Abstract
The cyclin-dependent kinase inhibitor p21/WAF1/CIP1 is an important regulator of cell cycle progression, senescence, and differentiation. Genotoxic stress leads to activation of the tumor suppressor p53 and subsequently to induction of p21 expression. Here we show that the tumor suppressor p53 cooperates with the transcription factor Sp1 in the activation of the p21 promoter, whereas histone deacetylase 1 (HDAC1) counteracts p53-induced transcription from the p21 gene. The p53 protein binds directly to the C terminus of Sp1, a domain which was previously shown to be required for the interaction with HDAC1. Induction of p53 in response to DNA-damaging agents resulted in the formation of p53-Sp1 complexes and simultaneous dissociation of HDAC1 from the C terminus of Sp1. Chromatin immunoprecipitation experiments demonstrated the association of HDAC1 with the p21 gene in proliferating cells. Genotoxic stress led to recruitment of p53, reduced binding of HDAC1, and hyperacetylation of core histones at the p21 promoter. Our findings show that the deacetylase HDAC1 acts as an antagonist of the tumor suppressor p53 in the regulation of the cyclin-dependent kinase inhibitor p21 and provide a basis for understanding the function of histone deacetylase inhibitors as antitumor drugs.
The tumor suppressor p53 can induce cell cycle arrest or apoptosis in response to a variety of stress signals, such as DNA damage, oncogenic stimuli, or hypoxia (reviewed in reference 49). Activation of p53 occurs by several mechanisms including protein stabilization and modification of the protein by phosphorylation and acetylation. p53 is a transcription factor that recognizes specific binding sites within numerous target genes including mdm2, cyclin G, bax, and p21/WAF1/CIP1 (for reviews see references 5 and 12). While multiple downstream targets are involved in the mediation of apoptotic effects, the main target for p53-induced cell cycle arrest seems to be the p21 gene.
p21 has been identified by virtue of its activation by p53 (13), its association with cyclin/cyclin-dependent kinase (CDK) complexes (23, 66), and its up-regulation during senescence (47). Furthermore, the p21 protein was shown previously to interact with the proliferating cell nuclear antigen (PCNA), thereby preventing DNA replication (10). Induction of p21 expression by genotoxic stress and its role during terminal differentiation of various cell types have been investigated intensively. While p21 is activated by p53-dependent mechanisms in response to DNA damage to ensure cell cycle arrest and repair, a variety of agents that promote differentiation, like phorbol ester or okadaic acid, can up-regulate p21 independently of p53 (for a review see reference 16). Similarly, the p21 gene can be activated by transforming growth factor β, Ca2+, lovastatin, or nerve growth factor (16).
Recently, a number of reports demonstrated the induction of p21 by inhibitors of histone deacetylases (HDACs), such as sodium butyrate (46), trichostatin A (TSA) (56), suberoylanilide hydroxamic acid (51), oxamflatin (32), MS-27-275 (52), apicidin (22), and trapoxin (54). The transcriptional activation of the p21 gene by these inhibitors is promoted by chromatin remodeling, following acetylation of histones H3 and H4 in the p21 promoter region (32, 54). This activation of p21 occurs in a p53-independent fashion, and therefore HDAC inhibitors are promising agents for cancer therapy, since they are operative in cells with mutated p53 genes, a hallmark of numerous tumors.
The promoter of the human p21 gene harbors six conserved GC boxes, binding sites for the transcription factor Sp1. The Sp1-Sp3 site between −87 and −72 from the transcription start site within the p21 promoter is essential for the activation of p21 by HDAC inhibitors (24, 51, 56). While Sp1 has been shown previously to be implicated in the activation of the p21 gene, studies of the role of the Sp1 homologue Sp3 report divergent results (15, 57, 64, 65). Members of the Sp1 transcription factor family are defined by the presence of three homologous C-terminal zinc finger motifs, enabling interactions with DNA, and are involved in the transcriptional regulation of numerous mammalian genes (59). In addition to its function as a transcriptional activator, Sp1 has been recently shown to act as a repressor by recruiting HDAC1 to the growth-regulated murine thymidine kinase gene (TK) promoter (11).
HDACs form a family of enzymes that catalyze the removal of acetyl moieties from acetylated proteins including histones, structural proteins, or transcription factors (25, 31). Together with their counterparts, the histone acetyltransferases, HDACs regulate the reversible acetylation of core histones and other proteins. Acetylation of histones results in a loosened chromatin structure and improves the accessibility of DNA for different factors, leading to a transcriptionally competent conformation. In addition, acetylation of transcription factors has been shown elsewhere to affect the stability and intracellular localization of proteins or to modulate the affinity for DNA or other proteins (34).
In this report we show that HDAC1 is a crucial regulator of p21 gene expression. The tumor suppressor p53 directly interacts with Sp1 and can compete with the transcriptional repressor HDAC1 for binding to the C terminus of Sp1, leading to histone acetylation and concomitant expression of p21. Activation of p53 efficiently counteracts deacetylase-mediated repression and induces cell cycle arrest by activating the p21 gene.
MATERIALS AND METHODS
Cell culture and transfection.
U2OS cells, Saos-2 cells, and 293 cells were grown in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal calf serum. Drosophila melanogaster SL-2 cells were maintained in Schneider's insect medium. Transient transfection of SL-2 cells and 293 cells was carried out by calcium phosphate coprecipitation as described previously (30).
Plasmid constructs.
Luciferase reporter constructs driven by the human p21 promoter were previously described (WWP-luc [56]). To construct p21-Pst and p21-Pstmt3, the 2.3-kbp PstI fragment (position −2501 to −209 relative to the transcription start site) was cloned into pWP101 to give p21-Pst and into pWP101-mtSp1-3, resulting in p21-Pstmt3. For protein expression in Drosophila SL-2 cells the open reading frames of human p53 and mouse HDAC1 were cloned into pPac. pPac expression vectors encoding Sp1 and Sp3 (20) and plasmids encoding glutathione S-transferase (GST)-Sp1 and the derived deletion constructs (11) were previously described. Vectors encoding GST-p53 fusion proteins or radiolabeled in vitro-translated p53 proteins were obtained by conventional PCR cloning. Specific sequence data are available on request.
Coimmunoprecipitations.
Whole-cell extracts were prepared as described previously (1), and equal amounts (500 μg) were incubated in 200 μl of extraction buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol [DTT], Boehringer's complete protease inhibitor cocktail) with 3 to 5 μl of the respective antibody for 1 h at 4°C. After addition of 20 μl of protein A-Sepharose bead suspension (10% [vol/vol]; Pharmacia), the mixture was further incubated under gentle shaking for 12 h at 4°C. After three washes with extraction buffer, the beads were resuspended in 50 μl of extraction buffer and 30-μl aliquots were examined for protein expression on Western blots. The remaining 20 μl was assayed for HDAC activity (see below).
The following antibodies were used in this study: HDAC1 was visualized on Western blots with a polyclonal rabbit antibody (Upstate Biotechnology). For detection of HDAC1 in Sp1 immunoprecipitates, a mouse monoclonal HDAC1 antibody (Upstate Biotechnology) was used. Hemagglutinin (HA)-tagged proteins were immunoprecipitated and detected with the monoclonal sera for the HA epitope (12CA5 and 16B12). Sp1 was immunoprecipitated and detected with a polyclonal rabbit antiserum raised against the full-length protein (20). Human p21 was detected with the polyclonal H-164 antibody (Santa Cruz), β-actin was visualized with a monoclonal antibody (AC-74; Sigma), and the p53 protein was immunoprecipitated with a polyclonal antibody (FL-393; Santa Cruz) and detected with the monoclonal DO-7 antibody (generous gift of B. Vojtesek).
HDAC assays and luciferase assays.
HDAC assays were done as described previously (7, 38). To measure enzymatic HDAC activity, 20 μl of immunoprecipitated proteins (one-third of a coimmunoprecipitation experiment) was incubated with 10 μl of [3H]acetate-labeled chicken erythrocyte histones in a total volume of 50 μl for 1 h at 30°C. The reaction was stopped by addition of 36 μl of 1 N HCl-0.4 M sodium acetate-800 μl of ethyl acetate. After centrifugation at 8,400 × g for 5 min, a 600-μl aliquot of the organic phase was counted in 3 ml of liquid scintillation cocktail.
For luciferase reporter assays, cells were grown in six-well plates and lysed 48 h after transfection in luciferase lysis buffer (100 mM potassium phosphate [pH 7.8], 0.2% Triton X-100). In mammalian cells luciferase activity and β-galactosidase activity (as a control for transfection efficiency) were assayed in parallel by using the Dual Light chemoluminescent reporter gene assay system (Tropix, Bedford, Mass.). An aliquot of each extract was analyzed on Western blots for the expression levels of coexpressed proteins. Cells were transfected in triplicate, and luciferase activities were shown as mean values with standard deviations.
GST pull-down assays.
Recombinant proteins were expressed in and purified from the Escherichia coli strain BL21 as described previously (11). Beads coated with GST fusion proteins (2 μg) were incubated in binding buffer (20 mM HEPES [pH 7.9], 1 mM MgCl2, 40 mM KCl, 0.1 mM EDTA, 0.1% Nonidet P-40) with 500 μg of whole-cell extract or in vitro-translated proteins or with 2 footprint units of purified human Sp1 (Promega) for 2 h at 4°C. After three washes with GST wash buffer (100 mM KCl, 20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.5 mM DTT), bound proteins were eluted by being boiled in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer, resolved by electrophoresis, and visualized by Western blotting. In vitro expression of radiolabeled proteins was performed in reticulocyte extracts (Quick Coupled transcription-translation system; Promega) in the presence of [35S]methionine. Labeled proteins were resolved by SDS-polyacrylamide gel electrophoresis, and gels were dried and exposed to X-ray films at −70°C. The signals of labeled proteins were quantified with a Molecular Dynamics Storm 840 scanner.
Chromatin immunoprecipitation experiments.
Chromatin immunoprecipitation assays were carried out as described previously (36) with a few modifications. Chromatin of U2OS cells or embryonic stem (ES) cells was cross-linked for 10 min by using formaldehyde. The resulting chromatin solution was diluted 1:10 and precipitated with acetyl-specific histone antibodies (Upstate Biotechnology), monoclonal HDAC1 antibodies (Upstate Biotechnology), or monoclonal DO-7 antibodies (p53). The following day chromatin-antibody complexes were isolated from the solution by incubation with 30 μl of protein A/G-Sepharose beads (50% slurry, 100 μg of salmon sperm DNA/ml, 500 μg of bovine serum albumin/ml) while being rocked at 4°C for 2 h. The beads were harvested and washed as described previously (36). Chromatin-antibody complexes were eluted from the A/G-Sepharose beads by addition of 2% SDS, 0.1 M NaHCO3, and 10 mM DTT to the pellet. Cross-linking was reversed by addition of 0.05 volume of 4 M NaCl and incubation of the eluted samples for 6 h at 65°C. The DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in water. Immunoprecipitated DNA was analyzed for p21 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene sequences by PCR.
PCR analysis of immunoprecipitated DNA.
All PCRs were performed on a Biometra D3 thermocycler with Promega PCR Master Mix. Mouse p21 primer pairs were described previously (36). The primer pairs 5′-CCA GCC CTT GGA TGG TTT-3′ and 5′-AGT CCG TGT CTT TCC TCC G-3′ (human p21 distal) or 5′-GGT GTC TAG GTG CTC CAG GT-3′ and 5′-GCA CTC TCC AGG AGG ACA CA-3′ (human p21 proximal) were used to analyze the human p21 promoter. Primers for the GAPDH promoter had been described in reference 6. The linear range for each primer pair was determined empirically with different amounts of genomic DNA. PCRs with increasing amounts of genomic DNA were carried out along with the immunoprecipitated DNA. PCR products were resolved on 2% agarose-Tris-acetate-EDTA gels and quantified with the ImageQuant program (Molecular Dynamics).
RESULTS
Activation of the p21 gene in U2OS and Saos-2 cells.
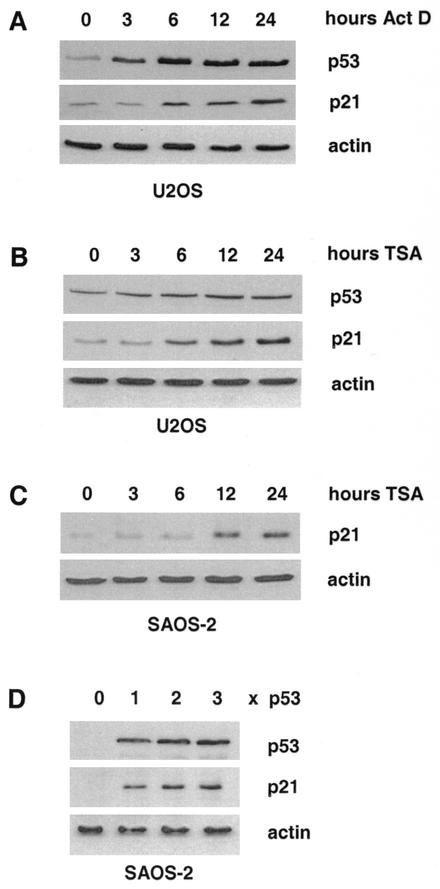
Induction of DNA damage by a variety of agents is known to activate the tumor suppressor p53, which in turn acts as transcriptional regulator of several target genes. One of the main targets is the gene encoding the CDK inhibitor p21. Treatment of the human osteosarcoma cell line U2OS with actinomycin D (30 ng/ml) led to strongly increased p53 protein levels (Fig. 1A, upper panel) and to a slightly delayed rise in p21 protein expression (Fig. 1A, middle panel). Similar changes in p53 and p21 expression were also observed with other stimuli such as UV irradiation (data not shown). In contrast, expression of β-actin was not affected by induction of DNA damage (Fig. 1A, lower panel).
FIG. 1.
Induction of p53 and p21 in response to DNA damage or HDAC inhibitor treatment. (A) Western blot analysis of protein extracts isolated from actinomycin D (30 ng/ml)-treated U2OS cells with antibodies to p53 and p21, respectively. (B) Same as for panel A, but cells were treated with the HDAC inhibitor TSA (50 ng/ml) instead of actinomycin D. (C) Western blot analysis of p21 expression in extracts prepared from Saos-2 cells after treatment with TSA for the indicated periods of time. (D) wt p53 protein was expressed in p53-deficient Saos-2 cells. Levels of ectopically expressed p53 and endogenous p21 were monitored by Western blot analysis. To control equal loading, the Western blots were reprobed with a β-actin antibody.
As depicted in Fig. 1B, p21 protein expression is also significantly enhanced in U2OS cells upon treatment with the HDAC inhibitor TSA, although the p53 protein is only mildly affected and β-actin protein levels do not change at all. In Saos-2 cells that lack endogenous p53 protein, TSA also enhanced p21 protein levels, indicating that the deacetylase inhibitor acts downstream of p53 (Fig. 1C). Expression of wild-type (wt) p53 in Saos-2 cells led to a dose-dependent rise in p21 protein expression without affecting β-actin protein levels. Thus, all regulatory factors required for p21 induction are present in this cell line (Fig. 1D). Taken together, these data suggest that HDACs repress the expression of the CDK inhibitor p21 in cycling cells. Activation of the tumor suppressor p53 or inhibition of HDACs by TSA can overcome this repression.
HDAC1 and p53 are antagonists in the regulation of the p21 promoter.
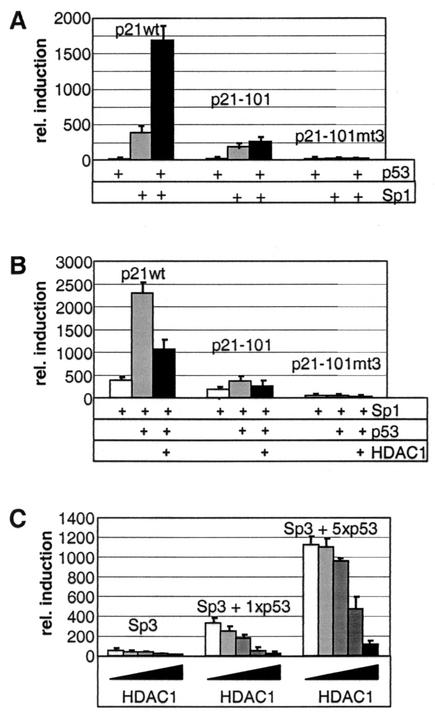
The human p21 promoter contains two p53-binding sites and six Sp1 motifs in the proximal part next to the TATA box (Fig. 2A). To examine the mechanisms regulating the expression of p21 in Saos-2 cells in more detail, we analyzed the effect of HDACs and the transcription factor p53 on p21 promoter activity in transient transfections. Luciferase reporter constructs containing different portions of the human p21 promoter illustrated in Fig. 2A were transfected into Saos-2 cells together with a control vector in the presence or absence of TSA or with a p53 expression vector. Amounts of expressed p53 protein were routinely monitored by Western blot analysis (data not shown). In accordance with the protein data shown in Fig. 1, the wt p21 promoter was strongly induced by both p53 and TSA (Fig. 2B). Deletion of the p53-binding sites (p21-BstX and p21-101) strongly impaired the responsiveness to p53 but had no consequence on the TSA effect. Removal of Sp1-binding sites 1 and 2 had little effect on the promoter activation by p53 or TSA (Fig. 2, p21-Pst).
FIG. 2.
Activation of the human p21 promoter by TSA and p53. (A) Schematic representation of p21 promoter deletion constructs used for luciferase reporter assays. (B) Saos-2 cells were transiently transfected with the indicated constructs together with a β-galactosidase reference vector and with or without a p53 expression plasmid. Where indicated, transfected cells were treated with TSA (50 ng/ml) for 16 h prior to harvesting. Luciferase activity was determined and depicted relative to the β-galactosidase activity.
Mutation of the Sp1-3 site completely abolished the activation of the p21 promoter by TSA in Saos-2 cells (Fig. 2B, p21-Pstmt3 and p21-101mt3). Importantly, the Sp1-3 site mutation in the p21-Pstmt3 construct also had a profound effect on the induction by p53 (Fig. 2B, compare p21-Pst and p21-Pstmt3), suggesting that the Sp1-3 site is important not only for the TSA effect but also for the activation of the p21 promoter by p53. These data are in accordance with previous findings demonstrating that Sp1 cooperates with p53 in the activation of the p21 gene in human hepatoma cells (33).
To investigate this cooperativity of p53 and Sp1 in more detail, we first analyzed the effect of both proteins on the p21 promoter in the Sp1-deficient cell line SL-2. Therefore, we transfected Sp1, p53, Sp1 plus p53, or a control vector together with p21 promoter constructs into SL-2 cells. Comparable expression levels for each protein were again confirmed by Western blot analysis (data not shown). While p53 expression alone had hardly any effect, Sp1 significantly activated the p21 wt promoter (Fig. 3A, p21wt). Coexpression of the two proteins led to synergistic induction of the p21 wt reporter. In accordance with the results obtained in Saos-2 cells, the basal p21 promoter p21-101, lacking the p53-binding sites, was responsive to Sp1 but not to p53 (Fig. 3A). Additional mutation of the Sp1-3 site completely abolished also the response of the reporter to Sp1 (p21-101mt3). Since the Sp1-related transcription factor Sp3 was also implicated in the regulation of the p21 gene (64, 65), we included this protein also in our study. Similar results were obtained in experiments in which Sp1 was replaced by Sp3 (data not shown).
FIG. 3.
The interplay among Sp1-Sp3, HDAC1, and p53 in the regulation of the p21 promoter in SL-2 cells. (A) Activities of different p21 promoter constructs, measured in the presence of p53 alone or Sp1 alone or with the two together. Induction of luciferase activity is shown relative to promoter activity in the absence of Sp1 and p53. (B) The effect of HDAC1 on the luciferase activity of p21 promoter constructs was tested in the presence of Sp1 alone or Sp1 and p53 together. Promoter activity is shown relative to luciferase activity in the absence of mammalian proteins. (C) Luciferase activity of the p21 wt promoter after transfection of Sp3 or Sp3 and two different concentrations of p53 together with increasing amounts of HDAC1. Induction of luciferase activity is shown relative to promoter activity in the absence of Sp3, p53, and HDAC1.
Recently, p21 expression was found to be increased in HDAC1-deficient ES cells, suggesting a role of the deacetylase in the regulation of the p21 gene (36). Therefore, we analyzed next the effect of HDAC1 on p53-mediated activation of the p21 promoter. Coexpression of HDAC1 with Sp1 and p53 led to a significant reduction of p53-dependent transcription from the p21 wt promoter and slightly repressed the activation of the promoter construct p21-101, which lacks the p53-binding sites (Fig. 3B). In a similar approach we tested the effect of increasing amounts of HDAC1 on the activating role of Sp3 on the p21 promoter. Coexpression of Sp3 with two different amounts of p53 led to dose-dependent induction of the p21 promoter (Fig. 3C). Additional expression of increasing amounts of HDAC1 counteracted the p53-mediated activation of the p21 reporter gene. Our data demonstrate a strong synergy between p53 and Sp1-Sp3 in the activation of the p21 gene. Furthermore, we provide evidence that p53 and HDAC1 are antagonists in the regulation of p21 promoter activity.
In vivo association of p53 with Sp1 and Sp3.
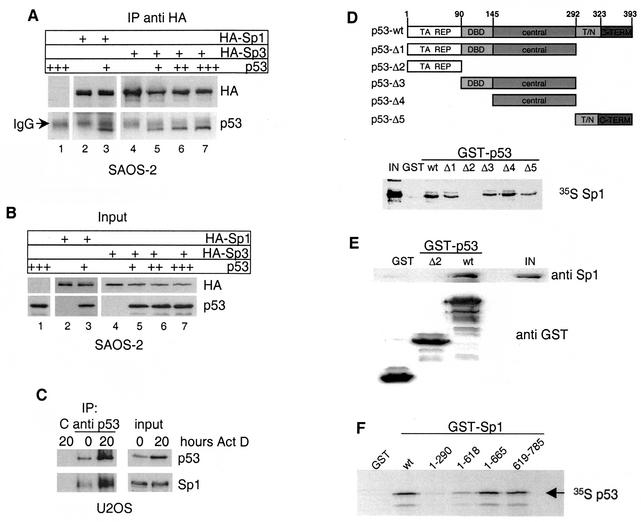
Next we asked whether p53 and Sp1 not only functionally but also physically interact in Saos-2 cells. Therefore, we analyzed the association of p53 with Sp1 or Sp3 in Saos-2 cells by immunoprecipitation experiments. HA-tagged versions of Sp1 or Sp3 (11) were coexpressed with different amounts of p53 in Saos-2 cells and precipitated with an HA-specific monoclonal antibody. The immunoprecipitates were analyzed for the presence of Sp1, Sp3, and p53. As shown in Fig. 4A, p53 was found in the precipitate in the presence of HA-Sp1 (lane 3). Expression of increasing amounts of p53 together with HA-Sp3 led to the dose-dependent coprecipitation of the tumor suppressor with Sp3 (lanes 4 to 7). Expression levels of the transfected proteins are shown in Fig. 4B. These results demonstrate that Sp1 and Sp3 are associated with p53 in transfected Saos-2 cells.
FIG. 4.
In vivo and in vitro interaction of p53 and Sp1-Sp3. (A) Western blot analysis of immunoprecipitates obtained with HA antibody from Saos-2 cells transfected with plasmids encoding either HA-Sp1 or HA-Sp3 alone or together with p53. The blot was subsequently probed with HA antibody to visualize Sp1 and Sp3 and p53 antibody to detect coimmunoprecipitated p53 protein. The signal from the cross-reaction with the heavy chain of HA antibody is indicated (IgG [immunoglobulin G]). IP, immunoprecipitation. (B) Western blot analysis of input protein levels of Sp1, Sp3, and p53. (C) U2OS cells were treated with 30 ng of actinomycin D/ml for 20 h, and p53 was immunoprecipitated before and after actinomycin D treatment. Immobilized 9E10 antibody was used as negative control (C). p53 and Sp1 were detected in immunoprecipitates and input extracts with the respective antibodies. IP, immunoprecipitation. (D) Upper panel: schematic representation of p53 deletion mutants used for GST pull-down experiments. Specific domains are labeled as follows: TA, transactivation; REP, repression; DBD, specific DNA-binding domain; central, central domain; T/N, tetramerization, nuclear localization; C-TERM, C terminus. Lower panel: GST pull-down assay done with the different GST-p53 constructs and in vitro-translated [35S]methionine-labeled Sp1. One hundred percent Sp1 protein input (IN) is shown as control. (E) GST pull-down assay with the indicated GST-p53 fusion constructs and purified Sp1 protein. p53-associated Sp1 and 20% of the input (IN) were detected by Western blot analysis. GST proteins were stained with Coomassie blue. (F) GST pull-down assay with the indicated GST-Sp1 constructs and in vitro-translated [35S]methionine-labeled human p53.
Next, we examined the interaction of endogenous Sp1 and p53 in U2OS cells. Expression of p53 was induced by actinomycin D treatment for 20 h, and p53 protein was immunoprecipitated from untreated and actinomycin D-induced cells. Low levels of p53 were expressed and precipitated in untreated U2OS cells (Fig. 4C). Correspondingly, low amounts of Sp1 were found in the precipitate obtained with the p53-specific antibody, while neither p53 nor Sp1 was precipitated by a control antibody (Fig. 4C). Induction of p53 by actinomycin D led to a significant increase both in precipitated p53 and in coprecipitated Sp1, indicating that induction of DNA damage by actinomycin D results in enhanced Sp1-p53 complex formation.
Direct binding of p53 to Sp1.
The p53 transcription factor contains several distinct functional domains (5) (Fig. 4D, upper panel). To investigate whether the association of p53 with Sp1 is direct, GST fusion proteins containing full-length p53 (wt) or different domains of the protein were incubated with radiolabeled in vitro-translated Sp1. Western blot analysis confirmed that the expression levels of all GST fusion proteins were similar (data not shown). To avoid a potential bridging of the two transcription factors by DNA, the pull-down assays were performed in the presence of 50 μg of ethidium bromide/ml. As shown in Fig. 4D (lower panel), wt p53 showed strong binding to Sp1. A similar in vitro association was observed for Sp3 (data not shown). The p53 deletion mutants Δ1, Δ3, and Δ4, which all contain the central domain, displayed significant interaction with Sp1. The C-terminal fragment encompassing amino acids 292 to 393 (Δ5) showed reduced binding, whereas the N-terminal transactivation domain of p53 (Δ2) did not bind at all to Sp1. These data suggest that the central domain and, to a minor extent, the C terminus of p53 are important for the interaction with Sp1.
To exclude a possible bridging by mammalian proteins present in the reticulocyte lysate, affinity-purified GST-p53 protein was incubated with purified Sp1 protein. The full-length p53 protein bound significant amounts of purified Sp1, whereas the N-terminal transactivation domain of p53 (Δ2) or the GST protein did not interact with Sp1 (Fig. 4E). Thus, p53 interacts directly with the transcription factor Sp1.
The C-terminal domain of Sp1 is crucial for the direct interaction with p53.
Next, we used the GST binding assay to identify the p53 interaction domain within the Sp1 protein. GST fusion proteins containing full-length Sp1 (wt) or different portions of the transcription factor were incubated with radiolabeled in vitro-translated p53. The binding assays were again performed in the presence of 50 μg of ethidium bromide/ml. Full-length Sp1 and the C-terminal fragment (residues 619 to 785) strongly interacted with p53, whereas an N-terminal fragment of Sp1 (residues 1 to 290) did not bind to the tumor suppressor protein (Fig. 4F). Deletion of the region encompassing amino acids 619 to 785 led to significantly reduced interaction, suggesting that the C terminus of Sp1 is crucial for binding to p53. Importantly, the same region (amino acids 619 to 785) was previously identified as an interaction domain of Sp1 required for association with the transcriptional repressor HDAC1 (11).
The tumor suppressor p53 can compete in vivo and in vitro with HDAC1 for binding to Sp1.
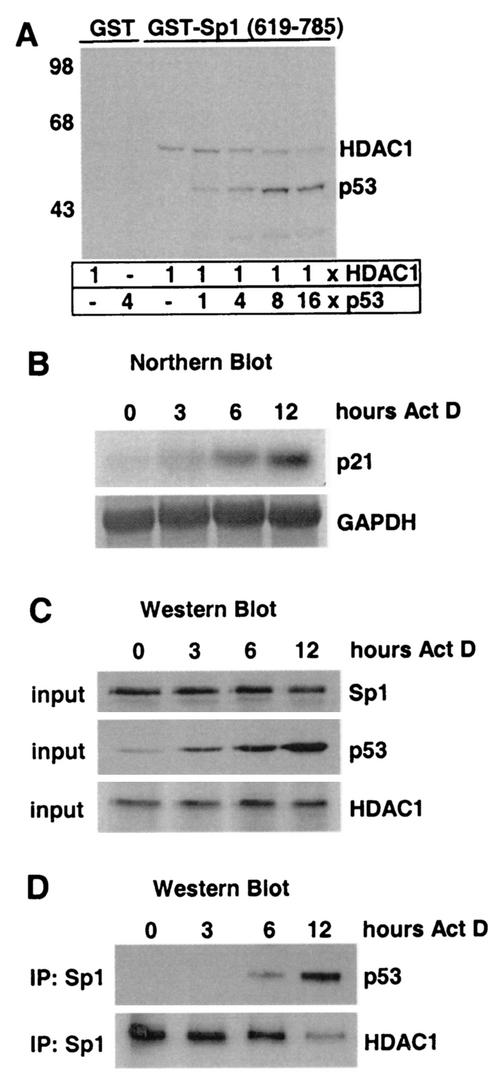
Since p53 and HDAC1 bind to the same portion of the transcriptional regulator Sp1, we analyzed whether p53 could compete for HDAC1 binding to Sp1. For this, we established an in vitro competition assay. GST-Sp1 (residues 619 to 785) was incubated with saturating amounts of radiolabeled HDAC1, and increasing amounts of radiolabeled full-length p53 were added to the binding reaction mixture. As shown in Fig. 5A, elevated binding of p53 to Sp1 resulted in a decrease in Sp1-associated HDAC1. This displacement was specific, since addition of the radiolabeled p53 deletion mutant Δ2, which is unable to bind Sp1 (Fig. 4D), had no effect on the association of HDAC1 with Sp1 (data not shown). These results demonstrate that p53 can compete with HDAC1 for binding to the C terminus of Sp1 in vitro.
FIG. 5.
The p53 protein competes with HDAC1 for binding to Sp1. (A) Autoradiogram of a GST pull-down assay with GST-Sp1 (residues 619 to 785) mixed with a constant amount of radiolabeled 35S-HDAC1 and increasing amounts of 35S-p53. GST protein was used as a control. Numbers at left are molecular masses in kilodaltons. (B) Northern blot analysis of RNA isolated from U2OS cells treated with actinomycin D for the indicated periods of time. The blot was sequentially hybridized with probes for p21 and GAPDH (as a loading control). (C) Western blotting of cell extracts isolated from actinomycin D-treated U2OS cells with antibodies specific for Sp1, p53, and HDAC1. (D) The extracts described for panel C were used for immunoprecipitation (IP) with Sp1 antibodies. Coprecipitated p53 and HDAC1 are detected by Western blot analysis.
We next asked whether Sp1 also interacts with HDAC1 and p53 during the activation of the p21 gene in vivo. To this end we induced p53 expression by actinomycin D treatment of U2OS cells. Cytosolic RNA was isolated at different time points and analyzed on a Northern blot. As shown in Fig. 5B, activation of p53 by actinomycin D led to a significant increase in p21 mRNA levels. Simultaneously with the RNA isolation, protein extracts were prepared and examined on Western blots. p53 was strongly induced in response to actinomycin D, whereas Sp1 and HDAC1 expression was not altered significantly (Fig. 5C). Sp1 was immunoprecipitated, and coprecipitated HDAC1 and p53 proteins were visualized on the same Western blot. Significant amounts of HDAC1 were found to be associated with Sp1 in untreated U2OS cells and in cells after 3 h of actinomycin D treatment (Fig. 5D). After 6 and 12 h of actinomycin D treatment p53 was found to be associated with Sp1. Simultaneously the amounts of Sp1-bound HDAC1 decreased in a p53-dependent manner. Taken together, competition between the transcriptional activator p53 and the repressor HDAC1 for binding to Sp1 occurs in vitro and in vivo.
The recruitment of HDAC1 to the p21 gene leads to reduced expression of the CDK inhibitor.
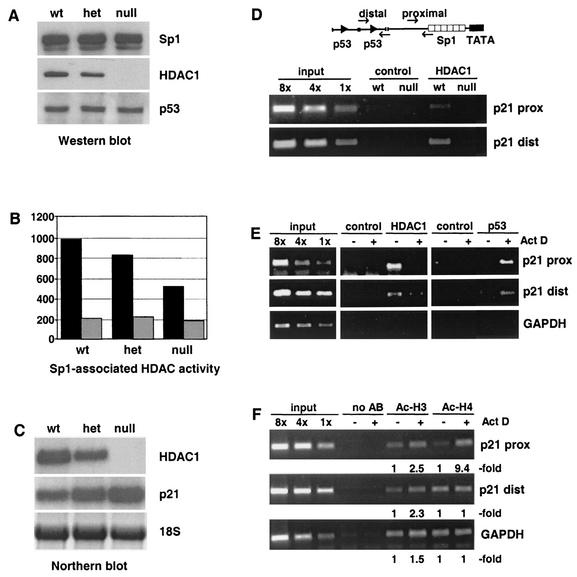
To prove a direct role of HDAC1 for the in vivo repression of the p21 gene, we compared the regulation of p21 expression in the presence and absence of HDAC1. It has recently been shown that loss of HDAC1 in mouse embryos and ES cells results in impaired cell proliferation and up-regulation of a subset of CDK inhibitors (36). To examine the role of HDAC1 for the regulation of p21 expression by Sp1, we first analyzed the protein levels of Sp1, HDAC1, and p53 in ES cells of different HDAC1 genotypes. As expected, HDAC1 expression was reduced in HDAC1-heterozygous cells and completely absent in HDAC1-null cells, whereas levels of Sp1 and p53 were not affected by disruption of one or two HDAC1 alleles (Fig. 6A). Sp1 immunoprecipitates were inspected for associated HDAC activity in the absence and presence of the deacetylase inhibitor TSA (Fig. 6B). In the absence of the deacetylase inhibitor, a significant reduction in Sp1-associated HDAC activity was observed in heterozygous and HDAC1-null ES cells (Fig. 6B, black bars). In the presence of TSA, Sp1-bound deacetylase activity in HDAC1-null cells was further reduced to background levels (Fig. 6B, gray bars), suggesting that in addition to HDAC1 another TSA-sensitive deacetylase is associated with Sp1. Nevertheless, these results indicate that HDAC1 is in part responsible for the Sp1-linked deacetylase activity (see also Discussion).
FIG. 6.
Presence of HDAC1 at the p21 promoter correlates with decreased p21 expression and hypoacetylated core histones. (A) Western blotting performed with whole-cell extracts isolated from HDAC1 wt, heterozygous (het), or deficient (null) ES cells. The blot was probed sequentially with antibodies to Sp1, HDAC1, and p53. (B) In parallel Sp1 and associated proteins were immunoprecipitated with Sp1 antibody from extracts described for panel A and analyzed for the HDAC assay in the absence (black bars) or presence (gray bars) of 200 ng of TSA/ml. The experimental data shown (in counts per minute) are representative of three independent assays. (C) Northern blot analysis with RNA isolated from HDAC1 wt, heterozygous (het), and null ES cell lines. The blot was probed with HDAC1 and p21. Methylene blue staining of 18S rRNA was used to confirm equal loading. (D) Chromatin immunoprecipitation analysis of the p21 promoter for the presence of HDAC1. Relative positions of PCR primers for the amplification of proximal or distal p21 promoter fragments are shown in a schematic drawing. Chromatin isolated from HDAC1 wt ES cells and HDAC1-null ES cells was precipitated with HDAC1-specific monoclonal antibodies or unspecific monoclonal antibodies (control). PCR primers to amplify PCR fragments corresponding to the distal and proximal p21 promoter region were used. Input levels are shown on the left part of the figure (input). (E) Chromatin immunoprecipitation analysis for the presence of p53 and HDAC1 at the p21 promoter. Cross-linked chromatin was isolated from U2OS cells before and after actinomycin D treatment (30 ng/ml for 6 h) and precipitated with specific antibodies for p53 or HDAC1 or unspecific antibodies (control). The human GAPDH gene was used as a control. (F) Chromatin immunoprecipitation performed with antibodies to acetylated histones H3 and H4 by using chromatin isolated from U2OS cells before and after actinomycin D treatment (30 ng/ml for 6 h). Primers for the human GAPDH gene were used as a control. The data shown are representative of three independent experiments. AB, antibody; prox, proximal; dist, distal.
Next we analyzed the expression of p21 mRNA in wt, heterozygous, and HDAC1-deficient ES cells. As shown in Fig. 6C a reduction in HDAC1 levels correlated with increased p21 expression. Previously, it was found that the p21 promoter is associated with hyperacetylated histones in HDAC1-null cells (36). To prove a direct involvement of HDAC1 in the repression of the p21 gene in HDAC1 wt cells, we analyzed the presence of the deacetylase on the p21 promoter by chromatin immunoprecipitation. Formaldehyde cross-linked chromatin from wt and HDAC1-null ES cells was immunoprecipitated with HDAC1-specific monoclonal antibodies. In parallel, chromatin immunoprecipitation assays were performed with unspecific monoclonal antibodies to control the specificity of the reaction. Two regions within the p21 promoter were investigated by PCR: a distal region containing the downstream p53-binding site and a proximal region near the Sp1-binding sites (Fig. 6D). In addition PCRs were carried out with standards comprising fixed amounts of genomic DNA as template to ensure that amplification reactions remained in the linear range. HDAC1 was found to be associated with the distal and the proximal part of the p21 promoter in HDAC1 wt cells. In contrast no p21-specific DNA fragments were immunoprecipitated with HDAC1 antibodies in HDAC1-null cells. Together with the previous finding that the p21 promoter is hyperacetylated in HDAC1-null cells (36), our data indicate that HDAC1 is directly involved in the regulation of p21 gene expression.
We have shown above that actinomycin D-induced activation of p53 leads to replacement of HDAC1 from the C terminus of Sp1 by the tumor suppressor (Fig. 5). In chromatin immunoprecipitation assays p53 was recently shown to bind to the p21 promoter in response to DNA damage (29). Therefore, we asked whether recruitment of p53 would affect the association of HDAC1 with the p21 promoter. Chromatin immunoprecipitation experiments showed that HDAC1 was present at the p21 promoter in logarithmically growing U2OS cells, while p53 was undetectable at both the distal and the proximal part of the promoter (Fig. 6E). Actinomycin D treatment led to nearly complete loss of HDAC1 binding and strongly increased association of p53.
To test whether actinomycin D-induced dissociation of HDAC1 from the p21 promoter affects local histone acetylation patterns, we performed immunoprecipitation experiments with chromatin isolated from U2OS cells before and after actinomycin D treatment. After cross-linking of cells with formaldehyde, sonicated chromatin was immunoprecipitated with antibodies directed against the acetylated isoforms of histone H3 and histone H4. In parallel, chromatin immunoprecipitation assays were performed in the absence of antibodies to control the specificity of the reaction. Primers specific for the human GAPDH gene were used as control. In addition PCRs were carried out with standards comprising fixed amounts of genomic DNA as template to ensure that amplification reactions remained in the linear range. As shown in the lower panel of Fig. 6F, acetylation of histone H3 was slightly increased at both the distal and the proximal part of the p21 promoter in response to actinomycin D treatment. Acetylation of histone H4 at the proximal p21 promoter was strongly induced upon activation of p53, whereas acetylation levels of histone H4 at the distal promoter were relatively high in the absence of actinomycin D and remained unchanged after induction of p53. Acetylation at the GAPDH locus showed no significant changes in response to actinomycin D, and no specific PCR products were amplified in the absence of specific antibodies. Our data demonstrate that, upon induction by actinomycin D, p53 can replace HDAC1 both at the Sp1 C terminus and at the p21 promoter. The replacement of the transcriptional repressor HDAC1 by the transcriptional activator p53 correlates with increased acetylation at the p21 promoter and activation of the p21 gene.
DISCUSSION
The role of p53 in the activation of the p21 gene.
In this report we showed that HDAC1 is recruited to the p21 gene and thereby represses the expression of the CDK inhibitor. Induction of p53 by DNA-damaging agents results in replacement of HDAC1 by the tumor suppressor at the C terminus of the transcription factor Sp1, increased histone acetylation at the p21 promoter, and transcriptional activation of the p21 gene.
Our results point towards a novel function of p53 as direct antagonist of the deacetylase HDAC1. Several previous findings have supported the idea of a link between p53 and chromatin-modifying enzymes. For instance, cooperation with acetyltransferases was shown previously to be crucial for transcriptional activation by p53 (3, 4, 14, 18, 55). On the other hand, interaction of p53 with the Sin3/HDAC1 repressor complex can mediate the negative regulation of a subset of target genes (44).
In addition, the p53 protein itself is a substrate for acetyltransferases and deacetylases. Acetylation of distinct lysine residues in the C terminus of p53 by P/CAF in vitro and by CBP/p300 in vitro and in vivo has been reported elsewhere (6, 27, 39, 53) and was linked previously to p53 stabilization (26, 27), the recruitment of cofactors (6), and increased DNA binding (17, 39, 53). Targeting of p53 by class I HDACs (26, 28, 41) and by class III deacetylases (37, 40, 62) was shown elsewhere to down-regulate p53 function.
The role of p53 acetylation in the regulation of the p21 gene is not completely clarified (discussed in reference 50). CBP/p300-mediated acetylation increased the affinity of p53 for short DNA fragments containing p53-binding sites (17, 39, 53) but had no effect on transcription factor binding to longer DNA fragments or to the chromatinized p21 promoter (14). In in vitro assays acetylation of p53 is not required for efficient transcription of the p21 gene (14). Acetylation-defective p53 mutants were shown elsewhere to bind the p21 promoter in vivo as well as the wt protein but were affected in their ability to recruit coactivator complexes (6). In contrast, Nakamura et al. (45) reported that acetylation of the p53 C terminus is more important for localization and MDM2-mediated degradation than for transcriptional activity. Loss of HDAC1 leads to histone hyperacetylation at the p21 gene (36) but has no significant effect on the acetylation level of p53 (S. Chiocca, unpublished results). These results suggest that the induced p21 expression in HDAC1-null ES cells is rather due to changes in the deacetylase-acetyltransferase balance on the p21 promoter than to changes in p53 acetylation. In agreement with this idea other known p53 target genes such as cyclin G and mdm2 are not affected in their expression levels by loss of HDAC1 (G. Lagger, unpublished data).
Taken together, these data demonstrate that the physical and functional interaction of p53 with acetylating and deacetylating enzymes is part of a complex regulatory network controlling the expression of p53 target genes. Future studies will show if these interactions are affected by naturally occurring p53 mutations in tumor cells.
Sp1 as a mediator of positive and negative signals.
The association of p53 with the transcription factor Sp1 is crucial for the activation of the p21 gene (33; this study) and several other genes (8, 19, 48, 60). Sp1 is considered a transcription factor required for the constitutive or induced expression of a variety of mammalian genes. This idea is supported by the fact that components of the basal transcription machinery and transcriptional activators can interact with Sp1 (59). In accordance with these findings Sp1-binding sites have been shown elsewhere to be crucial for the effect of HDACs, suggesting that these sequences are necessary for the recruitment of acetyltransferases (21, 24, 57, 64). Indeed the acetyltransferase p300 was shown elsewhere to collaborate with Sp1 and Sp3 in the activation of the p21 gene in response to TSA (65).
Recently published data indicate that certain promoters are negatively regulated by the targeting of Sp1 by HDACs (11, 42, 63, 67). In the case of the mouse TK gene, HDAC1 competes with the transactivating factor E2F1 for binding to Sp1 (11, 30). HDAC1-Sp1 complexes repress the TK gene promoter, whereas interaction with E2F1 and release of HDAC leads to activation of the TK promoter. Data presented in this report suggest that Sp1 has a similar function on the p21 promoter. Thus, Sp1 seems to be required for the effect of positive regulators such as p53 and acetyltransferases as well as for negative regulation by HDACs and might function as an anchor protein for acetylating and deacetylating activities on specific mammalian promoters. Interestingly, Sp1-associated proteins such as p53 and E2F1 have been shown previously to be modified by acetyltransferases (34), suggesting that Sp1 might participate not only in the control of the reversible acetylation of core histones but also in the modification of promoter-associated transcription factors.
The regulatory role of the Sp1-related transcription factor Sp3 is disputed (discussed in reference 59). In several studies Sp3 has been shown to act as a positive regulator of p21 promoter activity (15, 57, 64). Our results indicate that Sp3 can interact with HDAC1 (11) and p53 (this study) and might therefore have a dual regulatory function, similar to Sp1. The divergent results obtained in different studies might in part be due to the existence of three Sp3 isoforms and to differences in the experimental systems (59). Recently Sp3 has been shown to be acetylated in mammalian cells, suggesting that the regulatory function of this transcription factor is also modulated by reversible acetylation (9). The fact that Sp3 preferentially associates with the phosphorylated form of HDAC2 (58) indicates that the interaction between Sp1-Sp3 and transcriptional activators and repressors is not only dependent on the abundance of the different factors but also modulated by posttranscriptional modifications.
The role of HDAC1 as repressor of the p21 gene.
Although histone deacetylation is considered a general mechanism for the repression in mammalian cells, only about 2% of all genes are induced upon deacetylase inhibitor treatment (61). Similarly, the expression of just a small subset of genes is affected by the loss of HDAC1 in mouse ES cells (G. Lagger, M. Posch, G. Zupkovitz, and C. Seiser, unpublished observation). One key target consistently found to be induced by deacetylase inhibitors in different cell systems is the p21 gene (43). However, the identity of the involved deacetylase(s) regulating p21 gene expression was not revealed.
The first evidence for an involvement of HDAC1 in the repression of the CDK inhibitor gene came from a recent study of HDAC1 gene disruption in mice (36). HDAC1-null cells displayed increased levels of p21 protein and specific hyperacetylation at the p21 promoter. The proximal p21 promoter region containing the Sp1-binding sites was targeted by HDAC1 and hyperacetylated in the absence of the deacetylase. In addition HDAC1 was also found to be recruited to the distal p21 promoter region. This part of the p21 promoter contains binding sites for a variety of transcriptional regulators including nuclear hormone receptors and C/EBPα (16). Therefore, it is likely that HDAC1 is recruited not only by Sp1 but also by other transcription factors that regulate p21 gene expression.
Our findings indicate that HDAC1 is a crucial component of the regulatory machinery controlling p21 expression. However, we cannot exclude the possibility that other deacetylases are also involved in p21 repression. As shown in Fig. 5, Sp1 protein immunoprecipitated from HDAC1-null cells is still associated with significant deacetylase activity. Since two other members of the class I family, namely, HDAC2 and HDAC3, are up-regulated in HDAC1-null cells (36), it is likely that these deacetylases can in part compensate for the loss of HDAC1. Indeed, HDAC2, but not HDAC3, was recently shown to interact with Sp1 and Sp3 in human breast cancer cells (58).
Depending on the cellular background, activation of the p21 gene by deacetylase inhibitors results in apoptosis, differentiation, or cell cycle arrest. Therefore, HDAC inhibitors such as phenylbutyrate, suberoylanilide hydroxamic acid, and FR901228 are currently in clinical trials as new cancer therapeutics (35, 43). The p21 gene was shown elsewhere to be important for the butyrate-induced cell cycle arrest of colon carcinoma cells (2) and seems be one of the crucial targets for deacetylase inhibitors. Our data indicate that the development of more specific inhibitors for relevant enzymes such as HDAC1 might be a useful strategy in cancer therapy.
Acknowledgments
We thank A. Matejowics for valuable help and C. Hauser for help with the chromatin immunoprecipitation assays, H. Nomura for the p21 reporter constructs, W. Krek for the initial idea and the p53 expression vector, G. Zupkovitz for Northern blot hybridization, and B. Vojtesek for p53 antibodies.
This project was supported by the Austrian FWF (grants P14909-GEN and P13068-GEN to C.S. and grants P10873-GEN and P13031-MOB to E.W.), Austrian National Bank grant 8414 to H.R., and the Herzfelder Stiftung to C.S.
REFERENCES
- 1.Adamczewski, J. P., J. V. Gannon, and T. Hunt. 1993. Simian virus 40 large T antigen associates with cyclin A and p33cdk2. J. Virol. 67:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, S. Y., S. Meng, A. Shei, and R. A. Hodin. 1998. p21(Waf1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. USA 95:6791-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ard, P. G., C. Chatterjee, S. Kunjibettu, L. R. Adside, L. E. Gralinski, and S. B. McMahon. 2002. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol. Cell. Biol. 22:5650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 5.Bargonetti, J., and J. J. Manfredi. 2002. Multiple roles of the tumor suppressor p53. Curr. Opin. Oncol. 14:86-91. [DOI] [PubMed] [Google Scholar]
- 6.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 7.Bartl, S., J. Taplick, G. Lagger, H. Khier, K. Kuchler, and C. Seiser. 1997. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol. 17:5033-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borellini, F., and R. I. Glazer. 1993. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J. Biol. Chem. 268:7923-7928. [PubMed] [Google Scholar]
- 9.Braun, H., R. Koop, A. Ertmer, S. Nacht, and G. Suske. 2001. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 29:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 11.Doetzlhofer, A., H. Rotheneder, G. Lagger, M. Koranda, V. Kurtev, G. Brosch, E. Wintersberger, and C. Seiser. 1999. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19:5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.el-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 14.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 15.Gartel, A. L., E. Goufman, F. Najmabadi, and A. L. Tyner. 2000. Sp1 and Sp3 activate p21 (WAF1/CIP1) gene transcription in the Caco-2 colon adenocarcinoma cell line. Oncogene 19:5182-5188. [DOI] [PubMed] [Google Scholar]
- 16.Gartel, A. L., and A. L. Tyner. 1999. Transcriptional regulation of the p21(WAF1/CIP1) gene. Exp. Cell Res. 246:280-289. [DOI] [PubMed] [Google Scholar]
- 17.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 18.Gu, W., X. L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387:819-823. [DOI] [PubMed] [Google Scholar]
- 19.Gualberto, A., and A. S. Baldwin, Jr. 1995. p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J. Biol. Chem. 270:19680-19683. [DOI] [PubMed] [Google Scholar]
- 20.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, J. W., S. H. Ahn, Y. K. Kim, G. U. Bae, J. W. Yoon, S. Hong, H. Y. Lee, Y. W. Lee, and H. W. Lee. 2001. Activation of p21(WAF1/Cip1) transcription through Sp1 sites by histone deacetylase inhibitor apicidin: involvement of protein kinase C. J. Biol. Chem. 276:42084-42090. [DOI] [PubMed] [Google Scholar]
- 22.Han, J. W., S. H. Ahn, S. H. Park, S. Y. Wang, G. U. Bae, D. W. Seo, H. K. Kwon, S. Hong, H. Y. Lee, Y. W. Lee, and H. W. Lee. 2000. Apicidin, a histone deacetylase inhibitor, inhibits proliferation of tumor cells via induction of p21WAF1/Cip1 and gelsolin. Cancer Res. 60:6068-6074. [PubMed] [Google Scholar]
- 23.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 24.Huang, L., Y. Sowa, T. Sakai, and A. B. Pardee. 2000. Activation of the p21WAF1/CIP1 promoter independent of p53 by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) through the Sp1 sites. Oncogene 19:5712-5719. [DOI] [PubMed] [Google Scholar]
- 25.Hubbert, C., A. Guardiola, R. Shao, Y. Kawaguchi, A. Ito, A. Nixon, M. Yoshida, X. F. Wang, and T. P. Yao. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417:455-458. [DOI] [PubMed] [Google Scholar]
- 26.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 29.Kaeser, M. D., and R. D. Iggo. 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. USA 99:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlseder, J., H. Rotheneder, and E. Wintersberger. 1996. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khochbin, S., and H. Y. Kao. 2001. Histone deacetylase complexes: functional entities or molecular reservoirs. FEBS Lett. 494:141-144. [DOI] [PubMed] [Google Scholar]
- 32.Kim, Y. B., K. H. Lee, K. Sugita, M. Yoshida, and S. Horinouchi. 1999. Oxamflatin is a novel antitumor compound that inhibits mammalian histone deacetylase. Oncogene 18:2461-2470. [DOI] [PubMed] [Google Scholar]
- 33.Koutsodontis, G., I. Tentes, P. Papakosta, A. Moustakas, and D. Kardassis. 2001. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J. Biol. Chem. 276:29116-29125. [DOI] [PubMed] [Google Scholar]
- 34.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer, O. H., M. Gottlicher, and T. Heinzel. 2001. Histone deacetylase as a therapeutic target. Trends Endocrinol. Metab. 12:294-300. [DOI] [PubMed] [Google Scholar]
- 36.Lagger, G., D. O'Carroll, M. Rembold, H. Khier, J. Tischler, G. Weitzer, B. Schuettengruber, C. Hauser, R. Brunmeir, T. Jenuwein, and C. Seiser. 2002. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21:2672-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye, S. Minucci, P. G. Pelicci, and T. Kouzarides. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechner, T., A. Lusser, G. Brosch, A. Eberharter, M. Goralik Schramel, and P. Loidl. 1996. A comparative study of histone deacetylases of plant, fungal and vertebrate cells. Biochim. Biophys. Acta 1296:181-188. [DOI] [PubMed] [Google Scholar]
- 39.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo, J., A. Y. Nikolaev, S.-I. Imai, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2′ promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 41.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53α modulates its effect on cell growth and apoptosis. Nature 408:377-381. [DOI] [PubMed] [Google Scholar]
- 42.Maehara, K., N. Uekawa, and K. Isobe. 2002. Effects of histone acetylation on transcriptional regulation of manganese superoxide dismutase gene. Biochem. Biophys. Res. Commun. 295:187-192. [DOI] [PubMed] [Google Scholar]
- 43.Marks, P. A., R. A. Rifkind, V. M. Richon, and R. Breslow. 2001. Inhibitors of histone deacetylase are potentially effective anticancer agents. Clin. Cancer Res. 7:759-760. [PubMed] [Google Scholar]
- 44.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura, S., J. A. Roth, and T. Mukhopadhyay. 2000. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol. 20:9391-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 47.Noda, A., Y. Ning, S. F. Venable, O. M. Pereira-Smith, and J. R. Smith. 1994. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res. 211:90-98. [DOI] [PubMed] [Google Scholar]
- 48.Ohlsson, C., N. Kley, H. Werner, and D. LeRoith. 1998. p53 regulates insulin-like growth factor-I (Igf-I) receptor expression and Igf-I-induced tyrosine phosphorylation in an osteosarcoma cell line: interaction between p53 and Sp1. Endocrinology 139:1101-1107. [DOI] [PubMed] [Google Scholar]
- 49.Oren, M. 1999. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274:36031-36034. [DOI] [PubMed] [Google Scholar]
- 50.Prives, C., and J. L. Manley. 2001. Why is p53 acetylated? Cell 107:815-818. [DOI] [PubMed] [Google Scholar]
- 51.Richon, V. M., S. Emiliani, E. Verdin, Y. Webb, R. Breslow, R. A. Rifkind, and P. A. Marks. 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. USA 95:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito, A., T. Yamashita, Y. Mariko, Y. Nosaka, K. Tsuchiya, T. Ando, T. Suzuki, T. Tsuruo, and O. Nakanishi. 1999. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc. Natl. Acad. Sci. USA 96:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambucetti, L. C., D. D. Fischer, S. Zabludoff, P. O. Kwon, H. Chamberlin, N. Trogani, H. Xu, and D. Cohen. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J. Biol. Chem. 274:34940-34947. [DOI] [PubMed] [Google Scholar]
- 55.Scolnick, D. M., N. H. Chehab, E. S. Stavridi, M. C. Lien, L. Caruso, E. Moran, S. L. Berger, and T. D. Halazonetis. 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 57:3693-3696. [PubMed] [Google Scholar]
- 56.Sowa, Y., T. Orita, S. Minamikawa, K. Nakano, T. Mizuno, H. Nomura, and T. Sakai. 1997. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem. Biophys. Res. Commun. 241:142-150. [DOI] [PubMed] [Google Scholar]
- 57.Sowa, Y., T. Orita, S. Minamikawa-Hiranabe, T. Mizuno, H. Nomura, and T. Sakai. 1999. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 59:4266-4270. [PubMed] [Google Scholar]
- 58.Sun, J. M., H. Y. Chen, M. Moniwa, D. W. Litchfield, E. Seto, and J. R. Davie. 2002. The transcriptional repressor Sp3 is associated with CK2 phosphorylated histone deacetylase 2. J. Biol. Chem. 277:35783-35786. [DOI] [PubMed] [Google Scholar]
- 59.Suske, G. 1999. The Sp-family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 60.Torgeman, A., N. Mor-Vaknin, E. Zelin, Z. Ben-Aroya, M. Lochelt, R. M. Flugel, and M. Aboud. 2001. Sp1-p53 heterocomplex mediates activation of HTLV-I long terminal repeat by 12-O-tetradecanoylphorbol-13-acetate that is antagonized by protein kinase C. Virology 281:10-20. [DOI] [PubMed] [Google Scholar]
- 61.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 62.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 63.Won, J., J. Yim, and T. K. Kim. 2002. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277:38230-38238. [DOI] [PubMed] [Google Scholar]
- 64.Xiao, H., T. Hasegawa, and K. Isobe. 1999. Both Sp1 and Sp3 are responsible for p21waf1 promoter activity induced by histone deacetylase inhibitor in NIH3T3 cells. J. Cell. Biochem. 73:291-302. [PubMed] [Google Scholar]
- 65.Xiao, H., T. Hasegawa, and K. Isobe. 2000. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 275:1371-1376. [DOI] [PubMed] [Google Scholar]
- 66.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277:33431-33438. [DOI] [PubMed] [Google Scholar]