Abstract
NPAT is an in vivo substrate of cyclin E-Cdk2 kinase and is thought to play a critical role in coordinated transcriptional activation of histone genes during the G1/S-phase transition and in S-phase entry in mammalian cells. Here we show that NPAT transcription is up-regulated at the G1/S-phase boundary in growth-stimulated cells and that the NPAT promoter responds to activation by E2F proteins. We demonstrate that endogenous E2F proteins interact with the promoter of the NPAT gene in vivo and that induced expression of E2F1 stimulates NPAT mRNA expression, supporting the idea that the expression of NPAT is regulated by E2F. Consistently, we find that the E2F sites in the NPAT promoter are required for its activation during the G1/S-phase transition. Moreover, we show that the expression of NPAT accelerates S-phase entry in cells released from quiescence. The inhibition of NPAT expression by small interfering RNA duplexes impedes cell cycle progression and histone gene expression in tissue culture cells. Thus, NPAT is an important E2F target that is required for cell cycle progression in mammalian cells. As NPAT is involved in the regulation of S-phase-specific histone gene transcription, our findings indicate that NPAT links E2F to the activation of S-phase-specific histone gene transcription.
Histone proteins facilitate the packing of eukaryotic DNA into chromatin fibers. Histones are not simply static structural components of the chromosomes; instead, they participate actively in the regulation of multiple cellular processes that involve chromosomal DNA, such as DNA replication, transcription, DNA repair, recombination, and chromosome segregation (22). The bulk biosynthesis of histones is tightly coupled with DNA replication during S phase of the cell cycle. It is known that both transcriptional regulatin and posttranscriptional regulation are involved in the production of the linker histone (H1) and the core histones (H2A, H2B, H3, and H4) in S phase (6, 14, 33, 44). However, the molecular mechanism(s) coordinating the expression of the multiple histone genes remains to be elucidated. Unbalanced or precocious expression of histones results in the loss of chromosomes and thus in the loss of yeast viability (25), underscoring the importance of coordinated histone synthesis in S phase.
We have shown that NPAT is an in vivo substrate of cyclin E-Cdk2, a cyclin-dependent kinase regulating the G1/S-phase transition (7, 21, 55). The protein concentration of NPAT is cell cycle regulated in mammalian cells, and the overexpression of NPAT promotes S-phase entry in transformed cells (55). NPAT protein associates with histone gene clusters in vivo, and the expression of NPAT activates the transcription of multiple histone genes, suggesting that NPAT plays a crucial role in the coordinated transcriptional activation of histone genes at the G1/S-phase transition. Furthermore, the phosphorylation of NPAT by cyclin E-Cdk2 regulates its ability to activate histone gene transcription (24, 56). Therefore, NPAT provides a link between the cell cycle machinery and the regulation of histone gene transcription.
The E2F transcription factors are key regulators of cell proliferation, development, differentiation, and apoptosis (8, 13, 18, 27, 28, 38, 45, 51). For regulating cellular proliferation, they are known to regulate the transcription of a number of genes involved in the G1/S-phase transition and DNA replication in mammalian cells. Functional E2F activity consists of an E2F subunit and a DP subunit. Six E2F proteins (E2F1 to E2F6) and two DP proteins (DP1 and DP2) are present in mammalian cells. All E2F proteins share a conserved DNA binding domain and a domain involved in dimerizing with the DP proteins. E2F1 to E2F5 contain a transactivation domain, and these E2F proteins activate transcription from promoters bearing E2F binding sites in transient transfection assays. It is believed, however, that E2F1 to E2F3 are potent transcriptional activators, while E2F4 and E2F5 function as transcriptional repressors by recruiting the pRB family of proteins (pRB, p107, and p130) to E2F sites in vivo. Unlike the other E2Fs, E2F6 lacks a transcription activation domain and does not interact with the pRB family of proteins. E2F6 may function as a transcriptional repressor, possibly thorough modifying chromatin structure (8, 13, 27, 31, 48). Genetic studies with mice deficient in E2F genes have confirmed that E2F proteins play essential roles in the regulation of the G1/S-phase transition (11, 15, 53).
To elucidate the molecular mechanism regulating NPAT expression and the contribution of NPAT expression to cell proliferation, we investigated the transcriptional regulation of NPAT and the effect of the inhibition of NPAT expression on cell cycle progression. Here we report that NPAT expression is regulated by the E2F transcription factors. In addition, we demonstrate that NPAT accelerates S-phase entry in cells released from growth arrest and that the inhibition of NPAT expression by small interfering RNA (siRNA) impedes progression through various phases of the cell cycle in mammalian cells. Finally, we show that, consistent with the notion that NPAT plays a critical role in histone gene expression, histone mRNA levels are greatly reduced when NPAT expression is inhibited by siRNA.
MATERIALS AND METHODS
Cell culture and cell cycle analysis.
Human WI38, U2OS, and mouse NIH 3T3 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, unless otherwise specified. Pools of early-passage WI38-HAER-E2F1 cells were generated by infection with pBabePuro-HAER-E2F1 (49) and selected in 1.5 μg of puromycin/ml. Distributions of cells in the cell cycle were analyzed by fluorescence-activated cell sorting (FACS) as previously described (55).
Plasmids.
The plasmids used for the expression of E2F proteins were previously described (23). NPAT promoter fragments were amplified by PCR from previously described cosmid clones containing genomic DNA sequences for the ATM and NPAT genes (16). Amplified NPAT promoter fragments were cloned into the pGL3-basic luciferase reporter plasmid (Promega). The nucleotide at the 5′ end of a cDNA clone which contains the most 5′ sequence of NPAT from among the isolated NPAT cDNA clones is designated position +1. pGL-N2750 contains nucleotides −2750 to +22 (which is 44 nucleotides upstream from the initiation codon of the NPAT gene). pGL-N536, pGL-N134, pGL-N86, and pGL-N30 contain nucleotides −536, −134, −86, and −30 to +22 of the NPAT upstream sequence, respectively.
Northern blot analysis.
NIH 3T3 cells were starved in 0.1% serum for 48 h and then stimulated with 20% serum for various times. Total RNA was isolated by using a Qiagen RNeasy kit as described by the manufacturer. Thirty to 40 μg of isolated RNA was separated on a 1.2% formaldehyde-agarose gel and transferred to a Hybond N+ membrane (Amersham). A hybridization probe was prepared from a mouse NPAT expressed sequence tag clone (AI642904). Hybridization was carried out by using Ambion Ultrasensitive Hybridization Buffer as described by the manufacturer.
We infected WI38 cells with a retrovirus (pBabe-ERE2F1) that expresses human E2F1 fused with the ligand binding domain of the estrogen receptor (ER-E2F1) to generate ER-E2F1-expressing cells. For analysis of NPAT mRNA in ER-E2F-expressing cells, ER-E2F-expressing WI38 clones were plated on the day before induction. The ER-E2F1 fusion protein was activated by adding 4-hydroxytamoxifen (OHT) to a final concentration of 300 nM, and samples were harvested 4 h after induction. Cycloheximide was added when needed at a final concentration of 10 μg/ml. Total RNA was isolated by the CsCl ultracentrifugation method (1). Poly(A) RNA was subsequently isolated by using an Amersham poly(A) RNA purification kit as described by the manufacturer. Four to 8 μg of poly(A) RNA was separated on a 1.25% formaldehyde-agarose gel, transferred to a Hybond N+ membrane, and probed as described previously (1).
For analysis of histone mRNAs from siRNA-treated cells (see below), the cells were cotransfected with a plasmid expressing a puromycin resistance gene (pBabePuro). At 24 h after transfection, puromycin (1.5 μg/ml) was added to the culture medium to select for puromycin-resistant (transfected) cells. At 48 h after the addition of puromycin, the cells were harvested, and total RNA was isolated by using an RNeasy kit as described above. Thirty micrograms of isolated RNA was used for analysis as described above. The hybridization probes for histone mRNAs were prepared from PCR-amplified DNA by using clone BAC, which contains histone genes from the human chromosome 6 cluster (56), as the template. The hybridization signals were quantitated by using a phosphorimager (Bio-Rad).
Luciferase reporter assays.
Cells (U2OS or NIH 3T3) were grown in six-well plates and transfected with a luciferase reporter construct and various expression plasmids. For normalization of transfection efficiency, the cells were also cotransfected with a β-galactosidase-expressing plasmid (pCMV-lacZ). Transfections were carried out by using FuGene6 (Roche) according to the manufacturer's protocol. At various times, the cells were lysed with Promega reporter lysis buffer, and the luciferase and β-galactosidase activities of the lysates were measured as suggested by Promega.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation assays were performed and analyzed essentially as described by Frank et al. (10). Briefly, asynchronously growing U2OS cells were cross-linked with 1% formaldehyde for 10 min, the reaction was stopped by the addition of glycine, and the cells were washed in Tris-buffered saline and harvested in sodium dodecyl sulfate buffer. Following centrifugation, the cells were resuspended in immunoprecipitation buffer and sonicated to yield fragments with an average length of 500 to 1,000 bp. The lysate was precleared and then immunoprecipitated at 4°C overnight with polyclonal antibodies specific for E2F1 (sc-193; Santa Cruz) or E2F4 (sc-866) or with an unrelated anti-Flag antibody (F3165; Sigma). To recover immune complexes, blocked protein A beads were added and incubated for 2 h at 4°C, the beads were washed thoroughly, the complexes were eluted, cross-links were reversed, and material was recovered by phenol-chloroform extraction and ethanol precipitation.
DNA was resuspended in 200 μl of water, and 7.5 μl was used for each quantitative PCR with 200 nM primers in 25 μl of SYBR Green reaction mix (Perkin-Elmer). Primers were designed with Primer Express 1.0 software (Applied Biosystems, Foster City, Calif.) under the manufacturer's suggested conditions. The primer pairs used were as follows: for NPAT, CAGGAGCTGTCGCTGTGTTTG and TTGTTACCCTCGGACGTAGCC; for p107, AGGCAGACGGTGGATGACAACAC and TCAGCGTGGGGCTTGTCCTCGAA; and for β-actin, AACTCTCCCTCCTCCTCTTCCTC and GAGCCATAAAAGGCAACTTTCGG. The p107 and β-actin primers were previously described by Takahashi et al. (47).
PCRs were performed by using an ABI Prism 7700 sequence detection system. For each PCR run with SYBR Green I detection, a melting curve analysis was performed to guarantee the specificity in each reaction tube (absence of primer dimers and other nonspecific products). Quantitation was performed and expressed as previously described (10).
Microinjection.
Rat1 cells were plated on coverslips and made quiescent by serum starvation for 48 h. The nuclei of the cells then were microinjected directly with 50 ng of expression plasmid/μl together with 2 μg of rabbit immunoglobulin G (Jackson Laboratories)/μl by using a Zeiss automatic injection system. Bromodeoxyuridine (BrdU) (100 μM) was added immediately after injection; the cells were fixed 12, 14, and16 h after injection; and the proportion of BrdU-positive microinjected cells was determined.
Immunofluorescence staining.
NPAT protein and BrdU were stained as previously described (56), except that images were captured by using a SPOT RT camera (Diagnostic Instrument Inc.) connected to a Zeiss Axiophot Photomicroscope.
RNA interference.
RNA interference was carried out by using two different methods. In the first method, synthetic siRNA duplexes were used as described by Elbashir et al. (9). A synthetic siRNA duplex (NP/KD1) corresponding to the NPAT mRNA sequence 5′-AAGGUUUGCUGGCAGUCAGAG-3′ was used to inhibit NPAT protein expression. A synthetic siRNA duplex (Luc/KD) corresponding to the firefly luciferase mRNA sequence 5′-AACUUACACGCUGAGUACUUCGA-3′ was used as a negative control. The siRNA duplexes were purchased from Dharmacon Research, Inc. (Lafayette, Colo.). U2OS cells were grown on coverslips in six-well plates and transfected with the siRNAs by using Lipofectamine 2000 (Invitrogen) essentially as described by Elbashir et al. (9). To monitor transfected cells, a green fluorescent protein (GFP)-expressing plasmid (pEGFP; 50 ng) was also cotransfected. BrdU (final concentration, 20 μM) was added to the culture medium 1 h before the cells were fixed. Immunofluorescence staining of NPAT protein and BrdU was performed as previously described (56).
In the second method, a DNA vector-based approach (46) was used. A double-stranded oligonucleotide corresponding to the human NPAT mRNA sequence 5′-GGGAGATTCTCACCCTGAGTCC 3′ was cloned into pBS/U6 (kindly provided by Yang Shi) to generate pBS/U6/NPAT. One microgram of pBS/U6 or pBS/U6/NPAT was transfected into U2OS cells by using FuGene6. To monitor transfected cells, 50 ng of pEGFP was also cotransfected. At 24 h after transfection, the cells were trypsinized and plated on coverslips. Treatment with BrdU and immunofluorescence staining were carried out as described above.
FACS analysis.
U2OS cells were transfected with either pBS/U6 or pBS/U6/NPAT as described above. For selection of transfected cells, 100 ng of pCMVCD20 expressing human CD20 was cotransfected. At various times, the cells were harvested and analyzed for cell cycle distribution by FACS as described previously (55).
To calculate the percentage of cells that had moved out of G1 or S phase at 48 to 53 h posttransfection, the following formulae were used: for calculating the percentage of cells that had moved out of G1 phase (percent exited G1), percent exited G1 = (percent G1 cells at 48 h − percent G1 cells at 53 h)/percent G1 cells at 48 h; for calculating the percentage of cells that had moved out of S phase (percent exited S), percent exited S = [(percent G1 cells at 48 h − percent G1 cells at 53 h + percent S cells at 48 h) − percent S cells at 53 h]/percent G1 cells at 48 h − percent G1 cells at 53 h + percent S cells at 48 h).
RESULTS
NPAT expression is transcriptionally up-regulated at the G1/S-phase boundary in cells stimulated to enter the cell cycle from quiescence.
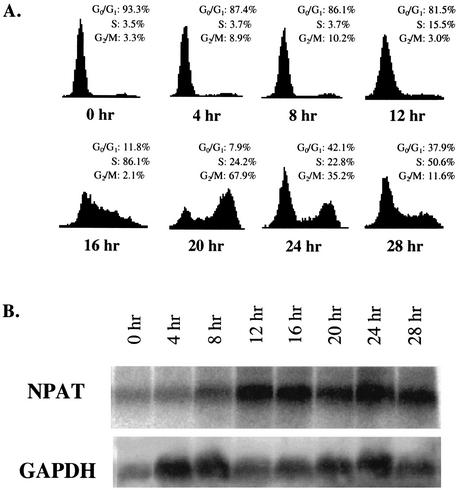
Zhao et al. previously showed that the NPAT protein concentration is low in early G1 phase and reaches the maximum level at the G1/S-phase boundary (55). We have also observed that the NPAT protein concentration diminishes in serum-starved WI38 cells and increases as cells reenter the cell cycle (56) (data not shown). These results indicate that NPAT expression is regulated during cell cycle progression. To determine whether the expression of NPAT is regulated at the transcriptional level, we monitored the expression of NPAT mRNA in NIH 3T3 cells after release from serum starvation. Figure1A shows that, as expected, serum starvation arrested NIH 3T3 cells at G0 phase, and the addition of serum to the medium stimulated the cells to reenter the cell cycle synchronously. The stimulated cells started to enter S phase at about 12 h after serum stimulation, and most cells reached S phase by 16 h after stimulation. NPAT mRNA levels were low in serum-starved and early-G1-phase NIH 3T3 cells. NPAT mRNA reached the maximum level at the G1/S-phase boundary (Fig. 1B). NPAT mRNA peaked at about the same time as cyclin E1 mRNA, which is known to be up-regulated during the G1/S-phase transition (2, 12, 32). Similarly, we have also observed the up-regulation of NPAT mRNA at the G1/S-phase boundary in human IMR90 cells released from serum starvation (data not shown). Consistent with our data, a recent cDNA microarray analysis of gene expression during the cell division cycle of human HeLa cells showed that NPAT transcription is up-regulated at the G1/S-phase boundary (52). Together, these results show that NPAT expression is regulated at the mRNA level during growth stimulation as well as during the cell division cycle.
FIG. 1.
NPAT is transcriptionally up-regulated in NIH 3T3 cells following growth stimulation. (A) Cell cycle distribution profiles for NIH 3T3 cells during quiescence and serum stimulation. NIH 3T3 cells were serum starved and restimulated to enter the cell cycle as described in Materials and Methods. At the indicated times after serum addition, the cell cycle distribution of the cells was analyzed by FACS. (B) Analysis of NPAT mRNA expression in NIH 3T3 cells treated as described for panel A. NPAT mRNA was analyzed by Northern blotting as described in Materials and Methods. The analysis of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in the same samples as a control is also shown.
Activation of the NPAT promoter by E2F.
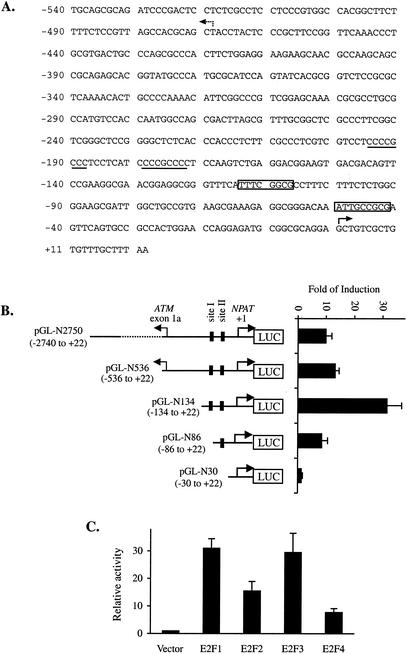
The human NPAT gene is localized on chromosome 11q22-q23. The 5′ end of the human NPAT gene is about 0.5 kb away from that of the ATM gene, and the two genes share a bidirectional promoter (3, 17). There are two putative E2F recognition sites in close proximity to the NPAT gene. The NPAT promoter also contains potential SP1 binding sites next to the putative E2F sites, and the promoter is TATA less (17) (Fig. 2A), a feature reminiscent of many known E2F-responsive genes (20). These observations, together with the finding that NPAT expression is regulated at the transcriptional level during growth stimulation and cell cycle progression, prompted us to examine whether the NPAT promoter is regulated by E2F. We cloned various DNA fragments from the 5′ end of the NPAT gene into a luciferase reporter plasmid and tested their activation by E2F1 in transient transfection assays (Fig. 2B). Several NPAT promoter constructs were strongly activated by E2F1. Deletion of nucleotides 5′ to the putative E2F binding sites in the NPAT promoter did not impede the activation of the NPAT promoter by E2F1. In contrast, a deletion that removes one putative E2F binding site (pGL-N86) significantly impaired the activation of the promoter by E2F1, and deletion of both putative E2F binding sites (pGL-N30) abolished the activation of the promoter by E2F1 (Fig. 2B). These results suggest that NPAT transcription is regulated by E2F, presumably through the E2F recognition sites in the promoter.
FIG. 2.
Activation of the NPAT promoter by E2F. (A) Sequence of the human NPAT promoter. The DNA sequence between the human ATM and NPAT genes is shown. The two putative E2F recognition sequences are boxed. Two potential SP1 sites are underlined. The +1 position for NPAT is indicated by the solid arrow. The broken arrow indicates the start site for the ATM gene (41). (B) Activation of the NPAT promoter by E2F1. A schematic representation of the NPAT promote-luciferase (LUC) reporter constructs is shown on the left. The nucleotide positions of the NPAT promoter fragments are indicated in parentheses. Site I and site II indicate the two E2F sites. U2OS cells were transfected with 1 μg of pCMV (vector) or 1 μg of pCMV-E2F1 together with 50 ng of the indicated NPAT promoter-luciferase reporter construct. For normalization of transfection efficiency among the different samples, 50 ng of pCMV-lacZ was also cotransfected. At 24 h after transfection, the cells were lysed and the activities of luciferase and β-galactosidase were assayed as described in Materials and Methods. Fold induction was calculated by comparing the normalized luciferase activity from E2F1-transfected cells with that from vector-transfected cells. The means and standard deviations from at least three independent experiments are shown. (C) Regulation of the NPAT promoter by E2F proteins. U2OS cells were transfected with 1 μg of pCMV (Vector) or 1 μg of the indicated E2F-expressing plasmid together with 50 ng of pGL-N134 and 50 ng of pCMV-lacZ. The samples were analyzed as described for panel B. The normalized luciferase activity from the cells transfected with the vector was set as 1.
To test whether the NPAT promoter can be regulated by other E2F proteins in addition to E2F1, we cotransfected the NPAT promoter-luciferase reporter (pGL-N134) with plasmids that express E2F1, E2F2, E2F3, or E2F4. As shown in Fig. 2C, the overexpression of E2F2, E2F3, and E2F4 can also activate the NPAT promoter. Thus, the NPAT promoter may respond to regulation by multiple E2F proteins.
The E2F recognition sites are required for the activation of the NPAT promoter by E2F proteins.
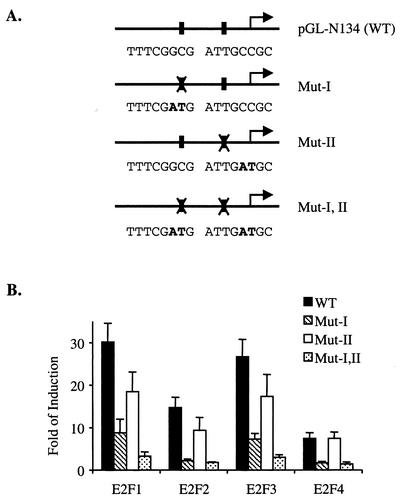
The results presented in Fig. 2B suggest that the putative E2F binding sites in the NPAT promoter mediate activation by E2F. To test directly the contribution of each of the two E2F sites to the activation of the NPAT promoter by the E2F proteins, we generated point mutations in these two E2F sites, either individually or in combination (Fig. 3A), and tested the activation of the mutant NPAT promoter by the overexpression of the E2F1, E2F2, E2F3, and E2F4 proteins. As shown in Fig. 3B, mutation of either one of the E2F sites partially decreases the activation of the NPAT promoter by E2F1, E2F2, and E2F3. Simultaneous mutation of both E2F sites almost completely abolishes the activation of the NPAT promoter by these E2F proteins, indicating that both E2F sites contribute to the regulation of the NPAT promoter by these three E2F proteins. Unlike the activation by E2F1, E2F2, and E2F3 overexpression, the activation of the NPAT promoter by E2F4 overexpression is abolished by mutation at site I, while mutation at site II had no effect on the activation of the NPAT promoter by E2F4 overexpression; these results suggest that E2F4 regulates NPAT expression through one E2F site (site I). In conclusion, these results show that E2F regulates the NPAT promoter directly through the E2F recognition sites in the promoter.
FIG. 3.
E2F recognition sites are required for activation of the NPAT promoter by E2F proteins. (A) Nucleotide sequences of E2F recognition sites in wild-type (WT) and mutant (Mut) NPAT promoters. The mutated nucleotides are shown in bold type. (B) Responses of wild-type and mutant NPAT promoters to activation by different E2F proteins. U2OS cells were transfected with the vector or the indicated E2F-expressing plasmid together with the indicated luciferase reporter constructs. The samples were analyzed as described in the legend to Fig. 2B.
Endogenous E2F proteins bind to the NPAT promoter in vivo, and the induction of E2F1 activates the expression of endogenous NPAT mRNA.
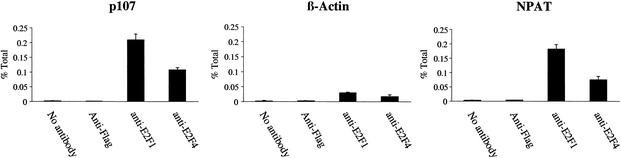
Having shown that the NPAT promoter can be activated by multiple E2F proteins in transient transfection assays, we sought to determine whether endogenous NPAT mRNA expression is regulated by E2F. We first examined whether endogenous E2F proteins bind to the NPAT promoter in vivo. For this purpose, we carried out chromatin immunoprecipitation assays by using antibodies specific for E2F1 and E2F4. To ensure that the E2F1 and E2F4 antibodies that we used can specifically immunoprecipitate the DNA fragments known to be bound by E2F1 and E2F4 in vivo (47), we tested the immunoprecipitation of p107 promoter sequences by the E2F1 and E2F4 antibodies. Both E2F antibodies precipitated promoter fragments from the p107 gene in U2OS cells (Fig. 4), but these two antibodies precipitated negligible amounts of promoter fragments from the actin gene, which is not regulated by E2F. These results demonstrate that the E2F1 and E2F4 antibodies that we used can detect specific interactions of the E2F1 and E2F4 proteins with the promoters regulated by these two proteins. Similar to the results obtained with the p107 promoter, the E2F1 and E2F4 antibodies also immunoprecipitated a DNA fragment from the NPAT promoter, while the control antibody failed to precipitate the same NPAT promoter fragment (Fig. 4). These results show that endogenous E2F proteins, at least E2F1 and E2F4, interact with the NPAT promoter in vivo, supporting the idea that NPAT expression is regulated by E2F in vivo.
FIG. 4.
E2F proteins bind the NPAT promoter in vivo. Chromatin immunoprecipitation analysis was performed with U2OS cell extracts by using the indicated antibodies. Immunoprecipitated promoter fragments were analyzed by quantitative PCR to determine the relative presence of E2F1 and E2F4 on the indicated promoters as described in Materials and Methods. All results are representative of at least two independent immunoprecipitations and are reported as means and standard deviations.
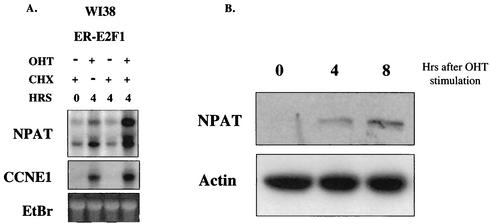
To examine directly whether the expression of E2F activates endogenous NPAT mRNA expression, we infected WI38 cells with a retrovirus (pBabe-ERE2F1) that expresses ER-E2F1 and selected clones that express ER-E2F proteins. Vigo et al. previously showed that ER-E2F fusion proteins are localized in the cytoplasm and are inactive in the absence of OHT. Upon stimulation by OHT, these proteins enter the nucleus and are activated (49). In addition, the expression of many previously characterized E2F target genes was demonstrated to be activated by these ER-E2F proteins (27, 49). As previously reported (26, 49), we found that the ER-E2F1 protein was localized in the cytoplasm in the absence of OHT and entered the nucleus rapidly when cells were treated with OHT (data not shown). The activation of ER-E2F1 significantly increased NPAT mRNA expression, even in the presence of the protein synthesis inhibitor cycloheximide (Fig. 5A). We also observed the induction of NPAT mRNA expression by activated ER-E2F2 and ER-E2F3 proteins (data not shown). These results suggest that NPAT transcription is regulated by E2F in vivo. Consistent with previous reports (5, 17), while a single band was detected for NPAT mRNA in mouse cells (Fig. 1B), two NPAT mRNA bands were observed in human cells (Fig. 5A). The origin of the two NAPT mRNA-specific bands detected in human cells remains to be determined. In addition to up-regulation at the mRNA level, the NPAT protein was also up-regulated upon activation of ER-E2F1 (Fig. 5B), most likely as a result of the increased NPAT mRNA level. Thus, the activation of E2F1 not only can induce the transcription of NPAT but also results in an increase in the NPAT protein concentration.
FIG. 5.
Activation of E2F1 results in increased expression of NPAT. (A) E2F1 up-regulates NPAT mRNA expression. ER-E2F1-expressing WI38 cells were treated with OHT and cycloheximide (CHX) or not treated for 4 h as indicated. mRNA was isolated and analyzed by Northern blotting. As previously reported (17), two NPAT mRNA bands were detected in human cells. As a positive control, the cyclin E1 (CCNE1) mRNA in the same samples was also analyzed. EtBr, ethidium bromide staining of the 28S RNA, used as a loading control. (B) Western blot analysis of NPAT protein in ER-E2F1-expressing cells following OHT treatment. The NPAT protein was detected by using a mouse monoclonal anti-NPAT antibody (56). The analysis of the actin protein, serving as a loading control, is also shown.
The E2F sites mediate NPAT promoter activation at the G1/S-phase transition.
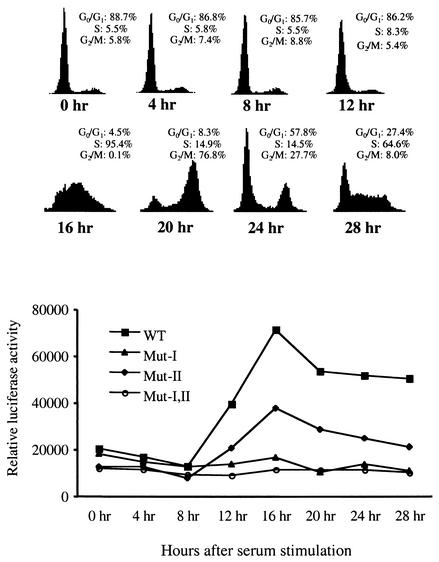
Given that NPAT mRNA levels reach a maximum at the G1/S-phase boundary after cells are stimulated to enter the cell cycle (Fig. 1) and that the overexpression of E2F activates the transcription of NPAT (Fig. 2 and 5A), it is possible that E2F regulates the expression of NPAT via the E2F sites in its promoter during growth stimulation. To test this possibility, we examined the activities of the wild-type and E2F site-mutated NPAT promoters in serum-starved cells as well as in cells stimulated to reenter the cell cycle. The NPAT promoter activity was low in serum-starved (G0) and early-G1-phase cells, and the promoter activity was markedly up-regulated when cells reached late G1/early S phase (Fig. 6). This observation is consistent with our analysis of endogenous NPAT mRNA expression in NIH 3T3 cells during serum stimulation (Fig. 1). Mutation of the 3′ E2F site (site II) reduced the activation of the promoter, while mutation of the 5′ E2F site (site I) completely abolished the activation of the promoter by serum stimulation. These results show that both E2F binding sites in the NPAT promoter contribute to the activation of the NPAT promoter at the G1/S-phase boundary during growth stimulation and that the 5′ E2F site (site I) appears to play a more important role than the 3′ E2F site (site II) in the up-regulation of NPAT expression during growth stimulation.
FIG. 6.
Activation of the NPAT promoter at the G1/S-phase boundary during growth stimulation depends on E2F recognition sites. NIH 3T3 cells were grown in six-well plates and transfected with 300 ng of wild-type (WT) or mutant (Mut) NPAT promoter constructs as indicated. For normalization of transfection efficiency, 200 ng of pCMV-lacZ was also cotransfected. At 18 h after transfection, the cells were washed and cultured in 0.1% serum. Cell proliferation was induced with 20% serum 48 h after serum starvation. At the indicated times, the cells were harvested and assayed for cell cycle distribution (top) and luciferase and β-galactosidase activities. The results shown are the means for triplicate samples. Similar results were also obtained in another independent experiment (data not shown).
The expression of NPAT accelerates S-phase entry in serum-stimulated cells.
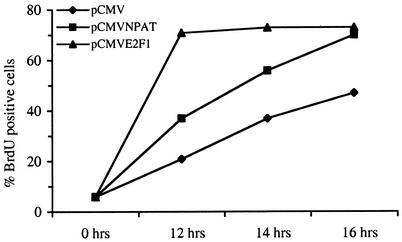
E2F1, E2F2, and E2F3 are potent transcriptional activators of E2F-responsive genes, such as Cdc6, cyclin E1, and Cdc25A, that are required for S-phase entry (8, 13, 27, 48). The overexpression of any of these E2F proteins is sufficient to induce quiescent cells to enter S phase (19, 23, 37). In addition, the overexpression of some of the E2F target genes has been shown to promote S-phase entry in cells released from quiescence (39, 42, 49). As the overexpression of NPAT promotes S-phase entry in asynchronously growing U2OS cells (55), it is possible that the expression of NPAT will accelerate S-phase entry in cells released from growth arrest. To test this possibility directly, we microinjected an NPAT-expressing plasmid into serum-starved Rat1 cells, released the cells from arrest by adding serum, and monitored the progression of the injected cells into S phase by measuring BrdU incorporation. As a positive control for this experiment, we also microinjected an E2F1-expressing plasmid. As shown in Fig. 7, cells injected with an NPAT-expressing plasmid incorporated BrdU at a higher rate than cells injected with the control vector at 12, 14, and 16 h after serum addition. Hence, the expression of NPAT accelerates the G1/S-phase transition in cells reentering the cell division cycle. The kinetics of S-phase entry in NPAT-injected cells are slower than those in E2F1-injected cells, consistent with reports that E2F regulates a variety of genes whose functions are critical for S-phase entry (8, 13, 45, 48).
FIG. 7.
NPAT accelerates S-phase entry in Rat1 cells released from quiescence. Serum-starved, quiescent Rat1 cells were injected with plasmid pCMV expressing E2F1 or NPAT as indicated. Cells injected with empty vector pCMV were used as a negative control. Serum was added, and at 12, 14, and 16 h DNA synthesis was assessed by measuring BrdU incorporation. Shown here are representative results from three independent experiments in which more than 200 cells were microinjected.
NPAT function is required for cell cycle progression.
Our studies have shown that NPAT is maximally expressed at the G1/S-phase boundary and that the overexpression of NPAT accelerates S-phase entry in asynchronously growing cells as well as in cells entering the cell cycle from quiescence (55) (Fig. 1 and 7). These results, together with the observation that mouse embryos deficient for NPAT fail to develop beyond the eight-cell stage (5), are consistent with the idea that NPAT plays a critical role in cell cycle progression. However, it remains to be determined whether NPAT function is essential for cell cycle progression in somatic cells. Recently, it was shown that siRNA consisting of duplexes of 21 to 23 nucleotides can induce highly efficient and gene-specific inhibition of the expression of a specific protein in mammalian cells (4, 9, 35, 36). To address whether NPAT is required for cell cycle progression, we used RNA interference assays with siRNA duplexes to inhibit the expression of NPAT protein in cultured human cells and examined the effect of the inhibition of NPAT expression on DNA synthesis in asynchronously growing U2OS cells.
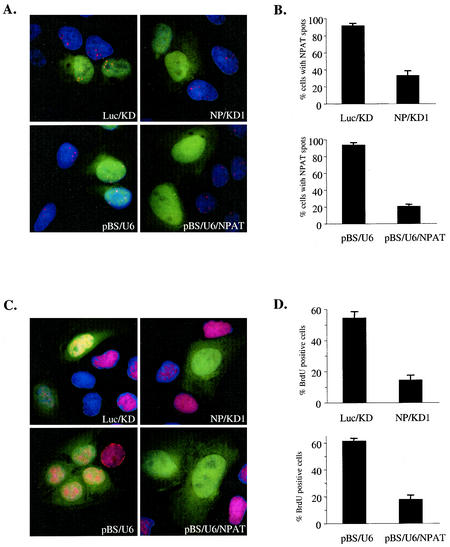
It was previously shown that NPAT protein concentrates at a few easily detectable nuclear spots that are associated with histone gene clusters. There are generally five to eight such NPAT spots in interphase U2OS cells (56). Therefore, we used immunofluorescence staining to assess the effect of siRNA on NPAT expression. Transfection of an siRNA targeting human NPAT mRNA (NP/KD1) but not a control siRNA targeting firefly luciferase mRNA (Luc/KD) into U2OS cells caused a dramatic decrease in NPAT protein levels, as judged by the disappearance of the NPAT spots (Fig. 8A, top panels). siRNA was highly efficient in eliminating NPAT protein, as the NPAT staining (spots) disappeared in most of the transfected cells (Fig. 8B, top panel).
FIG. 8.
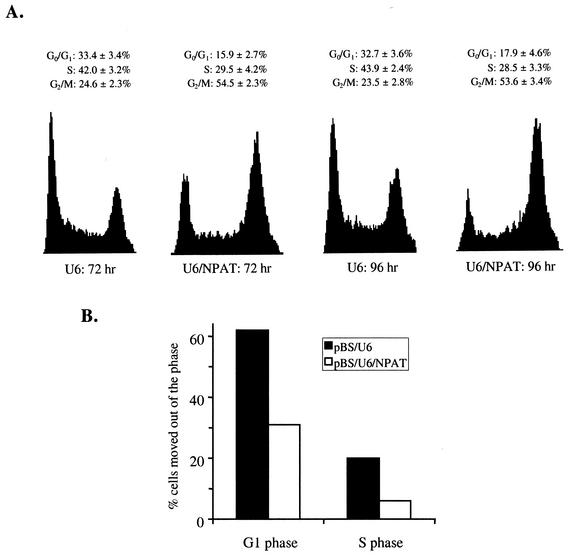
NPAT is required for cell cycle progression. (A) Inhibition of NPAT expression by siRNAs targeting NPAT. U2OS cells were transfected with the indicated synthetic siRNA (Luc/KD or NP/KD) or plasmid (pBS/U6 or pBS/U6/NPAT), together with a GFP-expressing plasmid to label the transfected cells (green). At 60 h (for Luc/KD and NP/KD) or 72 h (for pBSU6 or pBS/U6/NPAT) after transfection, the cells were fixed and stained with an anti-NPAT antibody (red) to assess the presence or absence of NPAT spots. The nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue). (B) Analysis of the inhibition of NPAT expression by siRNAs. U2OS cells were transfected and stained as described for panel A. GFP-positive (transfected) cells were examined for the presence or absence of NPAT spots. Cells with more than two detectable NPAT spots were scored as NPAT spot-positive cells. Shown are the means and standard deviations from three independent experiments. More than 200 GFP-positive cells were analyzed in each experiment. (C) Inhibition of BrdU incorporation by siRNAs targeting NPAT. U2OS cells were transfected as described for panel A. BrdU was added to the culture medium and incubated for 1 h. Cells were then fixed and stained with an anti-BrdU antibody (red). The nuclei were stained with DAPI. (D) Analysis of the effects of siRNAs on BrdU incorporation. U2OS cells were treated as described for panel C, and GFP-positive cells were analyzed for BrdU staining. The data represent the means and standard deviations from three independent experiments. More than 200 GFP-positive cells were analyzed in each experiment.
To provide additional evidence that the decrease in NPAT protein levels caused by siRNA results from specific RNA interference rather than from nonspecific effects of the chemically synthesized RNA duplexes, we carried out RNA interference assays with a DNA vector-based approach in which an siRNA duplex is expressed from a DNA vector in vivo (46). We constructed a plasmid (pBS/U6/NPAT) that carries the sequence for an siRNA duplex targeting a human NPAT sequence different from that targeted by the above-discussed synthetic siRNA. Similar to the transfection of the synthetic siRNA, the transfection of this plasmid (pBS/U6/NPAT) efficiently inhibited the expression of NPAT protein in U2OS cells, while the control vector (pBS/U6) had no effect on NPAT protein expression (Fig. 8A and B, lower panels). Thus, NPAT expression was specifically inhibited by siRNAs targeting NPAT sequences.
To examine the effect of the inhibition of NPAT expression on cell cycle progression, we monitored the incorporation of BrdU in U2OS cells transfected with either synthetic siRNA duplexes or plasmid pBS/U6/NPAT. As shown in Fig. 8C and D, both chemically synthesized siRNA and pBS/U6/NPAT that specifically targeted NPAT sequences resulted in dramatic decreases in BrdU incorporation in transfected cells, while the control siRNA and control vector pBS/U6 had no effect on BrdU incorporation. These results demonstrate that NPAT function is essential for cell cycle progression.
The inhibition of DNA synthesis by the NPAT-specific siRNAs could result from a block of S-phase entry or from an inhibition of progression through other phases of the cell cycle. In order to distinguish between these possibilities, we examined the cell cycle distribution of cells treated with NPAT-specific siRNA by FACS analysis. The siRNA-treated cells were distributed over all phases of the cell cycle, with an accumulation in G2/M phase (Fig. 9A). These results, together with the observation that BrdU incorporation was inhibited in siRNA-treated cells (Fig. 8C and D), suggest that NPAT function is required for transition through multiple phases of the cell cycle, including G2/M-phase progression. To test directly whether progression through the G1 and S phases was inhibited by NPAT-specific siRNA, we compared the rates of G1- and S-phase progression in siRNA-treated cells with those in control cells. pBS/U6/NPAT- or control vector pBS/U6-transfected cells were treated with nocodazole to block the reentry of the cells into G1 phase (55). Subsequently, the fractions of cells passing through G1 and S phases during a certain period of time were measured. As shown in Table1 and Fig. 9B, progression through both G1 and S phases was considerably slower in siRNA-treated cells than in control cells. Thus, it appears that NPAT plays a role in cell cycle progression at multiple control points.
FIG. 9.
Effects of inhibition of NPAT expression on cell cycle progression. (A) U2OS cells were transfected with control vector pBS/U6 or pBS/U6/NPAT. At the indicated time posttransfection, cells were harvested and the cell cycle distributions of the transfected cells were analyzed by FACS as described in Materials and Methods. The data represent the results from four independent experiments. (B) Inhibition of NPAT expression attenuates G1- and S-phase progression. The percentages of transfected cells that had moved out of G1 or S phase at 48 to 53 h posttransfection were calculated from the data presented in Table 1 as described in Materials and Methods.
Histone gene expression is inhibited in cells treated with NPAT-specific siRNA.
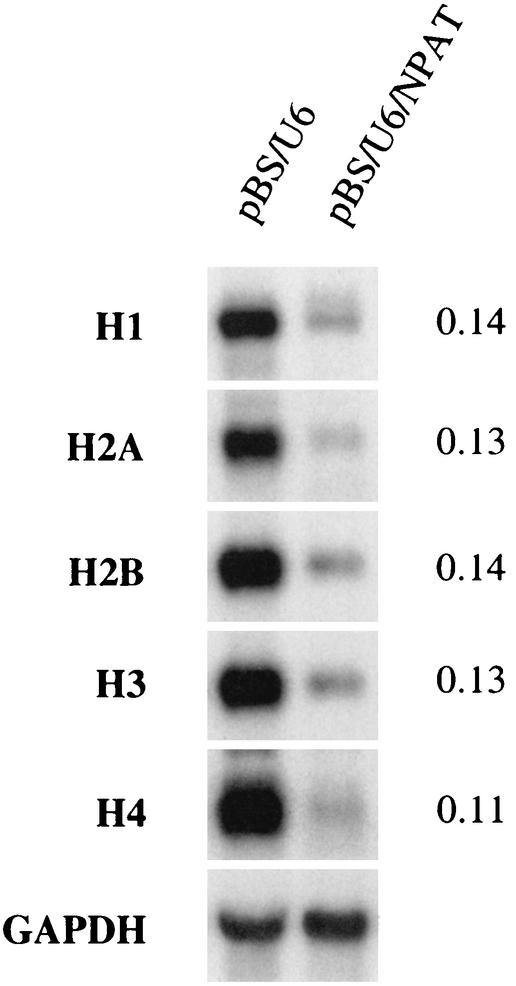
It has been shown that NPAT associates with the promoters of replication-dependent histone genes in vivo and that the overexpression of NPAT activates the histone promoters (24, 56). We have also observed that induced expression of NPAT activates the transcription of multiple endogenous histone genes (data not shown). These results indicate that NPAT regulates histone gene expression. If NPAT is required for histone gene transcription, then one would predict that the inhibition of NPAT expression should result in a reduction in the levels of histone mRNAs in vivo. To examine this idea, we inhibited NPAT expression by using siRNA and examined the levels of histone mRNAs in the treated cells by Northern blot analysis. While glyceraldehyde-3-phosphate dehydrogenase mRNA was not affected by NPAT-specific siRNA, the expression of the mRNAs of all five subtypes of histones was greatly reduced in siRNA-treated cells (Fig. 10). These results, together with the results from the above-mentioned previous studies, strongly support the notion that NPAT plays an essential role in the expression of replication-dependent histone genes.
FIG. 10.
Inhibition of histone expression in cells treated with NPAT-specific siRNA. U2OS cells were transfected with control vector pBS/U6 or pBS/U6/NPAT and treated as described in Materials and Methods. Total RNA isolated from transfected cells was analyzed by Northern blotting. The relative levels of histone mRNAs in pBS/U6/NPAT-transfected cells compared with those in pBS/U6-transfected cells are shown on the right. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
NPAT is an in vivo substrate of cyclin E-Cdk2 kinase and regulates coordinated transcriptional activation of the replication-dependent histone genes (24, 55, 56). Here we show that the expression of NPAT is regulated by the E2F transcription factor. Thus, NPAT links both cyclin E-Cdk2 and E2F to the regulation of histone gene expression. The fact that the expression of NPAT function is regulated at both transcriptional and posttranslational levels by key cell cycle regulators underscores the notion that NPAT plays a pivotal role in cell cycle progression. Indeed, our studies demonstrated that the expression of NPAT promotes S-phase entry in cells released from quiescence as well as in cycling cells. In addition, we show that NPAT function is required for progression through multiple phases of the cell cycle.
Several lines of evidence presented in this study indicate that NPAT is a bona fide E2F target gene. First, in transient transfection experiments, the NPAT promoter is activated by E2F proteins through the E2F recognition sequences in the promoter. Second, mutations of the E2F sites impair transcriptional activation of the NPAT promoter at the G1/S-phase boundary during growth stimulation. Third, endogenous E2F proteins associate with the NPAT promoter in vivo, and the activation of E2F induces the expression of the endogenous NPAT gene. Thus, the increased expression of NPAT at the G1/S-phase boundary likely results, at least partly, from transcriptional regulation by E2F.
E2F proteins interact with proteins in the pRB family that can elicit transcriptional repression. Thus, E2F proteins can mediate transcriptional repression as well as transcriptional activation. It has been proposed that E2F1, E2F2, and E2F3 act as potent transcriptional activators, whereas E2F4 and E2F5 mediate transcriptional repression (8, 13, 45, 48). Some studies have suggested that a major role of E2F in cell cycle progression is to mediate transcriptional repression of the E2F target genes (40, 54), while genetic studies with cells deficient in E2F1 to E2F3 have indicated that these E2F proteins are required for the transcriptional activation of a number of E2F target genes whose functions are essential for cell cycle progression (53). Thus, it appears that E2F-mediated transcriptional activation and repression are both crucial for cell cycle progression. When overexpressed, all four E2F proteins tested in our experiments can activate the NPAT promoter. However, it remains to be determined whether the activities of these E2F proteins are required for NPAT expression in vivo. Given that endogenous E2F1 and E2F4 proteins are associated with the NPAT promoter in vivo (Fig. 4), it is likely that at least these two E2F proteins participate in the regulation of NPAT expression. E2F1 may positively regulate the NPAT promoter, whereas E2F4 may function as a repressor of the promoter in G1 phase. The observed activation of the NPAT promoter by E2F4 overexpression may result from sequestration of the pRB family of proteins.
Mutation of the E2F sites in the NPAT promoter either attenuates or abolishes the activation of the NPAT promoter at the G1/S-phase boundary during growth stimulation (Fig. 6), suggesting that E2F regulates transcriptional activation of the NPAT gene during the G1/S-phase transition. The E2F site(s) may also mediate transcriptional repression of the endogenous NPAT promoter, although we did not detect such repression in our current assays (Fig. 6).
The inhibition of NPAT expression affected cell cycle progression at multiple stages (Fig. 8 and 9 and Table 1), suggesting a role for NPAT in more than one cellular process. The observation that the inhibition of NPAT expression attenuates S-phase entry indicates that NPAT regulates a rate-limiting step in the G1/S-phase transition. This idea is consistent with our finding that the overexpression of NPAT promotes G1/S-phase progression (55) (Fig. 7). It remains to be determined, however, whether histone synthesis is the rate-limiting step in NPAT-mediated S-phase entry. In addition to activating histone expression, NPAT may also perform other functions that are essential for S-phase entry. Our data also indicate that the rate of S-phase progression is reduced when NPAT expression is inhibited. Given the observation that NPAT plays a critical role in histone gene expression (24, 56) (Fig. 10) and the recent report that the inhibition of histone expression by HIRA overexpression causes cell cycle arrest in S phase (29), the observed inhibition of S-phase progression by NPAT-specific siRNA may result from reduced histone expression. Our present study has revealed that, in addition to being critical for G1/S-phase progression, NPAT function is also involved in G2/M-phase progression (Fig. 9A). Further studies are needed to elucidate the underlying molecular mechanism. Recent studies have shown that E2F function is also required throughout the cell cycle (13, 45, 53). It is possible that some of the E2F functions are executed through the activities of NPAT.
TABLE 1.
Effect of inhibition of NPAT expression on G1- and S-phase progressiona
| Phase | % of cells at the following h after transfection with the indicated plasmid:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 48
|
55
|
61
|
72
|
|||||
| U6 | U6/NPAT | U6 | U6/NPAT | U6 | U6/NPAT | U6 | U6/NPAT | |
| G1 | 35.4 | 22.9 | 13.4 | 15.7 | 10.7 | 10.1 | 4.4 | 5.8 |
| S | 43.3 | 30.1 | 52.1 | 35.0 | 10.9 | 24.3 | 3.6 | 10.8 |
| G2/M | 21.3 | 46.9 | 34.5 | 49.4 | 78.4 | 65.7 | 92.0 | 83.5 |
U2OS cells were transfected with pBS/(U6) or pBS/U6/NPAT (U6/NPAT) as described in Materials and Methods. Nocodazole (50 ng/ml) was added to the culture medium 48 h after transfection. Cells were harvested at the indicated times posttransfection, and the distribution of the transfected cells in the cell cycle was analyzed by FACS. The mean results from two independent experiments are shown.
During S phase, cells synthesize large quantities of histone proteins to complex with newly replicated DNA. This is accomplished by both transcriptional activation of histone genes in S phase and posttranscriptional regulation of histone transcripts (6, 14, 33, 44). Although E2F regulates the expression of a number of genes whose products are involved in DNA replication, the promoters of most histone genes do not contain E2F sites and appear not to be directly regulated by E2F (34). As NPAT regulates histone gene transcription, the observations in this study suggest that E2F indirectly regulates the expression of histone genes through its regulation of NPAT expression. Interestingly, the histone stem-loop binding protein, which is involved in histone pre-mRNA processing (50), has also been identified as an E2F target in DNA microarray analyses of genes induced by E2F (18, 27). Thus, E2F promotes S-phase entry not only by regulating the expression of proteins involved in DNA synthesis but also by regulating the expression of proteins involved in histone synthesis.
E2F activity is negatively regulated by tumor suppressor pRB and related proteins p107 and p130, which in turn are inactivated by cyclin D-dependent kinase Cdk4/Cdk6. The activity of Cdk4/Cdk6 itself is regulated by the INK4 cyclin-dependent kinase inhibitor family of proteins. This pRB/E2F pathway is deregulated in virtually all human cancers (30, 43). In view of our observation that NPAT is a critical target of E2F and can promote S-phase entry in mammalian cells under various conditions, it is tempting to speculate that the deregulated expression of NPAT may also contribute to tumorigenesis in human cancers. Further studies are needed to determine whether the regulation of NPAT may be altered in human cancers.
Acknowledgments
G. Gao and A. P. Bracken contributed equally to this work.
We thank Dirk Bohmann and Hartmut Land for helpful discussions and critical reading of the manuscript. We are grateful to Peter Keng for help with FACS analysis. We also thank Yang Shi for providing plasmid pBS/U6.
This work was supported by an award from the Howard Hughes Medical Institute to the University of Rochester School of Medicine and Dentistry under the Biomedical Research Support Program for Medical Schools, in part by NIH grant RO1 GMO65814 to J.Z., and by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) and Fondazione Italiana per la Ricerca sul Cancro (FIRC) to K.H.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology, p. 4.2.1-4.3.6. Greene Publishing Associates & Wiley-Interscience, New York, N.Y.
- 2.Botz, J., K. Zerfass-Thome, D. Spitkovsky, H. Delius, B. Vogt, M. Eilers, A. Hatzigeorgiou, and P. Jansen-Durr. 1996. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol. Cell. Biol. 16:3401-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd, P. J., P. R. Cooper, T. Stankovic, H. S. Kullar, G. D. Watts, P. J. Robinson, and M. R. Taylor. 1996. A gene transcribed from the bidirectional ATM promoter coding for a serine rich protein: amino acid sequence, structure and expression studies. Hum. Mol. Genet. 5:1785-1791. [DOI] [PubMed] [Google Scholar]
- 4.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Fruscio, M., H. Weiher, B. C. Vanderhyden, T. Imai, T. Shiomi, T. A. Hori, R. Jaenisch, and D. A. Gray. 1997. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol. Cell. Biol. 17:4080-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 7.Dulic, V., E. Lees, and S. I. Reed. 1992. Association of Human cyclin E with a periodic G1-S phase protein kinase. Science 257:1958-1961. [DOI] [PubMed] [Google Scholar]
- 8.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 10.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 12.Geng, Y., E. N. Eaton, M. Picon, J. M. Roberts, A. S. Lundberg, A. Gifford, C. Sardet, and R. A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12:1173-1180. [PubMed] [Google Scholar]
- 13.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 14.Heintz, N. 1991. The regulation of histone gene expression during the cell cycle. Biochem. Biophys. Acta 1088:327-339. [DOI] [PubMed] [Google Scholar]
- 15.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690-703. [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, T., T. Sugawara, A. Nishiyama, R. Shimada, R. Ohki, N. Seki, M. Sagara, H. Ito, M. Yamauchi, and T. Hori. 1997. The structure and organization of the human NPAT gene. Genomics 42:388-392. [DOI] [PubMed] [Google Scholar]
- 17.Imai, T., M. Yamauchi, N. Seki, T. Sugawara, T. Saito, Y. Matsuda, H. Ito, T. Nagase, N. Nomura, and T. Hori. 1996. Identification and characterization of a new gene physically linked to the ATM gene. Genome Res. 6:439-447. [DOI] [PubMed] [Google Scholar]
- 18.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, D. G., J. K. Schwarz, W. D. Cress, and J. R. Nevins. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365:349-352. [DOI] [PubMed] [Google Scholar]
- 20.Kel, A. E., O. V. Kel-Margoulis, P. J. Farnham, S. M. Bartley, E. Wingender, and M. Q. Zhang. 2001. Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J. Mol. Biol. 309:99-120. [DOI] [PubMed] [Google Scholar]
- 21.Koff, A., A. Giordano, D. Desai, K. Yamashita, W. Harper, S. Elledge, T. Nishimoto, D. Morgan, R. Franza, and J. Roberts. 1992. Formation and activation of a cyclin E-CDK2 complex during the G1 phase of the human cell cycle. Science 257:1689-1694. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 23.Lukas, J., B. O. Petersen, K. Holm, J. Bartek, and K. Helin. 1996. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol. 16:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeks-Wagner, D., and L. H. Hartwell. 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44:43-52. [DOI] [PubMed] [Google Scholar]
- 26.Moroni, M. C., E. S. Hickman, E. L. Denchi, G. Caprara, E. Colli, F. Cecconi, H. Muller, and K. Helin. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3:552-558. [DOI] [PubMed] [Google Scholar]
- 27.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, H., and K. Helin. 2000. The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta 1470:M1-M12. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, D. M., X. Ye, C. Hall, H. Santos, T. Ma, G. D. Kao, T. J. Yen, J. W. Harper, and P. D. Adams. 2002. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22:7459-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 32.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146-12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osley, M. A. 1991. The regulation of histoene synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 34.Oswald, F., T. Dobner, and M. Lipp. 1996. The E2F transcription factor activates a replication-dependent human H2A gene in early S phase of the cell cycle. Mol. Cell. Biol. 1889-1895. [DOI] [PMC free article] [PubMed]
- 35.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paddison, P. J., and G. J. Hannon. 2002. RNA interference: the new somatic cell genetics? Cancer Cell 2:17-23. [DOI] [PubMed] [Google Scholar]
- 37.Qin, X.-Q., D. M. Livingston, W. G. J. Kaelin, and P. Adams. 1994. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA 91:10918-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Resnitzky, D., M. Gossen, H. Bujard, and S. I. Reed. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell. 2:55-65. [DOI] [PubMed] [Google Scholar]
- 41.Savitsky, K., M. Platzer, T. Uziel, S. Gilad, A. Sartiel, A. Rosenthal, O. Elroy-Stein, Y. Shiloh, and G. Rotman. 1997. Ataxia-telangiectasia: structural diversity of untranslated sequences suggests complex post-transcriptional regulation of ATM gene expression. Nucleic Acids Res. 25:1678-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sexl, V., J. A. Diehl, C. J. Sherr, R. Ashmun, D. Beach, and M. F. Roussel. 1999. A rate limiting function of cdc25A for S phase entry inversely correlates with tyrosine dephosphorylation of Cdk2. Oncogene 18:573-582. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 44.Stein, G. S., J. L. Stein, A. J. Van Wijnen, and J. B. Lian. 1996. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol. Int. 20:41-49. [DOI] [PubMed] [Google Scholar]
- 45.Stevaux, O., and N. J. Dyson. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14:684-691. [DOI] [PubMed] [Google Scholar]
- 46.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 48.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 49.Vigo, E., H. Muller, E. Prosperini, G. Hateboer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 19:6379-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Z. F., M. L. Whitfield, T. C. Ingledue III, Z. Dominski, and W. F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 51.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitfield, M. L., G. Sherlock, A. J. Saldanha, J. I. Murray, C. A. Ball, K. E. Alexander, J. C. Matese, C. M. Perou, M. M. Hurt, P. O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13:1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, J., B. Dynlacht, T. Imai, T. Hori, and E. Harlow. 1998. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 12:456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]