression of HMGI-Y in these cells was markedly reduced compared to that in cells from control subjects, and this defect paralleled the decrease in IR promoter function in vivo in intact cells. Forced expression of HMGI-Y in these cells led to a significant increment in CAT activity, and this enhancement was specific, since no stimulation of CAT activity was observed when the pCAT-C2 reporter vector was transfected into cells in the presence of an expression vector containing HMGI-Y cDNA in the antisense orientation. CAT activity in cells with forced expression of HMGI-Y was also impaired by the Sp1 antisense effector plasmid and/or by cotransfection of a cis element decoy against the C/EBPβ binding site.
FIG. 8.
IR promoter activity and 125I-labeled insulin binding in human EBV-transformed lymphoblasts. EBV-transformed lymphoblasts were transfected with CAT reporter plasmids (5 μg) containing the C2 IR promoter element, in the absence or presence of an effector plasmid (10 μg) containing the HMGI-Y cDNA in either the sense (s) or antisense (as) orientation or Sp1 antisense oligonucleotides (5 μM). For decoy experiments, cells were cotransfected with a double-stranded oligonucleotide containing the wild-type C/EBPβ binding site sequence and CAT activity was measured 72 h later. CAT activity in each cell line is expressed as the factor of increase above the level of CAT activity obtained in transfections with the promoterless pCAT-basic vector alone (5 μg), which is assigned an arbitrary value of 1. Data from eight and six cell lines from normal and diabetic control subjects, respectively, were averaged; assays of cells from the diabetic patient with reduced expression of HMGI-Y were performed in triplicate in each transfection experiment. Results are the means ± standard errors (SE) for three separate transfections. open bar, mock (no DNA). Western blots of Sp1, C/EBPβ, and HMGI-Y are shown in the autoradiograms. Right, specific 125I-labeled insulin binding to EBV-transformed lymphoblasts subjected to HMGI-Y sense or antisense oligonucleotide treatment. Binding is expressed as the percentages of total insulin bound per 107 cells. Binding activity in the absence of HMGI-Y effectors refers to controls. Results are the means ± SE for three separate experiments. Transfection efficiency in the reporter gene analysis and hormone binding study was normalized to the expression of β-galactosidase from a separate reporter plasmid. Gray bar, healthy control subjects (n = 8); white hatched bars, patient with type 2 diabetes and reduced expression of HMGI-Y (n = 1); gray hatched bars, control subjects with type 2 diabetes and normal expression of HMGI-Y (n = 6).
The vast majority of patients with the common form of type 2 diabetes have normal IR levels. In studies of EBV-transformed lymphoblasts from six nonobese subjects with type 2 diabetes having normal IR expression, we found that HMGI-Y expression is similar to that in lymphoblasts from eight nondiabetic control individuals (Fig. 8). Transcriptional regulation of the IR gene in EBV-transformed lymphoblasts from these subjects, in which endogenous activity of either HMGI-Y, Sp1, or C/EBPβ was specifically reduced by antisense and decoy strategies, was also investigated. As shown in Fig. 8, the HMGI-Y antisense expression plasmid inhibited the activity of pCAT-C2, as judged by the decrease in CAT activity in transfected cells. Inhibition by the HMGI-Y antisense plasmid was specific, since no inhibition of CAT activity was observed when pCAT-C2 was transfected into cells in the presence of an expression vector containing HMGI-Y cDNA in the sense orientation. To ensure that inhibition by the HMGI-Y antisense plasmid was not related to a nonspecific effect, we transfected the pCAT-Promoter vector (a positive control having the simian virus 40 gene promoter upstream of the CAT coding region) (1, 3) into cultured lymphoblasts and exposed the cells to the HMGI-Y antisense expression plasmid. No inhibition of this CAT reporter plasmid was observed (data not shown). Once again, CAT activity was impaired in cells cotransfected with the Sp1 antisense effector plasmid and/or the C/EBPβ decoy element.
In previous studies of EBV-transformed lymphoblasts from several individuals with type 2 diabetes in which expression of the insulin receptor protein was normal, Brunetti et al. found that 125I-labeled insulin binding was similar to that in lymphoblasts from nondiabetic control subjects (2). 125I-insulin binding performed on intact EBV-transformed lymphoblasts from the diabetic patient with reduced expression of both insulin receptor and HMGI-Y proteins revealed a significant decrease in insulin binding compared to that in lymphoblasts from nondiabetic and diabetic control subjects. In concert with the findings above, forced expression of HMGI-Y in transfected lymphoblasts from the diabetic patient with reduced levels of HMGI-Y efficiently restored insulin binding capacity, as demonstrated by measuring 125I-insulin binding to intact cells (Fig. 8, right). Conversely, perturbation of the HMGI-Y-Sp1-C/EBPβ protein complex in vivo, by reducing HMGI-Y protein levels in cells transfected with the HMGI-Y antisense expression vector, had inhibitory effects on insulin binding capacity, as demonstrated by the reduction of 125I-insulin binding to intact EBV-transformed lymphoblasts from diabetic control subjects. 125I-insulin binding to these cells correlated with HMGI-Y protein expression, as shown by Western blot analysis of nuclear proteins from transfected cells (Fig. 8, right).
DISCUSSION
In eukaryotic cells, single regulatory elements are rarely sufficient to promote gene transcription during development and differentiation. Usually, activation of transcription by RNA polymerase II results from the synergistic effects of two or more cis-acting DNA motifs which are arranged in distinct sets around the site of transcription initiation and which bind specific transcription factors. The enhanceosome model for transcriptional activation provides one of the best-understood examples of how combinatorial interactions between distinct regulatory proteins and the transcriptional apparatus can lead to a highly specific activation of gene transcription in higher eukaryotes (9, 22, 25). Enhanceosome formation involves the creation of a stereospecific multiprotein-DNA complex that often includes the architectural transcription factor HMGI-Y and other transcription factors necessary for transcription initiation. Recently, we have shown that IR gene expression in eukaryotic cells is positively regulated by HMGI-Y (3). In the present study, we performed experiments designed to explore the biochemical mechanisms involved in HMGI-Y activation of IR gene transcription in HepG2 human hepatoblastoma cells, a cell line readily expressing IRs. We provide compelling evidence that transcriptional activation of the IR gene in these cells requires the assembly of a transcriptionally active multiprotein-DNA complex involving, in addition to the HMGI-Y nuclear protein, the ubiquitously expressed transcription factor Sp1 and C/EBPβ. We demonstrate for the first time that HMGI-Y induces transcriptional activation of the IR gene by potentiating the recruitment and binding of Sp1 and C/EBPβ to the IR promoter. Mutational interference with the AT-rich HMGI-Y site in both C2 and E3 elements abolishes the binding of HMGI-Y, adversely affects interactions of Sp1 and C/EBPβ with DNA in vitro, and blunts the transactivation of C2- or E3-containing reporter constructs by Sp1 and C/EBPβ in vivo. Antisense oligonucleotide-mediated inhibition of HMGI-Y protein synthesis demonstrated that binding of Sp1 to the IR promoter was adversely affected and, as a consequence, constitutive IR promoter activity was very low. Similarly, knockout of Sp1 with antisense oligonucleotides markedly impaired activation of the IR promoter, indicating that HMGI-Y-Sp1 interactions may play a central role in regulating IR promoter activity.
However, Sp1 binding sites within the promoter region of the IR gene are unlikely to be the major determinant of the tissue-specific expression of the IR gene since the distribution of Sp1 in tissue does not reflect that of the IR and since Sp1 expression in the classical insulin target tissues, muscle, liver, and fat, is low (33). Therefore, it appears likely that, in these tissues, additional factors are needed for transcriptional initiation even in the presence of low levels of Sp1. C/EBPβ plays an important role in the regulation of gene expression in the liver and other insulin-responsive tissues (7, 13). Studies of cell cultures and knockout mouse models indicate that C/EBPβ contributes to the regulation of hepatic glucose production and plays an important role in the regulation of metabolic processes (17). Herein, we demonstrate that C/EBPβ is required for full activation of IR gene transcription in HepG2 cells and that this transactivation appears to be specifically supported and strongly potentiated by Sp1. Evidence for cooperative interactions between C/EBPβ and Sp1 in the transcriptional regulation of the rat CYP2D5 cytochrome P-450 gene has been previously reported (16). In addition, a functional interplay among C/EBPβ and Sp1 family factors has been demonstrated in the context of the CD11c integrin gene promoter (19). On the other hand, functional cooperation between HMGI-Y and C/EBPβ for the transcriptional activation of the leptin promoter has been recently reported (24). However, cooperation between HMGI-Y and C/EBPβ occurs differently in the cases of the leptin gene and the IR gene. There are no HMGI-Y-DNA binding sites on the leptin promoter, and physical interaction between the two factors contributes to efficient functional cooperation in the transactivation of this gene. In contrast, the binding of HMGI-Y to the IR promoter is a prerequisite for the stimulation of the binding of C/EBPβ to its DNA binding site. Functional cooperation between HMGI-Y and C/EBPβ in the transactivation of the IR promoter could be mediated by HMGI-Y-induced changes in DNA structure that could enhance the affinity of C/EBPβ for its target DNA. In this scenario, HMGI-Y could facilitate the interaction between C/EBPβ and Sp1 and perhaps among other DNA-binding proteins that bind in the immediate vicinity, thereby promoting the formation of an active transcription complex. Mutations affecting either HMGI-Y or Sp1 DNA binding markedly impaired transactivation of the IR promoter by C/EBPβ, indicating that the binding of HMGI-Y and Sp1 to IR DNA is of crucial importance for both basal and induced IR gene expression.
Our observations consistently support the hypothesis that defects in the expression of these nuclear binding proteins may cause decreased IR expression and induce insulin resistance. Defects in a nuclear regulatory protein either identical to or highly related to the architectural transcription factor HMGI-Y in patients with insulin resistance and type 2 diabetes mellitus have been previously reported by our group (2). We report herein having found a defect in a protein from one patient with the usual features of type 2 diabetes. In EBV-transformed lymphoblasts from this patient, IR gene transcription was significantly impaired despite the fact that the IR gene was normal. In this patient we found that the expression of HMGI-Y was markedly reduced, suggesting that this defect may induce insulin resistance and type 2 diabetes. Consistent with this, overexpression of HMGI-Y in transfected lymphoblasts from this patient significantly increased IR gene transcription and efficiently restored cell surface expression of the IR and insulin-binding capacity. The identification of this defect in a patient with a common form of diabetes is consistent with the notion that tissue-specific alterations of IR levels might be associated with insulin resistance, an early feature of type 2 diabetes.
There has been considerable progress over the past few years in unraveling the molecular defects that give rise to insulin resistance, an early feature of type 2 diabetes mellitus. Nevertheless, there are many gaps in our understanding of the pathophysiology underlying insulin resistance. Our studies demonstrate that the bulk of IR gene expression in liver cells depends on functional and physical interactions among HMGI-Y, Sp1, and C/EBPβ. Additionally, these observations consistently support the hypothesis that defects in the expression of these nuclear binding proteins may cause decreased IR expression and induce insulin resistance. Together, these new findings provide further insight into the molecular processes regulating IR gene expression and open up new avenues for understanding the causes of insulin resistance syndromes and other pathological states in humans with IR dysfunction and impairment of insulin signaling and action.
Acknowledgments
This work was supported by Telethon-Italy grant E.613 and in part by a grant from MURST, protocollo 2002062899_002, Italy.
We thank K.-S. Chang, M. A. Lazar, G. Manfioletti, T. Maniatis, T. M. Penning, H. Rotheneder, G. Suske, D. Thanos, R. Tjian, and E. Wintersberger for providing reagents. Special thanks go to L. Levintow for critical reading of the manuscript.
REFERENCES
- 1.Brunetti, A., D. Foti, and I. D. Goldfine. 1993. Identification of unique nuclear regulatory proteins for the insulin receptor gene, which appear during myocyte and adipocyte differentiation. J. Clin. Investig. 92:1288-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunetti, A., L. Brunetti, D. Foti, D. Accili, and I. D. Goldfine. 1996. Human diabetes associated with defects in nuclear regulatory proteins for the insulin receptor gene. J. Clin. Investig. 97:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunetti, A., G. Manfioletti, E. Chiefari, I. D. Goldfine, and D. Foti. 2001. Transcriptional regulation of human insulin receptor gene by the high-mobility group protein HMGI-Y. FASEB J. 15:492-500. [DOI] [PubMed] [Google Scholar]
- 4.Bruning, J. C., D. Gautam, D. J. Burks, J. Gillette, M. Schubert, P. C. Orban, R. Klein, W. Krone, D. Muller-Wieland, and C. R. Kahn. 2000. Role of brain insulin receptor in control of body weight and reproduction. Science 289:2122-2125. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, M., and R. Reeves. 1996. High-mobility-group proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acids Res. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 6.Faggioli, L., C. Costanzo, M. Merola, E. Bianchini, A. Furia, A. Carsana, and M. Palmieri. 1996. Nuclear factor kB (NF-kB), nuclear factor interleukin-6 (NFIL-6 or C/EBPβ) and nuclear factor interleukin-6β (NFIL6-β or C/EBPδ) are not sufficient to activate the endogenous interleukin-6 gene in the human breast carcinoma cell line MCF-7. Eur. J. Biochem. 239:624-631. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, A. D., W. H. Landschulz, and S. L. McKnight. 1989. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in culture hepatoma cells. Genes Dev. 3:1314-1322. [DOI] [PubMed] [Google Scholar]
- 8.Giddings, S. J., and L. R. Carnaghi. 1992. Insulin receptor gene expression during development: developmental regulation of insulin receptor mRNA abundance in embryonic rat liver and yolk sac, developmental regulation of insulin receptor gene splicing, and comparison to abundance of insulin-like growth factor I receptor mRNA. Mol. Endocrinol. 6:1665-1672. [DOI] [PubMed] [Google Scholar]
- 9.Grosschedl, R. 1995. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr. Opin. Cell Biol. 7:362-370. [DOI] [PubMed] [Google Scholar]
- 10.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. Kel, O. Kel, E. Ignatieva, E. Ananko, O. Podkolodnaya, F. Kolpakov, N. Podkolodny, and N. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 26:364-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, M. S., and M. D. Lane. 2000. Sequential repression and activation of the CCAAT enhancer-binding protein-α (C/EBPα) gene during adipogenesis. Proc. Natl. Acad. Sci. USA 97:12519-12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, P. F., and S. C. Williams. 1994. CCAAT/enhancer binding (C/EBP) proteins, p. 771-795. In F. Tronche and M. Yaniv (ed.), Liver gene expression. R. G. Landes Co., Austin, Tex.
- 14.Kahn, C. R. 1985. The molecular mechanisms of insulin action. Annu. Rev. Med. 36:429-451. [DOI] [PubMed] [Google Scholar]
- 15.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, Y. H., S. C. Williams, M. Baer, E. Stemeck, F. J. Gonzalez, and P. F. Johnson. 1997. The ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol. Cell. Biol. 17:2038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, S., C. Croniger, C. Arizmendi, M. Harada-Shiba, J. Ren, V. Poli, R. W. Hanson, and J. E. Friedman. 1999. Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPβ gene. J. Clin. Investig. 103:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Y., X. Zhong, W. Li, M. G. Brattain, and S. S. Banerji. 2000. The role of Sp1 in the differential expression of transforming growth factor-β receptor type II in human breast adenocarcinoma MCF-7 cells. J. Biol. Chem. 275:12231-12236. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Rodriguez, C., L. Botella, and A. L. Corbi. 1997. CCAAT-enhancer binding proteins (C/EBP) regulate the tissue specific activity of the CD11c integrin promoter through functional interactions with Sp1 proteins. J. Biol. Chem. 272:29120-29126. [DOI] [PubMed] [Google Scholar]
- 20.Mamula, P. W., A. R. McDonald, A. Brunetti, Y. Okabayashi, K. Y. Wong, B. A. Maddux, C. Logsdon, and I. D. Goldfine. 1990. Regulating insulin-receptor-gene expression by differentiation and hormones. Diabetes Care 13:288-301. [DOI] [PubMed] [Google Scholar]
- 21.Mandrup, S., and M. D. Lane. 1997. Regulating adipogenesis. J. Biol. Chem. 272:5367-5370. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon β enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 23.Martin, B., A. Vaquero, W. Priebe, and J. Portugal. 1999. Bisanthracycline WP631 inhibits basal and Sp1-activated transcription initiation in vitro. Nucleic Acids Res. 27:3402-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melillo, R. M., G. M. Pierantoni, S. Scala, S. Battista, M. Fedele, A. Stella, M. C. De Biasio, G. Chiappetta, V. Fidanza, G. Condorelli, M. Santoro, C. M. Croce, G. Viglietto, and A. Fusco. 2001. Critical role of the HMGI-Y proteins in adipocytic cell growth and differentiation. Mol. Cell. Biol. 21:2485-2495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 26.Milos, P. M., and K. S. Zaret. 1992. A ubiquitous factor is required for C/EBP-related proteins to form stable transcription complexes on an albumin promoter segment in vitro. Genes Dev. 6:991-1004. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 28.Morishita, R., T. Sugimoto, M. Aoki, I. Kida, N. Tomita, A. Moriguchi, K. Maeda, Y. Sawa, Y. Kaneda, J. Higaki, and T. Ogihara. 1997. In vivo transfection of cis element “decoy” against nuclear factor-kappa B binding site prevents myocardial infarction. Nat. Med. 3:894-899. [DOI] [PubMed] [Google Scholar]
- 29.Quandt, K., K. Frech, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Queen, C., and D. Baltimore. 1983. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell 33:741-748. [DOI] [PubMed] [Google Scholar]
- 31.Reeves, R. 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277:63-81. [DOI] [PubMed] [Google Scholar]
- 32.Reeves, R., D. D. Edberg, and Y. Li. 2001. Architectural transcription factor HMGI-Y promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 21:575-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saffer, J. D., S. P. Jackson, and M. B. Annarella. 1991. Regulation of Sp1 during mouse development. Mol. Cell. Biol. 11:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saltiel, A. R., and C. R. Kahn. 2001. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414:799-806. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, S. I. 1999. Deconstructing type 2 diabetes. Cell 97:9-12. [DOI] [PubMed] [Google Scholar]
- 36.Thanos, D., and T. Maniatis. 1992. The high mobility group protein HMGI-Y is required for NF-kappa B-dependent virus induction of the human IFN-β gene. Cell 71:777-789. [DOI] [PubMed] [Google Scholar]
- 37.Virkamäki, A., K. Ueki, and C. R. Kahn. 1999. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Investig. 103:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegner, M., and F. Grummt. 1990. Netropsin, distamycin, and berenil interact differentially with a high-affinity binding site for the high mobility group protein HMG-I. Biochem. Biophys. Res. Commun. 166:1110-1117. [DOI] [PubMed] [Google Scholar]
- 39.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMGI-Y in the assembly and function of the IFN-β enhanceosome. EMBO J. 18:3074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, X. L., Y. Gui, K. A. Sharkey, and M. D. Hollenberg. 1999. Differential induction of nitric oxide synthase in rat gastric and vascular smooth muscle tissue: distinct tissue distribution and distinctive signaling pathways. J. Pharmacol. Exp. Ther. 289:632-640. [PubMed] [Google Scholar]