Abstract
The large subunit of Saccharomyces cerevisiae DNA polymerase ɛ, Pol2, comprises two essential functions. The N terminus has essential DNA polymerase activity. The C terminus is also essential, but its function is unknown. We report here that the C-terminal domain of Pol2 interacts with polymerase σ (Pol σ), a recently identified, essential nuclear nucleotidyl transferase encoded by two redundant genes, TRF4 and TRF5. This interaction is functional, since Pol σ stimulates the polymerase activity of the Pol ɛ holoenzyme significantly. Since Trf4 is required for sister chromatid cohesion as well as for completion of S phase and repair, the interaction suggested that Pol ɛ, like Pol σ, might form a link between the replication apparatus and sister chromatid cohesion and/or repair machinery. We present evidence that pol2 mutants are defective in sister chromatid cohesion. In addition, Pol2 interacts with SMC1, a subunit of the cohesin complex, and with ECO1/CTF7, required for establishing sister chromatid cohesion; and pol2 mutations act synergistically with smc1 and scc1. We also show that trf5Δ mutants, like trf4Δ mutants, are defective in DNA repair and sister chromatid cohesion.
Sister chromatid cohesion is the process by which newly replicated DNA duplexes are held together in the period between the end of S phase and the beginning of mitosis to ultimately ensure faithful transfer of parent genes to daughters. During DNA replication, cohesion is established via the formation of bridges thought to be composed of a multiprotein complex known as cohesin (12, 24, 45, 56, 58). Saccharomyces cerevisiae cohesin consists of at least four distinct proteins, Scc1/Mcd1, Scc3, Smc1, and Smc3; however, Scc2 and Scc4 are required for association with chromatin, and Pds5 may also be associated with the complex (23, 57, 60, 64). Smc1 and Smc3 are members of a family of coiled-coil ATPases known to be required for recombination, cohesion, and condensation from bacteria through humans. Their structure suggests that they perform the bridging role, assisted by Scc1 and Scc3 (44). Association of the cohesins with chromosomes occurs at late G1-early S phase at specific sites, called cohesion assembly regions, spaced at about 10-kb intervals along chromosome arms and more closely at centromeres (7, 36).
Several lines of evidence suggest that cohesion is established as the cohesion assembly regions emerge from the replication fork. First, when Scc1 was overexpressed after S phase, it could associate with individual chromosomes but was unable to establish interchromatid cohesion (23). Second, Eco1/Ctf7 (a protein acetylase), PCNA (the replication clamp), Ctf18, Ctf8, and Drc1 (members of a clamp loader complex), and Ctf4 (a DNA polymerase α [Pol α] binding protein) are all important for establishing cohesion (26, 34, 40, 57, 64). The closest link has been the demonstration that Pol σ, formerly Pol κ, is essential for efficient cohesion (10, 69, 70).
Pol ɛ is one of four essential nuclear DNA polymerases (Pol α, δ, ɛ, and σ) found in yeast (11, 54, 70). Most models of replication propose that Pol α initiates replication, that Pol δ is responsible for further synthesis of the lagging strand, and that Pol ɛ is responsible for leading strand synthesis. However, this role for Pol ɛ is far from proved, and this enzyme remains the most enigmatic of all the polymerases. The catalytic subunit of Pol ɛ, Pol2, houses a polymerase activity and 3′-5′ exonuclease proof reading activity at the N terminus, and point mutations in the active site are lethal (18, 43). However, several early observations suggested that the C terminus of the large protein provided a second essential function unrelated to the polymerase (9, 42, 47). This became strikingly clear when it was shown that the polymerase domain could be deleted from the protein without loss of cellular viability, suggesting that the only nonredundant essential function of the POL2 gene is encoded in the C-terminal half of the protein and is independent of the polymerase (18, 33). Mutations in the C terminus inactivate a checkpoint connecting aberrant or incomplete DNA replication to the initiation of anaphase and result in failure to induce transcription of RNR3 after treatment of cells with the replication inhibitor hydroxyurea (HU) (3, 5, 17-19, 46, 47, 49). The C terminus is also important for the structural integrity of the four-subunit holoenzyme (2, 3, 5, 17-20, 49). A third role for the C-terminal region is nucleation of a complex of the core holoenzyme with several additional proteins required both for initiation of replication and for the S-phase checkpoint (4, 17, 39, 68). Two of these proteins, Dpb11 and Sld2 (synthetically lethal with dpb11), may load the polymerase onto the (prereplication complex (preRC) (38). Mutations in the C terminus also confer DNA damage sensitivity on yeast strains. Despite this wealth of descriptive biochemistry, the function of POL2 that is independent of the polymerase remains unexplained. One analytical approach to identifying this role is to look for other proteins that interact with Pol2, through whose function one can deduce additional pathways requiring Pol2. We report on the outcome of one such study here, which suggests an additional role for Pol ɛ, namely in proper sister chromatid cohesion.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strains used are listed in Table 1. Plasmids and oligonucleotides used in strain construction are found in Tables 2 and 3. Plasmids containing fusions for two-hybrid assays were constructed as follows: for PHS7, TRF4 was cloned in frame at the SmaI/SacI site of pACT2; for PHS2, TRF4 was cloned at the SmaI/PstI site of pBTM116; for PHS9, DPB11 was cloned in frame at the EcoRI/NcoI site of pACT2; for PHS4, DPB11 was cloned at the EcoRI/SmaI site of pBTM116; for PHS8, TRF5 was cloned in frame at the NcoI/BamHI site of pACT2; for PHS3, TRF5 was cloned at the BamHI/PstI site of pBTM116; for PHS6, ECO1 was cloned in frame at the EcoRI/XhoI site of pACT2; and for PHS5, SMC1 was cloned in frame at the SmaI/XhoI site of pACT2.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| AFS479 | MATaura3-1 ade2-1 can1-100 trp1-1 leu2-3,112::lacO-256 (pAFS59)-LEU2+ his3-11,15::HIS3 [-GFP-Lac1(pAFS144)-HIS3+] | A. Straight |
| L40 | MATahis3Δ200 trp1-901 leu2-3,112 ade2 lys2-801am URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3 | Hollenberg, 1995 |
| BY4741 | MATahis3-D1 leu2-D0 met15D0 ura3D0 | Research Genetics |
| 1145 | BY4741 trf5Δ | Research Genetics |
| 6265 | BY4741 trf4Δ | |
| BY-pol2-12 | MATahis3-D1 leu2-D0 met15D0 ura3D0 pol2-12 | This study |
| BY-pol2-11 | MATahis3-D1 leu2-D0 met15D0 ura3D0 pol2-11 | This study |
| BY-trf4Δpol2-11 | MATa6265 trf4Δpol2-11 | This study |
| BY-trf5Δpol2-12 | MATa1145 trf5Δpol2-12 | This study |
| BY-trf5Δpol2-11 | MATa1145 trf5Δpol2-11 | This study |
| 985-7C | MATamcd1-1 trp1 ura3 bar1 gal1 | D. Koshland, 1997 |
| 955-9C | MATα mcd1-1 trp1 ura3 bar1 gal1 | D. Koshland, 1997 |
| YPH102 | MATα ura3-52 lys2-801 ade2-101 his3-200 leu2-1 | P. Heiter |
| Y414 | YPH102 pol2-12 | This study |
| 3aAS273 | YPH102 smc1-2 | V. Guacci, 1997 |
| 3aAS273a | MATaura3-52 lys2-801 ade2-101 his3-200 leu2-1 smc1-2 | This study |
| CY1243 | MATatrf4ts896-HIS3 trf5::LEU2 (his3-11,15) (leu2-3,112) trp1-1 ade2-1 ura3-1 | Castano and Christman, 1996 |
| TC102-2-12 | MATaleu2 ura3-52 can1 pol2-12 | Budd and Campbell, 1993 |
| SS111 | MATatrp1-289 ura3-1,2 ade2-101 gal2 can1 his3 | Budd and Campbell, 1993 |
| SS111-2-12 | SS111 pol2-12 | Budd and Campbell, 1993 |
| SOG3 | MATatrp1-289 ura3-1,2 ade2-101 gal2 can1 lacO-256 (pAFS52)-TRP1+ his3-11,15::HIS3 [-GFP-Lac1(pAFS144)-HIS3+] | This study |
| 12OG2 | SOG3 pol2-12 | This study |
| HKYDNA2 | MATadna2-2::LEU2 ura3-1 ade2-1 can1-100 trp1-1 leu2-3,112 | H. Klien |
| JCYDNA2 | HKYDNA2 lacO-256(pAFS52)-TRP1+ his3-11,15::HIS3 [-GFP-Lac1(pAFS144)-HIS3+] | This study |
| ts370 | MATaade1 ade2 ura1 his7 tyr1 gal1 cdc2-1 | Budd and Campbell, 1993 |
| JCYT59 | SOG3 trf5Δ | This study |
| JCY122 | AFS479 pol2-12 | This study |
| JCY123 | JCY122 × 3aAS273a | This study |
| JCY124 | JCY122 × 955-9c | This study |
| JCY125 | JCY122 × CY1243 | This study |
| JCYPD18 | ts370 lacO-256(pAFS52)-TRP1+ his3-11,15::HIS3[-GFP-Lac1(pAFS144)-HIS3+] | This study |
TABLE 2.
Plasmids
| Plasmid | Relevant genotype | Base plasmid | Reference |
|---|---|---|---|
| PMB12 | URA3 pol2-12 | pR306 | Budd and Campbell, 1993 |
| PDLT4 | URA3 GAL1-10-TRF4 | pSEY18 | This study |
| PDLT5 | URA3 GAL1-10-TRF5 | pSEY18 | This study |
| pSEY18-DPB2 | URA3 GAL1-10-DPB2 | pSEY18 | Dua and Campbell, unpublished |
| pSEY18pII | URA3 GAL1-10-POL2 | pSEY18 | Budd and Campbell, 1993 |
| PHS1 | TRP LexA-POL2 (aa 1265-2222) | pBTM116 | Dua and Campbell, 2000 |
| PHS2 | TRP LexA-TRF4 | pBTM116 | This study |
| PHS3 | TRP LexA-TRF5 | pBTM116 | This study |
| PHS4 | TRP LexA-DPB11 | pBTM116 | This study |
| PHS5 | LEU2 GAL4-SMC1 | pACT2 | This study |
| PHS6 | LEU2 GAL4-ECO1 | pACT2 | This study |
| PHS7 | LEU2 GAL4-TRF4 | pACT2 | This study |
| PHS8 | LEU2 GAL4-TRF5 | pACT2 | This study |
| PHS9 | LEU2 GAL4-DPB11 | pACT2 | This study |
| PHS10 | TRP LexA-DPB2 | pBTM116 | Dua and Campbell, 2000 |
| PD24 | LEU2 GAL4-DPB2 | pACT2 | Dua and Campbell, 1998 |
| PD14 | LEU2 GAL4-POL2 (aa 1265-2222) | pACT2 | Dua and Campbell, 1998 |
| PD18 | LEU2 GAL4-POL2 (aa 2163-end) | pACT2 | Dua and Campbell, 1998 |
| pAFS52 | 256 lacO TRP1 | YIplac204 | Straight and Murray, 1996 |
| pAFS59 | 256 lacO LEU1 | YIplac128 | Straight and Murray, 1996 |
| pAFS135 | GFP12-LacI-I12 | pRS303 | Straight (unpublished) |
| pAFS144 | GFP13-LacI-I12 | pRS303 | Straight (unpublished) |
TABLE 3.
Primers
| Gene | Insertion plasmid | Primers |
|---|---|---|
| TRF4 | pSEY18 | 5′ GTGATGTACAGTTGCATGCATCATTTTAACAAAAAG, |
| 5′ CAAGGGAACTATAGTCGACATATGGGGGCAAAG | ||
| TRF5a | pSEY18 | 5′ GGCGTTTAGCATGCCAGTAGTCCCTTATCGTTTGAG, |
| 5′ GGAGTTTATTGGATCCTCATGACAAGGCTC | ||
| TRF4 | pBTM116 | 5′ TATACAGTTCACTGCAGCATTTTATCTAAAA, |
| 5′ GCAAGGGAACTACCCGGGAAATATGGGGGC | ||
| TRF5 | pBTM116 | 5′ TAGAGAGCCTCCAGTCCCTTATCGTTTGAG, |
| 5′ GGAGTTTATTGGATCCTCATGACAAGG | ||
| DPB11 | pBTM116 | 5′ ATCCGTAGCATGAATTCCACTATGAAGCCC, |
| 5′ TATAAAATTACGCCCCGGGTTTCAAGAATC | ||
| SMC1 | pACT2 | 5′ GGCACAAGGCCCGGGAATGGGACGT, |
| 5′ GTTGGTTCTCGAGATTATTCTGCGTAATTGC | ||
| ECO1 | pACT2 | 5′ GCAGTAACCAGAATTCATATGAAAGCTAGG, |
| 5′ TCTTTTCCTCGAGGTCATATGTATACC | ||
| TRF4 | pACT2 | 5′ GCAAGGGAACTACCCGGGAAATATGGGG, |
| 5′ CAGTGATGTAGAGCTCAGTGCATCATT | ||
| TRF5 | pACT2 | 5′ GCGGAGTTTATTCCATGGTCATGACAA, |
| 5′ TAGAGAGCCAGGGATCCCTTATCGTTTGAG | ||
| DPB11 | pACT2 | 5′ ATCCGTAGCATGAATTCCACTATGAAGCCC, |
| 5′ ATTACGGCCATGGTTTCAAGAATCTATAAA | ||
| trf5Δ | Genomic PCR product | 5′ CCTGGCCTTTAGAGAGCCAGTAGTCCCTTCTCGTTTGAGCATCTA, |
| 5′ TTTTTCAAATAAACAAACGAGGGCGGAGTTTATTGGGTCGTCATG | ||
| TRF4-A | pVL1393 | 5′ GTACTCTAGACCTATAAATATGCATCATCATCATCATCACGGGGCAAAGA GTGTAACAGCCTCTTCTTCAA, |
| 5′ GATCGCGGCCGCTTAAAGGGTATAAGGATTATATCCATCTTC | ||
| TRF4-B | pVL1393 | 5′ GTACTCTAGACCTATAAATATGGGGGCAAAGAGTGTAACAGCCT, |
| 5′ GGATATAATCCTTATACCCTTCATCATCATCATCATCACTAAAGATCTGAT | ||
| TRF5-A | pET24a(+) | 5′ GTTTATTGGGAGCTCATGACAAGGC, |
| 5′ CTCGTTTGAGCTCGAGCTGTCTTAG | ||
| TRF5-correction | pET24a(+) | 5′ GGAGTTTATTGGCTCGAGATGACAAGGCTC, |
| 5′ CTATAAGAGTCCTCGAGAAGCTTAAGAGCCTGGCC |
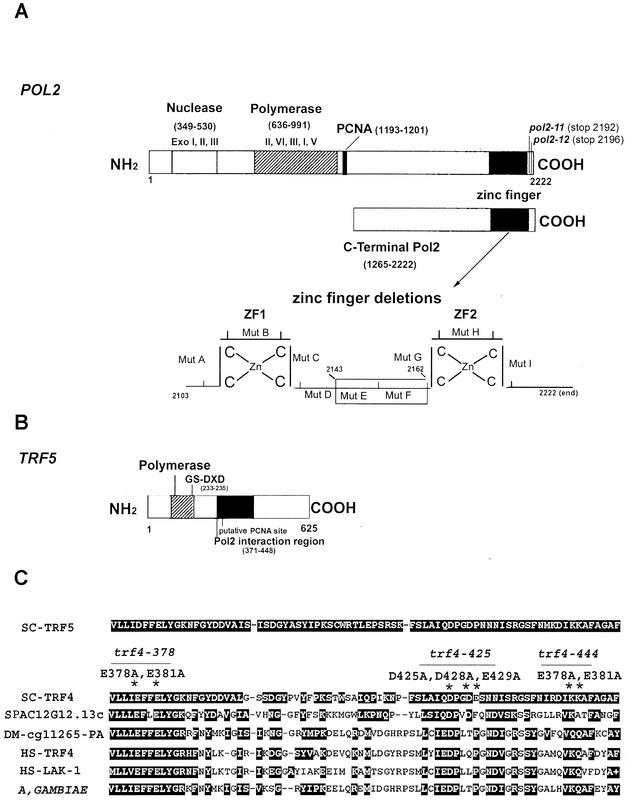
We have determined the Trf5 sequence from several strains in our laboratory as well as from the original Trf5 clone isolated by Castano et al. (14). Our sequence is inconsistent with those previously reported in reference 1 and GenBank (accession number U47282). Base 2070 is G instead of C, making amino acid 354 Met instead of Ile; base 2071 is C instead of G, making amino acid 355 His instead of Asp. In addition, an insertion of a G at base 2847 resulted in a frameshift, making amino acids 614 to 642 GQDEKSPLETKTVDAQTRRDYWLSKGQAL. A correction has been submitted to GenBank. The sequence from the Saccharomyces Genome Database also contains errors.
Two-hybrid screening protocol.
A two-hybrid library screen was performed using protocols found at Elsevier Trends Journals Technical Tips Online (http://www.biomednet.com/db/tto) (protocols P01616, P01713, and P01714 by D. L. Parchaliuk et al.). The C terminus of POL2 (amino acids [aa] 1236 to 2222) was previously cloned into pBTM116, a LexA binding domain vector (LexA-BD). The S. cerevisiae library used was a genomic DNA library prepared by partial restriction enzyme digestion and fractionation by gel electrophoresis. DNAs with a projected range of 500 to 3,000 bp were inserted into pGAL424 a GAL4 activation domain (GAL4-AD), and the resulting library was designated Y2HL-C3 (21, 31). pBTM116-POL2-C-terminal-BD was transformed into the yeast L40 strain containing reporter genes HIS3 and lacZ both fused downstream of multiple copies of the LexA operators and was maintained in SD (synthetic medium with dextrose)-Trp (29). The Y2HL-C3-AD library was then introduced, and positives were selected at 30°C on synthetic medium-Leu-Trp-His plus 1 mM 3-amino-1,2,4-triazole over 3 weeks. Coverage of the library was greater than 99%, with 106 colonies generated. Forty-six hundred transformants were screened for β-galactosidase activity using a filter lift assay (Clonetech). Each positive clone was purified and screened twice more. This procedure resulted in 203 total interacting clones of which 197 (Genotech) were recovered after isolation and transformation into Escherichia coli KC8-leuB (Clonetech).
Plasmid stocks (197) for gene identification were propagated in E. coli DH5α and then sequenced using a primer (5′ TACCACAATGGA) complementary to pGAL424. Gene identification was made using the Sacchasomyces Genome Database (SGD) and National Center for Biotechnology Information databases. Genes of interest were cotransformed into strain L40 with the pBTM116-POL2 C terminus or with pBM116 alone as a final screen for interaction and/or autoactivation. No transcriptional self-activators were identified. A 78-amino-acid fragment of Trf5 was isolated from the yeast library via interaction with the C terminus of POL2.
Two-hybrid interactions and mapping of Trf5 interaction within Pol2.
The two-hybrid strain L40 (Table 1) was cotransformed with the binding domain and activation domain fusion constructs using the polyethylene glycol-lithium method cited above. Transformants were selected on synthetic medium-Leu-Trp-His with 1 mM 3-amino-1,2,4-triazole incubated at 30°C for 3 to 5 days. Colonies were tested for β-galactosidase activity using a filter lift assay and then quantified for β-galactosidase activity using a high-sensitivity kit from Stratagene. The mutant genes pol2-A-pol2-I encode 10 amino acid deletions mapping at the C terminus of Pol2 (Fig. 1). pol2-A [pol2(Δ2103-2112)], pol2-B [pol2(Δ2113-2122)], pol2-E [pol2(Δ2143-2152)], pol2-F [pol2(Δ2153-2162)], pol2-H [pol2(Δ2173-2182)], pol2-I [pol2(Δ2183-2192)], and pol2-11 are also described elsewhere (17, 19). pol2-13 represents pol2 (Δ2163-end), a deletion of the second zinc finger. BD-TRF5 is described above.
FIG. 1.
(A) Schematic diagram of the POL2 gene describing conserved exonuclease domains and polymerase domains. The C-terminal portion of Pol2 used for the two-hybrid library screen includes aa 1265 to 2222. Ten amino acid deletions in the zinc finger region of Pol2 were used to map the interaction of Trf5; the zinc finger deletion region is shown enlarged. (B) Schematic diagram of the TRF5 gene. The conserved nucleotidyl transferase domain occurs between aa 178 and 241. The principle conserved motif is from aa 217 to 235. Aspartates 233 and 235 in Trf5 correspond to aspartates 236 and 238 in the DXD motif in Trf4, and since mutations in these residues reduce the DNA polymerase activity of Trf4, we have labeled this the putative active site (6, 70). (C) The 78-aa POL2 interaction region of TRF5 homologous to TRF genes in yeast, human, and fly. The alignment was made using BLAST data from the SGD and GenBank. SC, S. cerevisiae; SP, Schizosaccharomyces pombe; DM, Drosophila melanogaster; HS, Homo sapiens. The corresponding amino acid numbers and accession numbers are SC-Trf5_1050861 (aa 371 to 448); SC-Trf4_950226 (aa 374 to 451); SP-C12G12.13C_2130260 (aa 1006 to 1081); DM-CG11265_22831959 (aa 461 to 538); HS-Trf4_5565687 (aa 105 to 182); HS-Lak-1_5139669 (aa 181 to 258); A. gambiae_21288943 (aa 460 to 536). Both identities and similarities are highlighted. The asterisks identify mutations in trf4 that cause lethality (trf4-378; trf4-425; trf4-444).
Construction of His-Trf4, Trf4-His, and Trf5-His transfer vectors and recombinant viruses.
The plasmid pVL1393 (Pharmingen, San Diego, Calif.) was used as a baculovirus vector for expression of Trf4 and Trf5. A putative viral ribosome binding site and the hexahistidine-tagged proteins were added to the vector. The TRF4 and TRF5 genes were verified by double-stranded sequencing using primers based on the polyhedrin promoter and 3′ flanking sequences of pVL1393. TRF4 and TRF5 were copied by PCR from S. cerevisiae genomic DNA (Novagen) as the template and primers pSEY18-TRF4, TRF4-B, and TRF5-A (Table 3). Construction of the pVL1393 N-terminal six-His-tagged Trf4 baculovirus expression vector required that TRF4 be subcloned in frame at the SalI/SphI site of pFastBac Htc (Invitrogen, Carlsbad, Calif.) from the pSEY18-Trf4 PCR product. Using primers TRF4-A (Table 3), the final TRF4 construct was then subcloned in frame at the XbaI/NotI site of pVL1393.
For construction of pVL1393 carrying C-terminally six-His-tagged Trf5, the TRF5 gene was copied by PCR from S. cerevisiae genomic DNA using primers TRF5-A (Table 3) and then subcloned into pET24a(+) (Novagen) at the XhoI/SacI site. A sequence correction was made (see Table 3, footnote a) with the primer TRF5-correction, and the final product, constructed using primers TRF5-B, was then subcloned in frame at the BamHI/NotI site of pVL1393. Likewise, for the construction of pVL1393 carrying C-terminally six-His-tagged Trf4, TRF4-B primers were used to copy Trf4 and the product was subcloned in frame at XbaI/BgIII (Table 3). Liposome-mediated cotransfection (Lipofectin; Invitrogen) of Spodoptera frugiperda (Sf9) cells was performed using 2.5 μg of the recombinant transfer plasmid and 0.5 μg of linearized viral DNA (Baculogold; Pharmingen). Titer of virus was determined by plaque assay (50), and the virus was used to infect cells for expression of recombinant proteins. The Trichoplusia ni cell line BTI-TN-5B1-4 (High Five; Invitrogen) was grown in spinner flasks at 27°C in Ex-Cell 400 (JRH Biosciences, Lenexa, Kans.) to a density of 2 × 106 cells/ml. The cells were collected by centrifugation and infected with a multiplicity of infection of 5 to 10. The infected cells were collected by centrifugation at 60 h postinfection, and the expressed recombinant proteins were isolated.
Expression of His-Trf4, Trf5-His, and FLAG-Pol2 in insect cells and immunoprecipitation.
The pFast-FLAG-Pol2 baculovirus expression vector is described elsewhere (20). FLAG-Pol2 and His-Trf4 or FLAG-Pol2 and Trf5-His were coexpressed in insect cells by infecting High Five cells (5 × 106) with empty virus, His-Trf4 virus, Trf5-His virus, both His-Trf4 and FLAG-Pol2 viruses, or both Trf5-His and FLAG-Pol2 viruses from recombinant baculovirus stocks for 72 h. The harvested cell pellets (1.5 × 106 cells) were resuspended in 0.5 ml of lysis buffer (20 mM Tris [pH 8], 200 mM NaCl, and 1% Nonidet P40) and left on ice for 30 min with occasional agitation by vortex. The lysed cells were centrifuged, and the supernatant was incubated with 40 μl of activated and equilibrated anti-FLAG M2 affinity agarose (Sigma) beads for 2 h. The beads were washed four times in lysis buffer, and the bound protein was released by boiling in sodium dodecyl sulfate (SDS) cracking buffer. The crude extract and the bound protein were then resolved by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting, using the Amersham ECL system. The expression of FLAG-Pol2 was verified by Western blot analysis with anti-FLAG antibody. His-Trf4 was detected with anti-Trf4 antibody (provided by Michael Christman, Boston University). Trf5-His was detected with anti-His antibody.
Purification of Trf4-His from insect cells.
C-terminal His-tagged Trf4 (Trf4-His) was expressed in High Five cells for 72 h in spinner flasks at a density of 2 × 106 cells/ml. The cell pellet was lysed by homogenization in buffer A (20 mM Tris [pH 8.0], 20 mM imidazole, 500 mM NaCl, 10% glycerol, 0.5% Tween 20, and 5 mM β-mercaptoethanol) plus EDTA-free protease inhibitor cocktail (Roche). The extracts were clarified by centrifugation, and the supernatant was incubated in Qiagen Superflow Ni-nitrilotriacetic acid resin for 2 h at 4°C. The beads were rinsed four times batchwise in buffer A and then loaded to a column. The column was washed with 15 ml of buffer A, and the protein was eluted in buffer A with 300 mM imidazole. The protein peak was dialyzed overnight against buffer B (20 mM Tris [pH 8.0], 10% glycerol, 1 mM dithiothreitol [DTT], 0.05% Tween 20) containing 250 mM NaCl to maintain solubility of the recombinant protein and then loaded onto an HR5/5 Mono Q column (Pharmacia). The column was washed with 15 ml of buffer B and eluted with a 10-ml gradient of buffer B containing 250 mM to 1.25 M NaCl. The fractions were dialyzed against buffer B in a microdialysis apparatus (Gibco BRL) overnight. Trf4 was identified by Western blotting with anti-Trf4 antibody. The routine yield was 35 to 50 μg of protein. Concentration was estimated on SDS-polyacrylamide gel electrophoresis using bovine serum albumin as a standard. The protein was stored in aliquots at −70°C.
Pol ɛ stimulation assays.
Assays containing Pol ɛ represent the holoenzyme comprised of Pol2, Dpb2, Dpb3, and Dpb4, and these were described previously (20). The Pol2 140-kDa protein was also described in that work (20).
(i) Primer extension assay.
Oligo(dT)12-18 was labeled using polynucleotide kinase and [γ-32P]ATP. Unincorporated [γ-32P]ATP was removed using Bio-Rad 6 spin columns. Poly(dA)300-564-oligo(dT)12-18 (1:20, template to primer chains) was annealed in 25 mM HEPES (pH 7.1)-60 mM KCl by heating it at 70°C for 3 min and slowly cooling it to room temperature over 1 h. A 20-μl reaction (50 mM Tris [pH 8.0], 10 mM MgCl2, 2 mM DTT, 20 mM NaCl, 20 mM KCl, 2.5% glycerol, 0.2-mg/ml bovine serum albumin, 1 mM concentration or indicated amount of dTTP, and 0.15 U [unless otherwise indicated] of Pol ɛ [specific activity of 25,000 U/mg]) was started by addition of 100 nM 3′-OH ends of annealed poly(dA)-oligo(dT). After 15 min at 37°C, the reactions were quenched (30 μl of formamide with 15 mM EDTA and 0.2 mg of xylene cyanol/ml), boiled for 5 min, and placed on ice. The product (10 μl of the reaction mixture) was resolved on a 6% denaturing polyacrylamide gel.
(ii) dTTP incorporation assay.
Poly(dA)300-564-oligo(dT)12-18 (1:20, template to primer chains) was annealed in 25 mM HEPES (pH 7.1)-60 mM KCl by 3 min of heating at 70°C and slow cooling to room temperature over 1 h. The 20-μl reaction mixture contained 50 mM Tris (pH 8.0), 10 mM MgCl2, 2 mM DTT, 20 mM NaCl, 20 mM KCl, 2.5% glycerol, 0.2-mg/ml bovine serum albumin, 0.1 mM [3H]dTTP (New England Nuclear) with a specific activity of 82 cpm/pmol, 100 nM poly(dA)-oligo(dT), and enzyme. The reaction mixture was incubated at 37°C for 15 min. The reaction (18 μl) was quenched on DE81 membranes (diameter, 2.4 cm; Whatman), washed six times with 0.5 M sodium phosphate (pH 7.0), (5 min per wash), washed two times with water, briefly rinsed in 95% ethanol, and dried, and 3H was measured by scintillation counting.
Sister chromatid cohesion assay.
Strains were constructed as follows: strain AFS479 (W303 wild type) (see Table 1) contains a green fluorescent protein (GFP)-tagged chromosome system constructed by Aaron Straight, using plasmids pAFS59 and pAFS144 (Table 2). The lacO array is located at LEU2, 23 kb from CEN3. Wild-type POL2 in strain AFS479 was replaced by transformation with the linearized plasmid, PMB12, digested with MluI. PMB12 is a 13-kb DNA fragment containing pol2-12 in pRS306 URA3 (8). Ura+ transformants were purified and restreaked on synthetic medium with 5-fluoro-orotic acid and incubated at 25°C; 400 single colonies were restreaked on a subsequent 5-fluoro-orotic acid plate; these were then replica plated on yeast extract-peptone-dextrose plates and incubated at 25 and 37°C, screening for the temperature-sensitive phenotype of pol2-12 mutants. Strains SS111, SS111-pol2-12, and HKYDNA2 were transformed with the linearized plasmids pAFS52 and pAFS135 (59), generating strains SOG3, 12OG2, and JCYDNA2, respectively (8). The lacO array is integrated at TRP1, proximal to CEN4, in these constructs. The plasmids pAFS135 and pAFS144 were also constructed by Aaron Straight (59).
The PCR product from primers encoding 45 bp upstream and 45 bp downstream (Table 3) of the TRF5 gene was used to copy the deletion cassette from strain 1145 (BY4741 trf5Δ; KAN insertion). The resultant deletion cassette was used to replace wild-type TRF5 in strain SOG3, creating strain JCYT59. Gene replacement was verified by growth on 200 mg of G418/ml and by PCR. Crossing strains ts370 and SOG3 generated JCYPD18 (cdc2-1; with GFP signal). Resulting spores were tested for temperature sensitivity at 37°C, arrest with the dumbbell morphology, and a GFP signal.
To determine the efficiency of sister chromatid cohesion, cells were grown overnight to log phase in YPD supplemented with histidine, tryptophan, and adenine. Cells were collected by centrifugation and induced for the GFP-Lac repressor protein in SD-His for 30 min, collected by centrifugation, and synchronized at G1 by incubation in YPD plus 10 μg of α-factor/ml for 2.5 to 3 h at 30°C. α-Factor was then removed, and samples were either released into YPD to continue through the cell cycle or released into YPD with 15 μg of nocodazole/ml to arrest at G2/M. Samples were taken every 15 min and fixed in 70% ethanol for flow cytometry (18), in 100% or 70% ethanol to observe the GFP signal, and in 3.7% formaldehyde for immunofluorescence. 4′,6-Diamidino-2-phenylindole (DAPI) (0.01 μg/ml) was added as needed to visualize the nucleus. For immediate sister chromatid GFP data collection, GFP samples were fixed in 70% ethanol and 0.01 μg of DAPI/ml and placed on ice for 15 min, centrifuged, and resuspended in sterile H2O. This is a modified protocol taken from the work of Straight et al. (59). GFP samples collected in 100% ethanol and flow cytometry samples were stored at −20°C until analysis. Some samples were induced for GFP expression as described above and then directly arrested at G2/M from an asynchronous log phase culture grown in YPD plus 15 μg of nocodazole/ml for 3 h at 30°C.
Samples fixed for antitubulin immunofluorescence (monoclonal Rat-Harlan SERA-LAB) were tumbled at room temperature for 1 h in 3.7% formaldehyde, washed with SP buffer (1.2 M sorbitol in 0.1 M potassium phosphate [pH 7.5]), and stored at 4°C. Further procedures have been described previously (19).
Cells were imaged on a Zeiss Axioscope microscope using an apochromat 100 × 1.40 oil immersion lens. Images were acquired with a Hamamatsu digital camera, and image acquisition, processing, and documentation were done using the Axiovision 2.05, a modular image analysis system. To quantify, the data were taken from 7 to 10 individually printed optical frames, each including differential interference contrast, DAPI, and GFP fluorescence or fluorescein for spindle fluorescence.
Genetic interactions.
JCY122 was mated with 3aAS271a or 955-9C, and Y414 was mated with CY1243, to measure synergistic interactions between pol2-12 and smc1-2 or mcd1-1/scc1-1 mutants or the trf4ts896 trf5Δ double mutant (Table 1). Resultant diploid strains were sporulated, and tetrads were dissected. Genotypes of haploid spores were then determined using nutritional selection or growth on tester lawns at the nonpermissive temperature. Tester lawns used were TC102-2-12, SS111-2-12, 985-7C, and 955-9C (Table 1).
Drug sensitivity determinations.
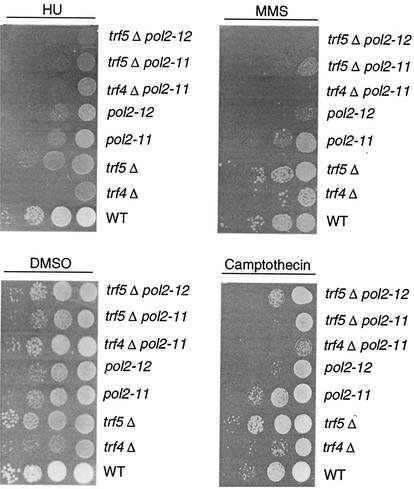
Hypersensitivity to camptothecin, HU, and methyl methanesulfonate (MMS) was measured as follows (see Fig. 4). Cells were grown to log phase and serially diluted, and 104, 103, 102, and 10 cells were plated on YPD plus or minus 10 μg of camptothecin/ml or 125 mM HU or 0.03% (vol/vol) MMS and incubated at 30°C for 3 days. Camptothecin plates were prepared by using the method of Walowsky et al. (67).
FIG. 4.
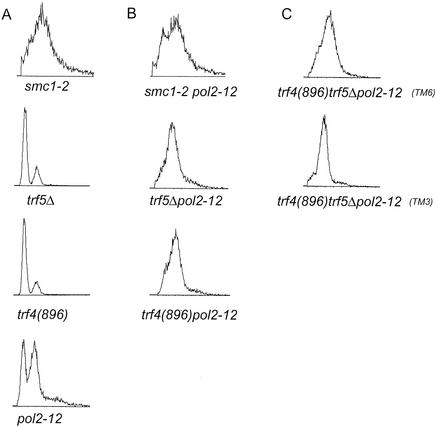
Effects of combining mutations in trf, pol2, and smc genes. (A) Flow cytometry profile of asynchronous cultures of exponentially growing pol2-12, trf4(896), trf5Δ, and smc1-2 single mutants. (B) Double mutants pol2-12 trf4(896) and pol2-12 trf5Δ. (C) Two pol2-12 trf4(896) trf5 triple mutants (TM6 and TM3). All strains were generated by the crosses described in Materials and Methods. Cells were grown to log phase (approximately 2 × 107 cells/ml) at 30°C; however, strains exhibiting an extended S phase were less dense.
RESULTS
A two-hybrid screen for genes that interact with the C terminus of Pol2.
To identify novel proteins that interact with the essential noncatalytic domain of Pol2, we performed a yeast two-hybrid screen with a fragment of the POL2 gene encoding aa 1265 to 2222 (end) as bait (Fig. 1A). Identification of five clones that encoded the second-largest subunit of Pol ɛ, DPB2, and two clones encoding SLD2, well-known Pol2-interactors, fostered confidence that the screen revealed functionally interacting proteins. Although many potentially interesting genes were identified and will be described elsewhere, one clone was of particular interest to us because it suggested a previously unappreciated role for Pol ɛ. This clone encodes an internal 78-amino-acid protein fragment of Trf5 (topoisomerase-related factor). Internal fragments are expected, since the library used in the screen was constructed with DNA fragments generated by partial restriction digest of genomic DNA with five different enzymes (21, 31).
Since TRF5 is thought to be a paralog of TRF4, which encodes Pol σ, we investigated whether Pol2 also interacts with Trf4. The nearly perfect conservation between the Pol2 interaction domain of Trf5 and the corresponding amino acids in Trf4 suggested that this would be the case (Fig. 1C); and, as shown in Table 4, Trf4 does interact with Pol2 in the two-hybrid assay.
TABLE 4.
Two-hybrid assaysa
| Protein | β-Galactosidase activity (U/mg)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AD | POL C-terminal-AD | TRF4-AD | TRF5-AD | DPB2-AD | DPB11-AD | SMC1-AD | ECO1-AD | |
| BD | 1.2 | − | − | − | − | − | − | − |
| POL2-C-terminal-BD | − | + | 22.3 | 5.8 | + | + | + | + |
| TRF4-BD | − | 1.1 | − | + | 0.6 | 2.3 | ND | ND |
| TRF5-BD | − | 33.4 | + | − | 16.0 | 36.0 | ND | ND |
| DPB2 | − | + | 0.8 | 9.7 | + | + | ND | ND |
Numerical values are based on quantitative assays of β-galactosidase activity (Stratagene; see Materials and Methods) and were measured in the same experiment and therefore are comparative. + indicates positive β-galactosidase activity measured using a filter lift assay only (Clontech). Activation of β-galactosidase activity in these samples was positive for blue color, indicating interaction of the genes tested. ND, not done; AD, activation domain (Gal4); BD, binding domain (LexA). Quantitative assays were not done with these clones. − indicates no β-galactosidase activity, represented by no color formation in a filter lift assay. Therefore, no interaction was observed.
The TRF5 78-amino-acid Pol2 interaction domain covers a region of the protein C-terminal to the conserved nucleotidyl transferase catalytic domain but coinciding with two regions of predicted structural similarity among members of the β-polymerase superfamily (Fig. 1B) (6). In addition, BLAST analysis of the 78-amino-acid TRF5 fragment (aa 371 to 448), using SGD and GenBank databases, revealed almost 78% amino acid sequence conservation between TRF4 and TRF5 in the Pol2-interacting region as well as significant homology with TRF-related genes in other organisms (Fig. 1C). One interesting feature of the 78-amino-acid Pol2 interaction domains of both TRF4 and TRF5 is a close match to a well-studied PCNA interaction motif (Fig. 1C) (71). Three trf4 mutants that map to the 78-amino-acid interaction domain (trf4-378, trf4-425, and trf4-444) (indicated by asterisks in Fig. 1C) have recently been shown to be lethal in the absence of TRF5, and two of these mutations cause sensitivity to MMS and camptothecin even in the presence of TRF5 (69). One of these lethal mutations occurs in the PCNA interaction motif. Another lethal mutation affects two lysine residues that fall outside of the region conserved in Pol β, but one of these residues is conserved in the closest Trf4 relative in S. pombe, SPAC12G12.13c (Fig. 1C). Since trf4 mutations falling in the Pol ɛ interaction domain are lethal, we propose that the interaction may be determined to be important for the essential, physiological function of Pol σ.
Mapping of the Trf5 interaction region in POL2 and demonstration of interaction of Trf5 with additional Pol ɛ-associated proteins, Dpb2 and Dpb11.
To map the Trf5 interaction region within Pol2 more closely (Table 5), a two-hybrid assay was conducted using a set of pol2 mutants carrying sequential 10-amino-acid deletions spanning the C-terminal zinc finger region, aa 2103 to 2222, used previously to study assembly of the Pol ɛ holoenzyme (Fig. 1) (17-20). This region contains essential amino acids, as well as motifs important for the putative checkpoint function and for avoidance of MMS-induced damage. As shown in Table 5, mutant pol2-B, in the first zinc finger region, which is highly sensitive to MMS, and pol2-E, in the interzinc region, which is inviable at all temperatures, essentially abolished interaction. pol2-F, which also showed reduced affinity, causes temperature-sensitive growth. pol2-H and pol2-I, which are viable at all temperatures, are sensitive to MMS at 37°C. The pol2-11 mutant, which has a stop codon at aa 2192, and the pol2-13 mutant, which has a deletion from aa 2163 to the end of the protein, also showed reduced interaction, and both caused temperature-sensitive growth. (We performed the two-hybrid assays at 30°C, a permissive temperature for growth of strains carrying these mutations; defects in binding could be greater at higher temperatures). These results suggest that mutations that affect the zinc finger domain reduce the Pol2-Trf5 interaction. The correlation between reduced interaction and significant defects in vivo suggests that the Pol2-Trf5 interaction is functional. Since the mutations also affect the stability of the holoenzyme (17, 19), we cannot be sure that this region represents an interface between Trf5 and Pol2.
TABLE 5.
Two-hybrid mapping of Trf5 interaction within Pol2
| Genotype or region | % β-Galactosidase activity for BD-TRF5a |
|---|---|
| AD | 0 |
| Pol2 C terminus | 100 |
| pol2-A | 30 |
| pol2-B | 10 |
| pol2-E | 0 |
| pol2-F | 45 |
| pol2-H | 58 |
| pol2-I | 69 |
| pol2-11 | 35 |
| pol2-13 | 28 |
One hundred percent activity is determined by the interaction of the activation domain (AD) fused to the Pol2 C terminus and the binding domain (BD) fused to Trf5 minus the interaction of the empty vector pAct2-AD and BD-Trf5. All fusion genes except that for BD-Trf5 were cloned into the activation domain vector pAct2. The values are an average of two different determinations and were further confirmed by colony filter-lift color assay.
We also checked the ability of Trf5 to interact with Dpb2 and Dpb11, since they both interact with the same region of Pol2 as Trf5 (17). As shown in Table 4, Trf5 interacts strongly with both Dpb2 and Dpb11 in the two-hybrid assay. This suggests that there are higher-order interactions (interactions between more than just two subunits) between the two polymerases, which has also been observed between Pol δ and Pol ζ (27). Trf4 does not interact with Dpb2 and Dpb11 in the two-hybrid assay; however, since Trf4 and Trf5 interact with one another, Trf5 may facilitate Trf4's interaction within the complex. This would suggest a possible synergy in the interaction between Trf4 and Trf5 and not just an overlapping function as suggested by homology.
Pol2 coimmunoprecipitates with Trf4 and with Trf5.
To investigate whether the two-hybrid reaction was reflected in a physical interaction between Pol ɛ and Pol σ, we carried out coimmunoprecipitation experiments. Since such interactions could be transient and therefore difficult to detect with proteins present at levels as low as those for replication proteins such as Pol ɛ, our experiments were carried out with the yeast proteins coexpressed in insect cells. TRF4 and TRF5 were each cloned into a baculovirus expression vector and coexpressed with Pol ɛ subunits, whose expression in insect cells we have described previously (17, 19, 20). Pol2 was fused to a FLAG epitope (20). When 6xHis-scTrf4 and FLAG-scPol2 were coexpressed in insect cells, 6xHis-scTrf4 bound to anti-FLAG beads more avidly in the presence than in the absence of coexpressed FLAG-tagged Pol2 (Fig. 2A, lanes 5 and 6), showing that Pol2 and Trf4 interact physically. The experiment was done in triplicate with the same result. Expression of FLAG-Pol2 and its efficient immunoprecipitation were verified by immunoblotting, and the expression levels of 6xHis-scTrf4 were shown to be the same in crude extracts expressing 6xHis-scTrf4 alone or coexpressing 6xHis-scTrf4 and FLAG-Pol2 (Fig. 2A, lanes 2 and 3). The anti-Trf4 antibody does not react with any endogenous insect cell or viral protein (Fig. 2A, lanes 1 and 4). Similar interactions were obtained with coexpression of Trf5 and Pol 2 (Fig. 2B). Thus, Pol2 and Pol σ interact physically.
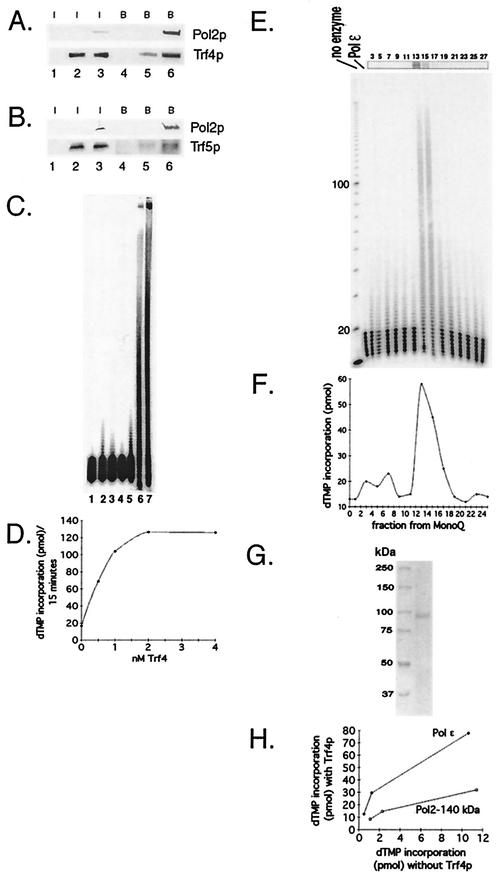
FIG. 2.
(A) Pol2 coimmunoprecipitates with Trf4. Coexpression of FLAG-Pol2 and His-Trf4 and immunoprecipitation protocols are described in Materials and Methods. Western blots of crude extract (labeled “I” for input protein) from insect cells expressing various combinations of Trf proteins and FLAG-Pol2 were probed with antibody against Trf4 or anti-FLAG Pol2 as indicated. Lane 1, no recombinant protein; lane 2, His-Trf4; lane 3, His-Trf4 plus FLAG-Pol2. These extracts were incubated with anti-FLAG beads. After washing of the beads, proteins that bound from extracts (labeled “B” for bound protein) were eluted by boiling. The proteins were analyzed on Western blots probed with antibody against Trf4 or anti-FLAG Pol2 as indicated on the right. Lane 4, no recombinant protein; lane 5, His-Trf4; lane 6, His-Trf4 plus FLAG-Pol2. (B) Pol2 coimmunoprecipitates with Trf5. Coexpression of FLAG-Pol2 and Trf5-His is described in Materials and Methods. Western blots of crude extract from insect cells are represented in the same order as those in Fig. 2A. (C) Recombinant Trf4 prepared in E. coli stimulates Pol ɛ holoenzyme. Trf4 was purified exactly as described previously (70). The oligo(dT)12-18 primer extension assay is described in Materials and Methods. Reaction mixtures contained 680 ng of Trf4, the amount required to observe Trf4 DNA polymerase activity (lanes 2 and 3), and/or 0.15 U of Pol ɛ, as indicated, are shown. The high level of Trf4 is saturating for stimulatory activity (see panel D). Lane 1, no protein; lane 2, Trf4-His with 0.1 mM dTTP; lane 3, Trf4-His with 1 mM dTTP; lane 4, Pol ɛ with 0.1 mM dTTP; lane 5, Pol ɛ with 1 mM dTTP; lane 6, Pol ɛ plus Trf4-His with 0.1 mM dTTP; and lane 7, Pol ɛ plus Trf4-His with 1 mM dTTP. (D) Titration of stimulatory activity of scTrf4 made in bacteria. The indicated amounts of scTrf4 were assayed for stimulation of [3H]dTMP incorporation by 0.15 U of Pol ɛ on an oligo(dT)-poly(dA) substrate as described in Materials and Methods. (E) scTrf4-His expressed in insect cell cochromatographs with Pol ɛ-stimulatory activity. scTrf4-His was expressed in insect cells. Silver staining of Trf4-His after purification through Ni2+-nitrilotriacetic acid and Mono Q columns, as described in Materials and Methods, and gel electrophoresis is shown at the top. Numbers refer to MonoQ fraction numbers. The same fractions are assayed for stimulation of primer extension by pol ɛ. Each fraction from the Mono Q column was dialyzed, and 2 μl of each fraction was used in a 20-μl reaction. Fraction 13 contained 17 ng of Trf4 protein (13 nM); but 2.7 nM Trf4 gave equivalent stimulation (not shown). The first lane shows no primer extension, the second lane shows activity of 0.15 U of Pol ɛ (0.5 nM) alone, and the subsequent lanes are the Mono Q fractions of the Trf4 purification assayed with 0.15 U of Pol ɛ (0.5 nm). The fraction numbers are identified above. (F) scTrf4-His and Pol ɛ-stimulatory activity copurify. [3H]dTMP incorporation assay: the same fractions from the Mono Q column were assayed for [3H]dTMP incorporation on an oligo(dT)-poly(dA) substrate as described in Materials and Methods in the presence of 0.15 U of Pol ɛ (0.5 nm). (G) Trf4 from the MonoQ column is highly purified. Coomassie-stained gel of Trf4 from fraction 14 of the MonoQ column. (H) scTrf4-His does not efficiently stimulate Pol2-140 lacking the C-terminal 1,000 amino acids. Three levels (0.075, 0.15, or 0.3 U) of either Pol ɛ (solid dots) or truncated Pol2 protein (open dots) were assayed with saturating amounts (40 ng) of Trf4-His purified from insect cells. Similar results were obtained with scTrf4 prepared in E. coli.
Trf4 and Pol ɛ holoenzyme act synergistically in DNA synthesis.
To determine if the Pol ɛ-Pol σ interaction was functional, the effect of Trf4 on Pol ɛ activity was determined. scTrf4 protein was expressed and purified from bacteria and characterized as previously described (70). We have recently described the Pol ɛ holoenzyme (Pol2, Dpb2, Dpb3, and Dpb4) preparation used for these studies (20). As shown in Fig. 2C, the two proteins showed significant synergy on a poly(dA)-oligo(dT) substrate, the optimal substrate for Pol ɛ (9, 25). As controls that the stimulation was not due to contaminating E. coli DNA polymerase I (the most abundant DNA polymerase in the bacterial extracts) in the scTrf4 preparation, we showed that DNA polymerase I of E. coli (Gibco BRL) did not stimulate Pol ɛ and antibody to E. coli Pol I (gift of S. Linn, U.C. Berkeley) did not inhibit the stimulation by Trf4 (not shown). Titration of Trf4, as shown in Fig. 2D, demonstrates that saturating amounts of Trf4 protein were measured in Fig. 2C. Therefore, we propose that we are observing stimulation of Pol ɛ by Trf4, though determining the exact extent to which each polymerase is stimulated will require further work.
To further increase confidence that the stimulation of Pol ɛ was due to scTrf4 protein rather than to a contaminating bacterial protein, the scTrf4 protein was tagged with six histidine residues at the C terminus as described in Materials and Methods, expressed in baculovirus-infected insect cells, and purified by affinity chromatography and ion exchange chromatography (Fig. 2E). The scTrf4-HIS protein showed DNA polymerase activity with the same specificity as Trf4 prepared from E. coli, for instance, sensitivity to dideoxy-TTP, a characteristic of β-type DNA polymerases (not shown). (Full enzymatic characterization will be reported elsewhere). Like scTrf4 protein produced in E. coli, the scTrf4-HIS protein stimulated Pol ɛ activity (Fig. 2E and F). N-ethylmaleimide levels that inhibit Pol ɛ but not Trf4 inhibited stimulation, and levels of ddTTP that inhibit Trf4 completely but only marginally affect Pol ɛ only marginally inhibited stimulation (data not shown). The Pol ɛ stimulation activity cochromatographed on the ion exchange column with the Trf4 protein and showed high purity (Fig. 2E, F, and G). Western blotting with anti-Trf4 antibody verified that the band at 90 kDa in Fig. 2E and G is Trf4. Thus, the Trf4 protein and Pol ɛ holoenzyme interact functionally.
We next addressed the extent to which the C-terminal domain of the Pol2p, i.e., the region sufficient for interaction with Pol σ, was necessary for stimulation. We have described a preparation of Pol2 protein that lacks a large fragment of the C terminus, Pol2-140, but that retains efficient polymerase activity (20). This Pol2-140 preparation contained 90% Pol2-140 protein but did retain about 10% full-length Pol2p (20). Nevertheless, as shown in Fig. 2H, this preparation showed substantially reduced stimulation by scTrf4-HIS, suggesting that physical interaction provides optimum stimulation. The residual stimulation could be due to the full-length Pol2p in the preparation or to the ability of Trf4 to interact with Pol ɛ in the presence of the DNA template. (The bacterially produced Trf4 protein also failed to stimulate Pol2-140; data not shown). Pol σ could also interact with other subunits of Pol ɛ, as seen with Dpb2 and Trf5 in the two-hybrid assay (Table 4).
Genetic interactions between POL2 and TRF genes suggest their interaction is important both for viability and for DNA repair.
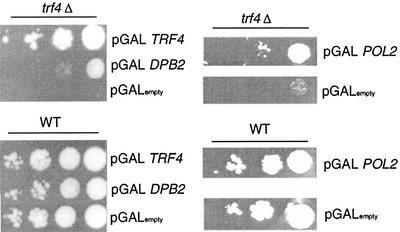
To determine if the biochemical interaction between Pol ɛ and Pol σ were physiologically significant, we first investigated possible mutual high-copy-number suppression. Deletion of TRF4 results in a cold-sensitive phenotype at 16οC (14). Overexpression of Pol2 and Dpb2 increased viability of the trf4Δ strain at 16°C, although only Trf4 was able to restore the wild-type growth rate and viability (Fig. 3). Conversely, overexpression of TRF4 or TRF5 did not suppress the temperature-sensitive phenotype of the pol2-12 or pol2-11 mutant, nor did overexpression of TRF4 or TRF5 suppress the temperature sensitivity of the pol2-16 mutant, which encodes only the C-terminal half of Pol2 and therefore lacks the Pol2 polymerase domain (not shown). Thus, Trf4 is probably not the polymerase compensating for loss of the Pol2 polymerase activity in the pol2-16 mutant (18).
FIG. 3.
Genetic interaction of POL2 and TRF4. POL2 and DPB2 suppress the cold sensitivity in a trf4Δ mutant. Plasmids PDLT4, pSEY18-II, pSEY18-DPB2, and pSEY18 (GAL1 and GAL10 expression vectors with inserts of TRF4, POL2, DPB2, or empty [no insert], respectively) were transformed into strains 6265 (BY4741-trf4Δ) and the wild type (BY4741) and selected for growth on synthetic medium with dextrose-Ura. The resultant colonies were purified, grown to log phase in liquid SRaff-URA, and serially diluted onto plates containing 2% galactose. Duplicate plates were incubated at 16°C for a week.
In addition, we tested the effect of the introduction of the pol2-12 mutation into various trf mutants. The mutation in pol2-12 is a stop codon at aa 2196, 15 aa downstream of the zinc finger region, and this truncation disrupts the zinc finger region, resulting in all of the same phenotypes as for the pol2-F mutant: checkpoint defects, defects in assembly of the holoenzyme, and temperature-sensitive growth that is suppressed by overproduction of Dpb2 (17-20, 47). As shown in Table 5, the mutant pol2-11, which is a stop codon in the amino acid adjacent to pol2-12, significantly reduces Pol2-Trf5 interaction by two-hybrid assay even at the permissive temperature. pol2-12 was used instead of pol2-F because it has proved impossible to integrate pol2-F into the chromosome. (pol2-E could not be used because it is lethal.) We introduced pol2-12 into trf4Δ, trf5Δ, trf4(896), and trf4(896)ts trf5Δ mutants. All double and triple mutants were viable at 23°C. The defects in pol2-12 trf4(896) trf5Δ and pol2-12 trf5Δ were synergistic at higher temperatures, having a lower maximum permissive temperature than either single mutant or the trf4(896) pol2-12 mutant (Table 6). Flow cytometry of asynchronous log phase cultures revealed an abnormal cell cycle distribution compared to single mutants for the pol2-12 trf4(896) trf5Δ mutant, pol-12 trf5Δ mutant, and also for the pol2-12 trf4(896) mutant at 30°C (Fig. 4). This could be due either to an extended S phase or to cell enlargement. Either explanation combined with a reduced permissive temperature would suggest that cells with diminished Pol2 function have an elevated requirement for Trf activity and vice versa, suggesting that these proteins mutually reinforce each other.
TABLE 6.
Genetic interaction between pol2 mutant and cohesion genes
| Mutation | Growth at temp (°C)a
|
|||
|---|---|---|---|---|
| 25 | 30 | 33 | 37 | |
| pol2-12 | +++ | +++ | +++ | − |
| trf4(896) | +++ | +++ | +++ | +++ |
| trf5Δ | +++ | +++ | +++ | +++ |
| pol2-12 trf4(896) | +++ | +++ | +++ | − |
| pol2-12 trf5Δ | +++ | +++ | − | − |
| trf4ts(896) trf5Δ | +++ | +++ | +++ | − |
| pol2-12 trf4(896) trf5Δ | +++ | ++ | − | − |
| smc1-2 | +++ | +++ | +++ | − |
| pol2-12 smc1-2 | +++ | ++ | − | − |
| scc1-1/mcd1-1 | +++ | +++ | +++ | − |
| pol2-12 scc1-1/mcd1-1 | +++ | − | − | − |
+++, normal growth; ++, slow growth; +, poor growth; −, no growth.
We next compared pol2 and trf mutants with respect to sensitivity to several DNA synthesis-modifying and DNA-damaging agents—HU, MMS, and camptothecin. Two double mutants, the pol2-11 trf5Δ and pol2-11 trf4Δ mutants, both of which grow more slowly than either single mutant and show reduced viability at 30°C, were also tested for sensitivity to these drugs (Fig. 5). HU is an inhibitor of DNA synthesis that blocks ribonucleotide reductase, and all of the mutants show increased sensitivity to 125 mM HU (compared to the wild type). The trf4Δ mutant itself was significantly sensitive to HU. The trf5Δ mutant was less sensitive, but the pol2-12 trf5Δ and pol2-11 trf5Δ strains showed 10- to 100-fold reductions in viability compared to single mutants on HU (125 mM). Camptothecin is a plant alkaloid whose sole target is TOP1. It is thought that the initial cleavable complex formed between the enzyme and DNA in the presence of the drug is converted upon replication to a double-strand break (DSB) (13). trf4Δ mutants are sensitive to camptothecin and to MMS (67). As shown in Fig. 5, trf4Δ mutants are 10- to 100-fold more sensitive to camptothecin than the wild type at 30°C. pol2-11 and pol2-12 mutants also showed sensitivity to camptothecin. The pol2-11 trf4Δ mutant was significantly more sensitive to camptothecin than either single mutant. The trf5Δ mutant was not sensitive to camptothecin. pol2 mutants are sensitive to 0.03% MMS (17), and trf4 mutants have also been shown to be MMS sensitive previously (67). As shown in Fig. 5, the trf5Δ mutant was only slightly sensitive to 0.03% MMS relative to the wild type. pol2-11 trf5Δ, pol2-12 trf5Δ, and pol2-11 trf4Δ mutants showed more sensitivity than single mutants. Thus, pol2 mutations have a more severe effect on ability to repair damage in cells in which TRF4 or TRF5 is also impaired, as expected of interacting proteins.
FIG. 5.
Sensitivity of pol2-11, pol2-12, trf4, and trf5 mutants and various combination mutants to DNA synthesis inhibitors and DNA damaging reagents. Strains were grown to log phase and serially diluted at 104, 103, 102, and 10 cells per row on YPD with dimethyl sulfoxide (DMSO) (control plates, DMSO was used to dilute camptothecin; see Materials and Methods), 10-μg/ml camptothecin, 125 mM HU, and 0.03% (vol/vol) MMS. Strains are isogenic with the following designations: WT, BY4741; trf4Δ, 6265; trf5Δ, 1145; pol2-11, BY-pol2-11; pol2-12, BY-pol2-12; trf4Δ pol2-11, BY-trf4Δpol2-11; trf5Δ pol2-11, BY-trf5Δpol2-11; and trf5Δ pol2-12, BY-trf5Δpol2-12 (see Table 1).
Genetic interactions between POL2 and genes involved in sister chromatid cohesion.
Since Trf4 is required for sister chromatid cohesion, the interaction between Pol ɛ and Pol σ suggested that Pol ɛ might be required for sister chromatid cohesion as well as for DNA repair (13, 14, 52, 67, 70). As an indirect probe of whether Pol2 might be involved in cohesion, we carried out a two-hybrid analysis with two additional cohesion proteins, Smc1 and Eco1/Ctf7, which interact with TRF4 (70). Eco1/Ctf7 is of particular interest as it is required for establishing but not for maintaining cohesion and because it interacts genetically with PCNA, a polymerase processivity factor. As shown in Table 4, Pol2 interacted with both of these proteins.
To test if the Pol2-Smc1 interactions represented functional interactions, double mutants were prepared by crossing pol2-12 mutants with smc1-2 or mcd1-1/scc1-1 cohesion mutants (23, 70). Each double mutant was recovered at 23°C. However, the maximum permissive temperature for growth was significantly decreased for the pol2-12 smc1-2 and pol2-12 scc1-1/mcd1-1 mutants (Table 6). The most defective double mutant, the pol2-12scc1/mcd1-1 mutant, failed to grow at 30°C. Introduction of a POL2-expressing plasmid restored growth at 30 and 33°C but not at 37°C (data not shown). These studies suggest, though do not prove, that Pol2 interacts with the cohesion apparatus. Flow cytometry of asynchronous log phase cultures revealed a dramatically extended S phase for these mutants as well, similar to the S phase of pol2 trf double mutants (Fig. 4).
Defects in sister chromatid cohesion in pol2 and trf5 mutants.
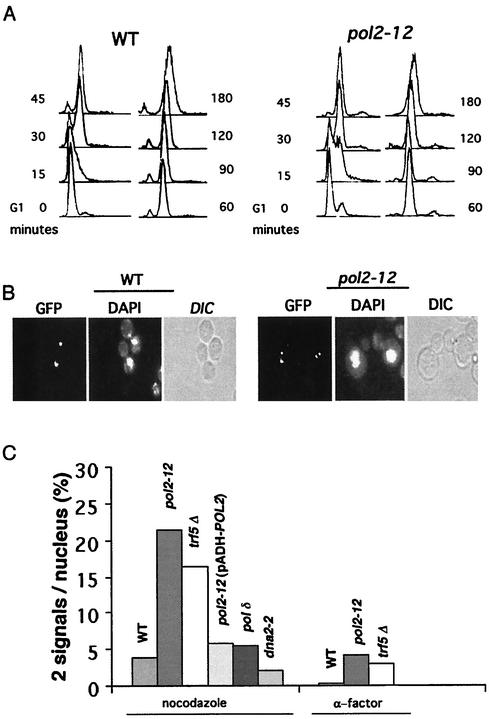
We next tested directly for a defect in sister chromatid cohesion in pol2-12 mutants. During the cell cycle, duplicated sister chromatids remain associated until the establishment of tension through kinetochore-spindle attachment in metaphase and dissolution of cohesins at the metaphase-anaphase transition. If cells are arrested before this transition by using a microtubule-depolymerizing agent such as nocodazole, normal cells will arrest with sister chromatids attached. The proficiency of sister chromatid cohesion can be measured by comparing the stability of cohesion in mutants relative to that for the wild type. Sister chromatid separation can be observed by fluorescence microscopy using GFP-marked chromosomes (59). In this system, an array of lac operators is integrated at the LEU2 or TRP1 locus, and a GFP-Lac repressor fusion protein is expressed. Cells can then be monitored microscopically throughout the cell cycle. One defined fluorescent signal per cell represents attached chromatids, and two signals indicates separated chromatids. Using this procedure, others have shown that deletion of TRF4 results in defective sister chromatid cohesion, even in the presence of TRF5 (70).
Since pol2-12 mutants fail to complete DNA replication at 37°C and synthesize no high-molecular-weight DNA (8), the following experiment was carried out at 30°C, where S phase is slow but eventually goes to completion and cells are viable (Fig. 6A and 7B). Mutant pol2-12 cells were incubated with α-factor for 3 h. G1-arrested cells were released into medium containing nocodazole, and samples were fixed at various times. Flow cytometry (Fig. 6A) and cellular morphology (Fig. 6B) verified G2 arrest. The same mutants carrying an HA-Pds1 gene in addition to the GFP signal system were found to contain intact Pds1 at the arrest point, indicating that the spindle checkpoint appeared to be intact during nocodazole arrest in these mutants (data not shown) (15, 16, 28). The fraction of cells with two dots (GFP signals) per nucleus is shown in Fig. 6C. More than 20% of the pol2-12 cells showed two dots, a fivefold increase relative to the wild type. This level of defect in cohesion is similar to that observed for the trf4Δ, ctf18Δ, ctf8Δ, and ctf4Δ cohesion mutants (26, 40, 70). Since nocodazole prevents formation of spindles, there is presumably no tension on the sister chromatids, and therefore this level of separated dots may be an underestimate of the defect associated with the pol2 mutation. The double signals were not due to aneuploidy, since they did not appear in the α-factor-arrested cells. The defect was observed with lac operators inserted at either LEU2, located on chromosome III, or at TRP1, located on chromosome IV. The cohesion defect was observed in two different strains carrying the pol2-12 allele (data not shown) and therefore is not due to strain background effects. Introducing the wild-type POL2 gene on a plasmid, pADH-POL2, restored cohesion to normal levels (Fig. 6C). The defect in cohesion is also observed at 23°C, and a shift to 37°C after nocodazole arrest did not increase the fraction of cells with two spots (not shown).
FIG. 6.
Defective sister chromatid cohesion observed in both a pol2-12 mutant and a trf5Δ mutant. (A) Flow-cytometric analysis of nocodazole-arrested pol2-12 mutant and an isogenic wild-type strain carrying the GFP signal. (B) Typical cells with attached sisters (one GFP dot per cell body) or separated sisters (two separated dots per cell body). (C) Defective sister chromatid separation was measured as total GFP dots, relative to double GFP dots (dd), for 500 to 700 cells in two separate experiments for each strain. Both pol2-12 and trf5Δ are defective. Replacing POL2 alleviates the defect. Isogenic controls for 12OG2(pADH-POL2) were wild type, SOG3, and 12OG2 (a pol2-12 mutant) (Table 1). These gave similar values of approximately 20% dd, similar to values observed with isogenic strains AFS479 and JCY122 shown here. Strains are designated as follows: AFS479 wild type (WT), JCY122 (pol2-12), JCYT59 (trf5Δ), 12OG2 [pol2-12(pADH-POL2)], JCYPD18 [cdc2-1 (Pol δ)], and JCYDNA2.
FIG. 7.
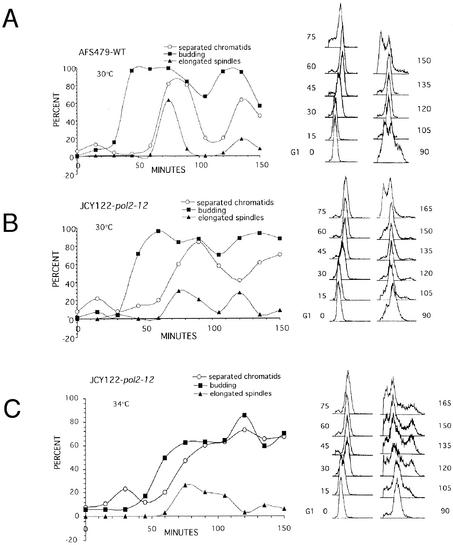
Sister chromatid separation in a pol2 mutant begins earlier in the cell cycle than in the wild type. Wild-type cells (AFS479) and pol2-12 cells (JCY122) were grown to log phase at 30°C, induced for GFP, and then arrested with α-factor for 3 h. Cells were then released in YPD at 30°C (A and B) or 34°C (C). Samples were taken every 15 min and analyzed for bud emergence by differential interference contrast (represented as budding index), timing of sister chromatid separation by GFP assay, spindle elongation by indirect immunofluorescence using an antitubulin antibody, DNA content by flow cytometry, and nuclear morphology by DAPI staining.
We also demonstrated that the cohesion defect in the pol2-12 mutant was not a general defect of DNA replication mutants (Fig. 6C). A mutation affecting the catalytic domain of Pol δ, cdc2-1, and a mutation affecting a replication helicase-nuclease, dna2-2, caused no increase in double dots compared to the wild type.
Since trf5Δ mutants had not previously been examined for sister chromatid cohesion defects, we included an isogenic trf5Δ strain in this study. As shown in Fig. 6C, the trf5Δ mutant showed defects in sister chromatid cohesion comparable to those of the pol2-12 mutant and to those previously reported for a trf4Δ mutant (70). The compromised cohesion in these mutants suggests overlapping functions for TRF4 and TRF5, in line with their high degree of homology and the fact that deletion of both is lethal (13). However, two additional results lead us to propose that Trf4 and Trf5 may also perform independent functions. First, two-hybrid results indicate that Trf5 interacts with Dpb2 and Dpb11 while Trf4 does not and that Trf5 interacts with Trf4, while neither self-interacts (Table 4). Second, we observe differences between trf4 pol2 and trf5 pol2 double mutants (Table 6 and Fig. 5).
pol2 mutants show premature sister chromatid separation and missegregation of chromosomes.
To investigate further if cohesion is compromised in the pol2-12 mutant, we followed sister chromatid cohesion through a synchronized cell cycle in the wild type and in the mutant at 30°C and compared the timing of sister chromatid separation with that of bud emergence and spindle elongation (Fig. 7). Analysis of cells by flow cytometry indicated that the wild type passed synchronously through two rounds of the cell cycle (Fig. 6A). The pol2-12 mutant passed more slowly through the cell cycle (Fig. 7B). Bud emergence was delayed, and bud growth lagged behind that of the wild type. In addition, there was a delay in cytokinesis, with only a small population of cells dividing concurrently with wild type during the first round of the cell cycle (90 min) (Fig. 7B). Spindle elongation was also extended over a greater interval for the pol2-12 mutant at 30°C.
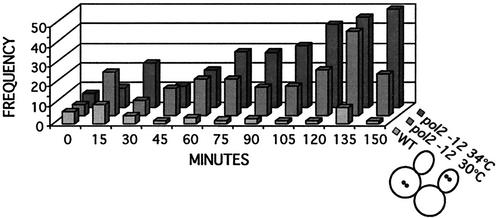
To determine whether separation of chromatids was premature in the mutant, we corrected for the delay in budding in the pol2-12 mutant {by applying the formula S (sister separation) = [U/(2 − U)] × (TD + 1) + TD, where, U = the fraction of unbudded cells and TD = the fraction of cells with two dots (41)}. This allowed us to estimate the cumulative fraction of cells that had separated sister chromatids at each time point (41). We found that wild-type cells separated their chromatids 45 min after budding, whereas the mutant cells began separating their chromatids 30 min after budding. Consistent with the calculated decrease in time separating budding from sister separation in the mutant compared to the wild type, separated sisters were seen in cells with small buds in the mutant but not in the wild type. In addition, Fig. 8 shows that the frequency of double dots in the same cell is dramatically higher in the mutant strain than in the wild type. In most cases this pattern coincided with unequal separation of the entire nuclear mass, as revealed by DAPI staining of the same cells, and with misalignment of spindles, as revealed by tubulin staining of cells at the same point in the cell cycle. At least 20% of the cells showed a nuclear mass in the daughter bud at 60 min, and at least 50% of the missegregated nuclei contained two GFP dots (see Fig. 8). The increased frequency of same-cell sisters would be a predicted consequence of premature failure of sister chromatid cohesion. It is also representative of chromosome loss, since increased missegregation of chromosomes would generate the considerable number of cells without chromosomes or with unequally distributed sisters that is observed. Such missegregation of chromosomes has also been reported for scc1 (cohesin) mutants. Despite missegregation, pol2-12 mutant cells continue to replicate their genomes, again like scc1 mutants (41). It is also apparent from the dramatic increase in the frequency of same-cell sisters after nuclear division that all faulty sites may not be readily detected in earlier time points of the α-factor-arrested cells.
FIG. 8.
The frequency of separated sister chromatids occurring in a single nucleus increases for the pol2 mutant. Cells that displayed two GFP dots in Fig. 6 were again scored for the percentage of these that displayed two signals in one nucleus.
We attempted to carry out the same experiment with the mutant at 34°C. At 34°C, the time between budding and sister chromatid separation was dramatically reduced for the mutant, to only a few minutes. The mutants were also severely retarded in spindle elongation and cytokinesis, probably due to replicative stress. In fact, at the later time points there is the appearance of a 4C DNA content cell population observed with flow-cytometric analysis (Fig. 7C). This is accompanied by the accumulation of large budded cells with two GFP dots in each cell or four fluorescent signals per nucleus, indication also of defective cohesion (Fig. 8). These four-dot cells were not seen in the wild type or in the mutant at 30°C.
DISCUSSION
In the course of our search for the essential function of the C terminus of yeast Pol ɛ, we isolated a 78-amino-acid protein fragment of Trf5 in a two-hybrid library screen. We have verified that this interaction reflects both a physical and a functional, that is, enzymatic, interaction. We have provided genetic evidence that the interaction is important for viability and for DNA repair by demonstrating synergy between mutations affecting pol2, trf4, and trf5. The requirement for Pol σ in establishing sister chromatid cohesion led us to also examine pol2 mutants for defects in sister chromatid cohesion. We have shown that POL2 is linked to the sister chromatid cohesion machinery by presenting two-hybrid evidence for interaction with two additional sister chromatid cohesion proteins and synergy between mutations in pol2 and in cohesin genes. Furthermore, mutation of the C-terminal region of pol2 itself causes a defect in sister chromatid cohesion (Fig. 6-8). Thus, we conclude that Pol ɛ has at least two essential functions. Its polymerase activity is required for normal DNA replication, as previously shown by using temperature-sensitive mutants (5). Its noncatalytic C-terminal half plays a role in mediating efficient sister chromatid cohesion and/or in an essential type of DNA repair.
The yeast TRFs (encoded by TRF4 and TRF5), now designated Pol σ, are members of the Pol β superfamily of nucleotidyl transferases and are essential for completion of S phase in yeast, for DNA repair, and for efficient sister chromatid cohesion (6, 10, 13, 70). Trf4 protein contains a Mg2+-dependent DNA polymerase activity and coimmunoprecipitates with a Mn2+-dependent poly(A) polymerase activity from yeast extracts (50, 64, 65; also unpublished data). Two of the six relatives of Trf4 for Schizosaccharomyces pombe, cid1+ and cid13+, lack DNA polymerase activity but have poly(A) polymerase activity and appear to be entirely cytoplasmic (51, 53). Despite primary sequence conservation in the nucleotidyl transferase active site, S. pombe cid1+ and cid13+ do not appear to be functional homologs of yeast Trf4 protein, which clearly has DNA polymerase activity, is nuclear, and associates with chromatin in a cell cycle-specific manner (67, 69). Further work will be required to determine if yeast Trf4 or Trf5 has cytoplasmic components and functions that require poly(A) polymerase activity in addition to their nuclear functions.
Several point mutations in the domain of Trf4 that are sufficient for interaction with Pol ɛ, Trf4 amino acids 374 to 451, are sensitive to DNA damage and lethal in the absence of Trf5 (69). We propose that both the essential and DNA repair functions of this noncatalytic domain of Pol σ are related to its ability to interact with Pol ɛ, and we predict that this Trf domain will be involved in sister chromatid cohesion (53). Two previous findings support this conclusion. First, mutations in the Trf4 catalytic domain do not completely disrupt sister chromatid cohesion (70). Second, one trf4 mutation just downstream of the Pol ɛ interaction domain (aa 491) is even more deficient in sister chromatid cohesion than the trf4Δ mutation (69). Thus, the nucleotidyl transferase activity of Trf4 is not the sole portion of the protein essential for sister chromatid cohesion. It is interesting that preliminary analysis shows that the putative active-site residue, amino acids 236 and 238, in the nucleotidyl transferase domain are not essential for stimulation of Pol ɛ by Trf4 (data not shown).
Our studies have uncovered some differences between Trf4 and Trf5. For instance, Trf4 does not interact with Dpb2 and Dpb11 in the two-hybrid assay, but Trf5 does. Also, the interaction between Pol2 and Trf5 is stronger than the interaction between Pol2 and Trf4. trf4Δ and trf5Δ mutants also show different sensitivities to various DNA-damaging agents, and pol2 trf4 and pol2 trf5 double mutants also show different synergies. Despite the fact that the trf4Δ or trf5Δ mutant is themselves viable and double mutants are inviable, nevertheless there may be some differentiation of function between the two proteins.
It has been proposed that when the replication apparatus reaches a site of cohesion assembly, it recruits Pol σ to the fork and there is a polymerase switch in which the primer is transferred to Pol σ before the site is replicated (12, 70). After bypass, the primer could be returned by the presumably nonprocessive Pol σ to Pol ɛ for further processive synthesis. The synergy that we observe could be analogous to the synergy seen between Pol III holoenzyme and Pol V in copying templates downstream of lesions (63). It might derive from increasing the affinity of Pol ɛ for the primer termini in the assay used here. Alternatively, Pol σ could increase the affinity of Pol ɛ for DNA in general or even act to stabilize Pol ɛ. Although this increase in affinity might not be necessary at every base, it might be important at sites of protein blocks, as might be envisioned during cohesion, or at sites of template damage. Thus, the stimulation suggests that the two polymerases act in concerted fashion but not necessarily that there is a switch. A possible role for Pol σ other than switching is also suggested by the fact that, as mentioned, we find that the nucleotidyl transferase active-site mutant of Trf4 efficiently stimulates pol ɛ in vitro (data not shown). Polymerase switching was first explored in pioneering biochemical studies aimed at elucidating the transition from Pol α-primase to Pol δ during replication initiation; RFC, the clamp loader, was shown to play a key role in loading the second polymerase (65, 66, 72). Other switches have recently been shown to occur when a replicative polymerase encounters abasic or UV-induced damage and other damage in the template. A polymerase switch model for replication of cohesion sites is supported by the demonstrated interaction between the polymerase clamp (PCNA) and Eco1/Ctf7 which is required for the establishment of cohesion, as well as the evidence that an alternative clamp-loader complex is required for proper cohesion (26, 34, 40). One component of the alternative clamp-loader subunit, Ctf18, has been shown to copurify with both Pol2 and Dpb2 during affinity purification of Ctf18 from yeast (22). It remains to be determined whether there is a switch at sites of cohesion and, if so, what induces it. Nevertheless, the fact that Pol ɛ associates with replication origins in G1 and remains associated with the other replication proteins at the replication fork (1) suggests that Pol ɛ is a component of the replisome and therefore might be involved in such a switch.
It has recently been proposed that cohesin forms a topological bond with chromatin by forming a protein ring around the DNA (24). Opening and closing of the cohesin ring might be essential to allow passage of the replisome. Alternatively, remodeling of the replisome through transient separation of leading and lagging strand complexes might facilitate traversing the cohesin. In any case, linking passage of the replication fork to formation of a cohesin ring around the sisters at 10-kb intervals might prevent rotation of the replisomes, which might otherwise lead to entangling of the sisters (61, 62). Topoisomerase might collaborate by disentangling sisters that do become intertwined; and, if so, this would account for the as yet unexplained synthetic lethality of trf4 and top1. How the two polymerases described here might interact with the cohesin complex, which both this and previous studies suggest they do (14), remains to be determined.
The Pol ɛ-Pol σ interaction may also be required for repair (Fig. 5). The role in repair could be related to the cohesion functions, since several lines of evidence suggest that cohesion is also required for recombinational repair. Sjogren and Nasmyth (55) showed directly that smc1, scc1, scc2, and pds5 mutants are as defective in repair of x-ray-induced damage as are rad54 mutants. They further demonstrated that cohesion, and not just the proteins themselves, is required. Since DNA polymerases are also required for DSB repair, Pol ɛ and Pol σ might be required at stalled replication forks, repaired by recombinational pathways (35). Pol ɛ has been purified with cohesion proteins SMC1 and SMC3 in a mammalian recombination complex (RC 1) that catalyzes cell-free DNA strand transfer and repair of gaps and deletions (32), and yeast Pol σ coimmunoprecipitates with Smc1 (13). POL2 has been shown to be required for gene conversion at an HO-induced DSB, along with several additional replication proteins (30). Pol σ mutants, like pol2 mutants (this work), are very sensitive to camptothecin, which results in DSBs during DNA replication that are presumably repaired by DSB repair pathways. Pol σ (Trf4 but not Trf5) also interacts with and is synthetically lethal with top1, which affects recombination (67). Finally, both Pol σ and Pol ɛ mutants are very sensitive to MMS. One mechanism by which the role in repair might occur is suggested by the fact that Pol ɛ dimerizes, probably through interaction with Dpb2 dimers (17). Substitution of Pol σ for Dpb2 might facilitate replication by Pol ɛ past abasic sites, as has been proposed for Rev1 protein in mutagenic bypass by Pol δ and Pol ζ (27). Rev1 is proposed to substitute for the dimerizing subunit of Pol δ, Pol32. Rev1, like Pol σ, possesses nucleotidyl transferase activity but is thought to perform a scaffolding role rather than to function in synthesis, since mutation of its active site does not affect its ability to act in mutagenic bypass synthesis (27, 48). Similarly, during establishment of cohesion, Trf4 might compete with Dpb2 for binding to Pol2p and thus help transiently remodel the fork for interaction with the cohesins.
In addition to a proposed role in establishing cohesion and in DNA repair, it has also been suggested that Pol ɛ might be part of the signaling apparatus that inhibits the metaphase-to-anaphase transition in response to DNA damage during S phase or replication defects. The conclusion that pol2-12 mutants are defective in metaphase/anaphase arrest is primarily based on observation of elongation of spindles in HU-treated pol2-12 cells. It is possible, however, that it is the defect in cohesion that allows premature spindle elongation at 37°C, due to lack of opposition to spindle-generated poleward traction rather than a lack of checkpoint signaling. That is, there is no substrate for the checkpoint pathway to act upon. The puzzling observation that RAD53 is fully activated in HU-treated pol2 mutants would be consistent with the latter proposal (37). A defect in cohesion would not explain the reduced induction of RNR3 in the presence of HU (47) or MMS in pol2 mutants (19). The effect on RNR3 might be explained, however, through a defect in interaction of the mutant pol2-11 protein with the Trfs. If, like cid13+ in S. pombe, Trf4 and/or Trf5 have cytoplasmic as well as nuclear functions, the Pol ɛ-Pol σ interaction might play a role in stabilizing RNR3 mRNA (53). If the defect in cohesion in pol2 mutants is responsible for the apparent checkpoint defect, then one might expect scc1 mutants to be defective in the S phase checkpoint. Cells depleted for scc1 have been reported to delay the cell cycle when treated with x-rays in late S phase, but they do show aberrant timing of spindle elongation (55).
In summary, the interaction between Pol σ and Pol ɛ suggests new roles for Pol ɛ. Studies reported here support a role in sister chromatid cohesion. In addition, the recent discovery that a Trf-related protein in S. pombe has poly(A) polymerase activity suggests that there may be additional outcomes of the Pol ɛ-pol σ interaction that remain to be characterized.
Acknowledgments
We are grateful to Michael Christman and members of his laboratory for strains, plasmids, and valuable advice. We thank R. D. Deshaies and G. Alexandru for comments on the manuscript and Ray Deshaies for the use of his microscope.
This work was supported by PHS GM25508. S. Edwards was supported by a PHS Research Supplement for Underrepresented Minorities.
REFERENCES
- 1.Aparicio, O. M., D. M. Weinstein, and S. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 2.Araki, H., R. D. Hamatake, L. H. Johnston, and A. Sugino. 1991. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 19:4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki, H., R. K. Hamatake, L. H. Johnston, and A. Sugino. 1991. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:4601-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki, H., S.-H. Leem, P. Amornrat, and A. Sugino. 1995. Dpb11, which interacts with DNA polymerase II(ɛ) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 92:11791-11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araki, H., P. A. Ropp, A. L. Johnson, L. H. Johnston, A. Morrison, and A. Sugino. 1992. DNA polymerase II, the probable homolog of mammalian DNA polymerase ɛ, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 11:733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravind, L., and E. Koonin. 1999. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. [DOI] [PubMed] [Google Scholar]
- 8.Budd, M. E., and J. L. Campbell. 1993. DNA polymerases δ and ɛ are required for chromosomal replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:496-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budd, M. E., K. Sitney, and J. L. Campbell. 1989. Purification of DNA polymerase II, a distinct polymerase, from Saccharomyces cerevisiae. J. Biol. Chem. 264:6557-6565. [PubMed] [Google Scholar]
- 10.Burgers, P. M. J., E. V. Koonin, E. Bruford, L. Blanco, K. C. Burtis, M. F. Christman, W. C. Copeland, E. C. Friedberg, F. Hanaoka, D. C. Hinkle, C. W. Lawrence, M. Nakanishi, H. Ohmori, L. Prakash, S. Prakash, C.-A. Reynaud, A. Sugino, T. Todo, Z. Wang, J.-C. Weill, and R. Woodgate. 2001. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 276:43487-43490. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, J. L. 1993. Yeast DNA replication. J. Biol. Chem. 268:25261-25264. [PubMed] [Google Scholar]
- 12.Carson, D. R., and M. F. Christman. 2001. Evidence that replication fork components catalyze establishment of cohesion between sister chromatids. Proc. Natl. Acad. Sci. USA 98:8270-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castano, I., P. Brzoska, B. Sadoff, H. Chen, and M. Christman. 1996. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 10:2564-2576. [DOI] [PubMed] [Google Scholar]
- 14.Castano, I., S. Heath-Pagliuso, B. Sadoff, D. Fitzhugh, and M. Christman. 1996. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 24:2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchnko, M. Mann, and M. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93:1067-1076. [DOI] [PubMed] [Google Scholar]
- 16.Cohen-Fix, O., and D. Koshland. 1997. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc. Natl. Acad. Sci. USA 94:14361-14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dua, R., S. Edwards, D. L. Levy, and J. L. Campbell. 2000. Subunit interactions within the Saccharomyces cerevisiae DNA polymerase epsilon (Pol ɛ) complex-demonstration of a dimeric pol ɛ. J. Biol. Chem. 275:28816-28825. [DOI] [PubMed] [Google Scholar]
- 18.Dua, R., D. Levy, and J. L. Campbell. 1999. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ɛ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274:22283-22288. [DOI] [PubMed] [Google Scholar]
- 19.Dua, R., D. L. Levy, and J. L. Campbell. 1998. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ɛ in DNA replication and the S/M checkpoint pathway. J. Biol. Chem. 273:30046-30055. [DOI] [PubMed] [Google Scholar]
- 20.Dua, R., D. L. Levy, C. Li, P. M. Snow, and J. L. Campbell. 2002. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase {epsilon} in insect cells: purification and characterization. J. Biol. Chem. 277:7889-7896. [DOI] [PubMed] [Google Scholar]
- 21.Fields, S., and O. K. Song. 1989. A novel genetic system to detect protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 22.Gavin, A.-C., M. Bösche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. M. R. J. Schultz, A.-M. Michon, C.-M. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M.-A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular Architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 25.Hamatake, R. K., H. Hasegawa, A. B. Clark, K. Bebenek, T. A. Kunkel, and A. Sugino. 1990. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae: identification of the catalytic core and a possible holoenzyme form of the enzyme. J. Biol. Chem. 265:4072-4083. [PubMed] [Google Scholar]
- 26.Hanna, J. S., E. S. Kroll, V. Lundblad, and F. A. Spencer. 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21:3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. J. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerses δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiloti, Z., Y. S. Chung, Y. Mochizuki, C. F. Hardy, and O. Cohen-Fix. 2001. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol. 11:1347-1352. [DOI] [PubMed] [Google Scholar]
- 29.Hollenberg, S. M., R. Sternblanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 31.James, P., J. Holladay, and E. A. Cragi. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jessberger, R., G. Chui, S. Linn, and B. Kemper. 1996. Analysis of the mammalian recombination protein complex RC-1. Mutat. Res. 350:217-227. [DOI] [PubMed] [Google Scholar]
- 33.Kesti, T., K. Flick, S. Keranen, J. E. Syvaoja, and C. Wittenberg. 1999. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3:679-685. [DOI] [PubMed] [Google Scholar]
- 34.Koshland, D. E., and V. Guacci. 2000. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 12:297-301. [DOI] [PubMed] [Google Scholar]
- 35.Kuzminov, A. 1999. Recombination repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laloraya, S., V. Guacci, and D. Koshland. 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151:1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marini, F., V. Paciotti, G. Lucchini, P. Plevani, D. F. Stern, and M. Foiani. 1997. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 16:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masumoto, H., S. Muramatsu, Y. Kamimura, and H. Araki. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415:651-655. [DOI] [PubMed] [Google Scholar]
- 39.Masumoto, H., A. Sugino, and H. Araki. 2000. Dpb11 controls the association between DNA polymerases alpha and varepsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer, M. L., S. P. Gygi, R. Aebersold, and P. Hieter. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7:959-970. [DOI] [PubMed] [Google Scholar]
- 41.Michaelis, C., R. Ciostk, and K. Nasmyth. 1997. Coheins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 42.Morrison, A., H. Araki, A. B. Clark, R. K. Hamatake, and A. Sugino. 1990. A third essential DNA polymerase in S. cerevisiae. Cell 62:1143-1151. [DOI] [PubMed] [Google Scholar]
- 43.Morrison, A., and A. Sugino. 1993. DNA polymerase II, the ɛ polymerase of Saccharomyces cerevisiae. Prog. Nucleic Acid Res. 46:93-120. [DOI] [PubMed] [Google Scholar]
- 44.Nasmyth, K. 2001. Disseminating the Genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 45.Nasmyth, K., J.-M. Peters, and F. Uhlmann. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379-1384. [DOI] [PubMed] [Google Scholar]
- 46.Navas, T. A., Y. Sanchez, and S. J. Elledge. 1996. RAD9 and DNA polymerase ɛ form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10:2632-2643. [DOI] [PubMed] [Google Scholar]
- 47.Navas, T. A., Z. Zhou, and S. J. Elledge. 1995. DNA polymerase ɛ links the DNA replication machinery to the S phase checkpoint. Cell 80:29-39. [DOI] [PubMed] [Google Scholar]
- 48.Nelson, J., P. Gibbs, A. Nowicka, D. Hinkle, and C. W. Lawrence. 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 37:549-554. [DOI] [PubMed] [Google Scholar]
- 49.Ohya, T., S. Maki, Y. Kawasaki, and A. Sugino. 2000. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase ɛ in Saccharomyces cerevisiae. Nucleic Acids Res. 28:3846-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Reilly, D. R., L. K. Miller, and V. A. Kuckow. 1994. Baculovirus expression vectors: a laboratory manual. Oxford University Press, New York, N.Y.
- 51.Read, R. L., R. G. Martinho, S.-W. Wang, A. M. Carr, and C. J. Norbury. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. USA 99:12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadoff, B., S. Heath-Pagliuso, I. Castano, Y. Zhu, F. Kieff, and M. Christman. 1995. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 141:465-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saitoh, S., A. Chabes, W. H. McDonald, L. Thelander, J. R. Yates, and P. Russell. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109:563-573. [DOI] [PubMed] [Google Scholar]
- 54.Sitney, K. C., and J. L. Campbell. 1990. The yeast DNA replication apparatus—genetic and biochemical analysis, p. 125-146. In E. P. R. Strauss and S. H. Wilson (ed.), The eukaryotic nucleus: molecular biochemistry and macromolecular assemblies, vol. 1. Telford Press, New York, N.Y.
- 55.Sjogren, C., and K. Nasmyth. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11:991-995. [DOI] [PubMed] [Google Scholar]
- 56.Skibbens, R. 2000. Holding your own: establishing sister chromatid cohesion. Genome Res. 10:1664-1671. [DOI] [PubMed] [Google Scholar]
- 57.Skibbens, R., L. Corson, D. Koshland, and P. Hieter. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skibbens, R., C. Rieder, and E. Salmon. 1995. Kinetochore motility after severing between sister centromeres using laser microsurgery—evidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci. 108:2537-2548. [DOI] [PubMed] [Google Scholar]
- 59.Straight, A. F., A. S. Belmont, C. C. Robinett, and A. W. Murray. 1996. GFP tagging of budding yeast chromosome reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599-1608. [DOI] [PubMed] [Google Scholar]
- 60.Strunnikov, A., V. Larionov, and D. Koshland. 1993. Smc1—an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 123:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sundin, O., and A. Varshavsky. 1981. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell 25:659-669. [DOI] [PubMed] [Google Scholar]
- 62.Sundin, O., and A. Varshavsky. 1980. Terminal stages of SV40 DNA replication proceed via multiple intertwined catenated dimers. Cell 21:103-114. [DOI] [PubMed] [Google Scholar]
- 63.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD'2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, R. A. Schleiffe, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 266:1950-1960. [PubMed] [Google Scholar]
- 66.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J. Biol. Chem. 266:1961-1968. [PubMed] [Google Scholar]
- 67.Walowsky, C., D. J. Fitzhugh, I. B. Castano, J. Y. Ju, N. A. Levin, and M. F. Christman. 1999. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J. Biol. Chem. 274:7302-7308. [DOI] [PubMed] [Google Scholar]
- 68.Wang, H., and S. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, Z., I. B. Castano, C. Adams, C. Vu, D. Fitzhugh, and M. F. Christman. 2002. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol sigma DNA polymerase. Genetics 160:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, Z., I. B. Castano, A. D. J. Penas, and C. Adams. 2000. Pol kappa, a DNA polymerase required for sister chromatid cohesion. Science 289:774-779. [DOI] [PubMed] [Google Scholar]
- 71.Warbrick, E., W. Heatherington, D. P. Lane, and D. M. Glover. 1998. PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res. 26:3925-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuzhakov, A., Z. Kelman, and M. O'Donnell. 1999. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96:153-163. [DOI] [PubMed] [Google Scholar]