Abstract
Human ras genes play central roles in coupling extracellular signals with complex intracellular networks controlling proliferation, differentiation, and apoptosis, among others processes. c-H-ras pre-mRNA can be alternatively processed into two mRNAs due to the inclusion or exclusion of the alternative exon IDX; this renders two proteins, p21H-Ras and p19H-RasIDX, which differ only at the carboxy terminus. Here, we have characterized some of the cis-acting sequences and trans-acting factors regulating IDX splicing. A downstream intronic silencer sequence (rasISS1), acting in concert with IDX, negatively regulates upstream intron splicing. This effect is mediated, at least in part, by the binding of hnRNP A1. Depletion and add-back experiments in nuclear extracts have confirmed hnRNP A1's inhibitory role in IDX splicing. Moreover, the addition of two SR proteins, SC35 and SRp40, can counteract this inhibition by strongly promoting the splicing of the upstream intron both in vivo and in vitro. Further, the RNA-dependent helicase p68 is also associated with both IDX and rasISS1 RNA, and suppression of p68 expression in HeLa cells by RNAi experiments results in a marked increase of IDX inclusion in the endogenous mRNA, suggesting a role for this protein in alternative splicing regulation.
A common mechanism for gene expression regulation in metazoa is the use of alternative splice sites (SS) to produce multiple protein-coding sequences from the same pre-mRNA. It was recently predicted that nearly 60% of all human genes undergo at least one process of alternative splicing (41). The ever increasing known examples of alternative pre-mRNA processing are often tissue type or developmental state specific, indicating that complex regulation is involved in the selection of SS pairs (for a review, see reference 47). However, little is yet known about the detailed mechanisms regulating alternative splicing in mammalian cells.
A number of RNA sequences that positively or negatively regulate the inclusion of alternative exons have been identified (12, 22, 24, 26, 32, 35, 38, 42, 61). Binding of certain sets of splicing factors to these regulatory sequences, together with the intrinsic strengths of the SS, dictates the specificity and efficiency of splicing, resulting in promotion or repression of each splicing event.
The SR protein family is a well-characterized class of proteins involved in both constitutive and alternative splicing (29, 58). Their mechanism of action involves binding to certain RNA sequences (commonly exonic enhancers) through their RNA recognition motifs and the recruitment of other splicing factors, stimulating the splicing efficiency of weak adjacent SS (1, 30, 33, 56). SR protein binding to an exon is known to increase the binding of U2AF65 to an upstream 3′SS (60) or that of U1 snRNP to a downstream 5′SS (6, 27, 36, 39). SR proteins can also act in a RNA binding-independent way, promoting the assembly of general splicing factors in the protein-protein network interactions that make up the mature spliceosome (5). Factors other than SR proteins are sometimes required for efficient splicing enhancement, and some cell- or tissue-specific splicing factors have been described (37, 54, 55, 63).
The heterogeneous nuclear ribonucleoprotein (hnRNP) family, which rapidly associate with nascent pre-mRNA transcripts and are involved in RNA metabolism, have also been implicated in splicing regulation (for reviews, see references 25 and 40). hnRNP proteins can hinder or assist the splicing machinery, thereby playing a role in SS selection.
Although the general antagonistic properties of SR proteins and hnRNP A1 have been evidenced in vivo and in vitro for a long time (9, 52, 53) and further analyzed (2, 28), all these analyses have been performed on chimeric model pre-mRNA or natural reporter genes which are not necessarily specific targets in vivo for these trans-acting factors. In fact, the specific involvement of SR proteins and hnRNP A1 has been well evidenced for alternative splicing of human immunodeficiency virus (HIV) pre-mRNA (49, 66), but presently, no equivalent specific targets among the mammalian genes was revealed, and this latter question will be the main point of this study.
Human c-H-ras proto-oncogene is subject to alternative splicing regulation, and Ras proteins are a central component of mitogenic signal transduction pathways (reviewed in references 50 and 62). Three ras genes (H-ras, K-ras and N-ras) encode very similar small GTPase proteins, which cycle between active GTP-bound and inactive GDP-bound states. K-ras pre-mRNA undergoes alternative splicing in its last intron, producing two mRNAs which differ only in their last coding exon 4 (E4). Similarly, the H-ras pre-mRNA can produce two mature RNAs, p19mRNA and p21mRNA, the former of which contains the alternative exon IDX (20) (Fig. 1A). p19mRNA is translated into a shorter protein, p19H-RasIDX, due to the presence of an in-frame stop codon (Guil et al., submitted). p19H-RasIDX presumably lacks some of the biochemical and functional characteristics of its counterpart, p21H-Ras. Here, we focus on the splicing of single-intron, two-exon H-ras transcripts and determine which cis-acting sequences and trans-acting factors are responsible for the repression of IDX inclusion, as a step towards further understanding of alternative splicing in human proto-oncogenes.
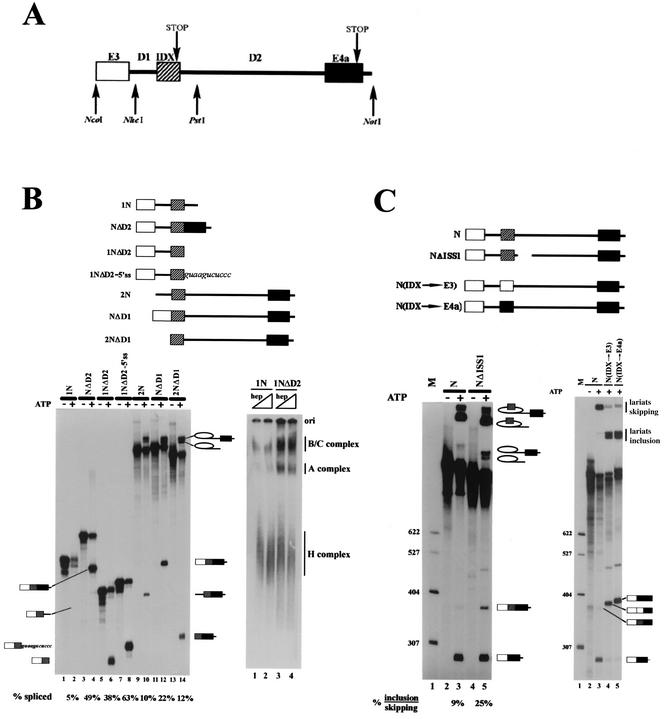
FIG. 1.
Intron D1 splicing is repressed by a downstream ISS. (A) Map of the region around alternative exon IDX, with exons shown as boxes and introns shown as lines. Restriction sites indicate the start or end of the different constructs. Pre-mRNA 1N, pre-mRNA 2N, and pre-mRNA N encompass the NcoI/PstI region, the NheI/NotI region, and the whole NcoI/NotI fragment, respectively. (B) Left, in vitro splicing assays of the constructs depicted in the upper diagram; odd lanes are negative controls without ATP. Band identity was assessed by extensive RT-PCR and/or primer extension of all lariats and products (drawings in the margins). % spliced, ratio of spliced RNA to initial pre-mRNA. Right, spliceosome complex formation on the 1N and 1NΔD2 substrates. The H, A, and B/C complexes are indicated on the right of the gel. Lanes 1 and 2, construct 1N in the presence of 0.8 and 1.6 ¨g of heparin (hep)/¨l as unspecific competitor. Lanes 3 and 4, construct 1NΔD2 in the presence of 0.8 and 1.6 mg of heparin/¨l. (C) Left, in vitro splicing assays of the N and NΔISS1 substrates depicted in the upper diagram. Lane 1, size markers (M) (plasmid pBR322 cut with HpaII and 59 end labeled); lanes 2 and 3, construct N; lanes 4 and 5, construct NΔISS1; lanes 2 and 4, negative controls without ATP. Right, in vitro splicing assays of N(IDX→E3) and N(IDX→E4a) depicted in the upper diagram. Lane 1, size markers (M); lanes 2 and 3, substrates N; lane 4, N(IDX→E3); lane 5, N(IDX→E4a). E3 and E4a are 92 and 101 nt long. Lane 2, negative control without ATP. Band identity is indicated in the right of each panel.
MATERIALS AND METHODS
Plasmid construction.
Oligonucleotides used for plasmid construction are listed in Table 1. Construct N is an NcoI-NotI fragment of the human c-H-ras gene made blunt and cloned into the Acc65I-SmaI sites of pBSSK+ (Stratagene). Its derivatives NΔD1, NΔD2, and NΔISS1 were obtained by PCR amplification of the whole plasmid and religation with rasIDXfor and 3′Ex3rev (NΔD1), 5′Ex4for and 3′IDXrev (NΔD2), and D2-2772for and D2-2720rev (NΔISS1). Constructs N(IDX→E3) and N(IDX→E4a) were obtained by digesting construct N with BglI and CfoI, blunt ending, and ligating to the fragments from exon 3 (amplified with oligonucleotides Ex3for and 3′Ex3rev) or from exon 4a (amplified with oligonucleotides 5′Ex4for and D2-848 and digested with HaeIII). Construct 2N is a NheI-NotI fragment of the H-ras gene, blunt ended and cloned into the Acc65I and SmaI sites of pBSSK+. Its derivative 2NΔD1 was obtained by PCR from 2N template with oligonucleotides 5′IDXfor and T7rev. Construct 1N is a NcoI-PstI fragment of H-ras cloned into the KpnI and NotI sites of pBSSK+. Construct 1N derivatives and the oligonucleotides used are as follows: 1NΔD2 (pBS-737for and 3′IDXrev), 1NΔD2-5′SS (pBS-737for, D2-2720rev), 1NISS1Δ1 (D2-2731for, D2-2720rev), 1NISS1Δ2 (D2-2741for, D2-2730rev), 1NISS1Δ3 (D2-2751for, D2-2740rev), 1NISS1Δ4 (D2-2761for, D2-2750rev), 1NISS1Δ5 (D2-2772for, D2-2760rev), 1NISS1Δ23 (D2-2751for, D2-2730rev), and 1NISS1Δ234 (D2-2761for, D2-2730rev).
TABLE 1.
Oligonucleotides used for plasmid construction
| Primer name | Sequence (5′→3′) | H-ras sequencea | Plasmid(s) generated |
|---|---|---|---|
| T7for | TAATACGACTCACTATAGGG | ||
| T7rev | CCAATTCGCCCTATAAGTGAG | pBS-2NΔD1 | |
| 5′IDXfor | GGCAGCCGCTCTGGCTCTAGCTCC | 2628-2651 (i) | pBS-NΔD1, pBS-2NΔD1, pBS-IDX |
| Ex3for | ACGCACTGTGGAATCTCGGC | 2449-2468 (i) | pBS-N(IDX→E3) |
| 3′Ex3rev | CTGCCGGGTCTTGGCCGAGG | 2514-2533 (c) | pBS-NΔD1, pBS-N(IDX→E3) |
| 5′Ex4for | GGAGTGGAGGATGCCTTCT | 3231-3249 (i) | pBS-NΔD2, pBS-N(IDX→E4a) |
| D2-848rev | AGTCCCCCTCACCTGCGTCA | 3348-3367 (c) | pBS-N(IDX→E4a) |
| 3′IDXrev | GCGAGGGGCCGCTCCCTCAC | 2687-2706 (c) | pBS-NΔD2, pBS-1NΔD2, pBS-IDX |
| D2-2772for | TGCAGTCCTGGGCCGACACAG | 2772-2792 (i) | pBS-NΔISS1, pBS-1NISS1Δ5 |
| D2-2720rev | GGGAGACTTACAGCGCG | 2704-2720 (c) | pBS-NΔISS1, pBS-1NΔD2-5′ss, pBS-1NISS1Δ1 |
| pBS-737for | GCGGCCGCCACCGCGGTGGA | pBS-1NΔD2, pBS-1NΔD2-5′ss | |
| D2-2731for | GGCAGTGAGGGAGGCGAGGG | 2731-2750 (i) | pBS-1NISS1Δ1 |
| D2-2741for | GAGGCGAGGGCCGGGGTCTG | 2741-2760 (i) | pBS-1NISS1Δ2 |
| D2-2730rev | CTGCCGTCCCGGGAGACTTA | 2711-2730 (c) | pBS-1NISS1Δ2, pBS-1NISS1Δ23, pBS-1NISS1Δ234 |
| D2-2751for | CCGGGGTCTGGGCTCACGCC | 2751-2770 (i) | pBS-1NISS1Δ3, pBS-1NISS1Δ23 |
| D2-2740rev | CCTCACTGCCCTGCCGTCCC | 2721-2740 (c) | pBS-1NISS1Δ3 |
| D2-2761for | GGCTCACGCCCTGCAGTCCT | 2761-2780 (i) | pBS-1NISS1Δ4, pBS-1NISS1Δ234 |
| D2-2750rev | CCCTCGCCTCCCTCACTGCC | 2731-2750 (c) | pBS-1NISS1Δ4 |
| D2-2760rev | CAGACCCCGGCCCTCGCCTC | 2741-2760 (c) | pBS-1NISS1Δ5 |
| D2-2711for | TAAGTCTCCCGGGACGGCAG | 2711-2730 (i) | pBS-ISS1, pBS-ISS1Δ23 |
| Ex4arev | CTGCCGGATCTCACGCACCA | 3256-3275 (c) | pBS-ISS1, pBS-ISS1Δ23 |
| D2-2771rev | GGGCGTGAGCCCAGACCCCG | 2762-2771 (c) | pBS-IDXISS1 |
| pSUPER-p68for | GATCCCCGAGCACCCTGATTTGGCTATTCAAGAGA TAGCCAAATCAGGGTGCTCTTTTTGGAAA | pSUPER-p68 | |
| pSUPER-p68rev | AGCTTTTCCAAAAAGAGCACCCTGATTTGGCTATC TCTTGAATAGCCAAATCAGGGTGCTCGGG | pSUPER-p68 |
Whether the sequence is identical (i) or complementary (c) to the c-H-ras gene is indicated. Nucleotide positions are numbered according to the numbering of the sequence with GenBank accession no. V00574.
rasISS1 and ISS1Δ23 RNAs used for gel shift assays and agarose bead coupling were obtained from PCR fragments performed with D2-2711for and Ex4arev on templates N and NΔISS1, respectively. The PCR band was cloned into the Ecl136I site of pBSSK+. Both RNAs were in vitro transcribed after digestion with PstI. The IDX fragment was amplified from construct N with 5′IDXfor and 3′IDXrev, cloned into the Ecl136I and PstI sites of pBSSK+, and digested with EcoRI before transcription. The IDXISS1 fragment was obtained similarly with oligonucleotides 5′IDXfor and D2-2771rev and was mutated in position 2537 (A→G) to eliminate the 5′SS.
pSUPER-p68 was obtained by hybridization of oligonucleotides pSUPER-p68for and pSUPER-p68rev, digestion with BglII and HindIII and cloning in plasmid pSUPER (Oligoengine) cut with the same enzymes.
In vitro pre-mRNA splicing assays.
Pre-mRNA substrates were prepared by standard in vitro transcription with T7 RNA polymerase in the presence of [α-32P]CTP. Template DNAs were linearized with BamHI (constructs 2N, N, and derivatives) or with ScaI (construct 1N and derivatives).
In vitro splicing was performed in 25-μl reaction mixtures containing 40% nuclear extract (provided by 4C, Computer Cell Culture Center S.A., Mons, Belgium), 0.5 mM ATP, 25 mM creatine phosphate, 1.5 mM MgCl2, and 20,000 cpm (∼10 fmol) of each pre-mRNA. Reactions were incubated at 30°C for 90 min (2 h for the depleted extracts) followed by phenol-chloroform extraction, precipitation, and 5% denaturing gel electrophoresis.
Transfection assays and RNA analysis.
Transfections in HeLa cells were performed by using the standard calcium phosphate coprecipitation procedure. At 48 h posttransfection, total RNA was extracted and reverse transcribed with oligonucleotide dT and Superscript II (GIBCO BRL). After 30 cycles of PCR amplification with oligonucleotide T7 and IDX3′rev, the DNA bands were resolved on 2% agarose gels. For RNA interference experiments, ∼8 × 106 HeLa cells were electropored at 250 V and 960 μF with 15 μg of plasmid, obtaining ∼90% transfection efficiency. Cells were harvested at appropriate times, and 5 μg of total RNA was used in each Northern blot analysis, following standard procedures.
Gel shift assays.
Samples (20,000 cpm; ∼30 fmol) of each RNA were incubated for 30 min at 30°C under splicing conditions with 0.16 μg of heparin/μl and run on 6% native acrylamide gel in 1× Tris-borate-EDTA. In competition assays, cold RNAs were preincubated for 10 min at 30°C in the presence of nuclear extract before addition of labeled RNA. Gel shifts performed with recombinant proteins were essentially the same, but 40% Roeder D was used instead of nuclear extract.
To analyze spliceosome assembly, splicing reaction mixtures with 10 fmol of 1N or 1NΔD2 pre-mRNAs were incubated for 30 min at 30°C. Reactions were stopped with heparin and separated on a 4% native acrylamide-bisacrylamide (80:1) gel in 1× Tris-borate-EDTA.
RNA affinity purification and Western blotting.
Substrate RNAs for bead immobilization were synthesized in vitro from the PstI linearized constructs pBS-ISS1 and pBS-ISS1Δ23 or the EcoRI linearized pBS-IDX by using T3 RNA polymerase. HIV-ESS RNA was synthesized in vitro by using T7 RNA polymerase and a DNA oligonucleotide template. After transcription, all RNAs were gel purified. Coupling to beads, affinity purification of factors, and nuclear extract depletion were performed as described previously (10). Proteins were separated on 12.5% (8% in Fig. 6) sodium dodecyl sulfate (SDS)-polyacrylamide gels and visualized by Coomassie blue staining or immunoblotted with anti-hnRNP A1 monoclonal antibody 4B10 (a gift from G. Dreyfuss, University of Pennsylvania), with anti-p68 monoclonal antibody PAb204 (a gift from Fuller-Pace, University of Dundee), with anti-hnRNP H/H' polyclonal antibody AM113 (a gift from Wilusz, University of Pittsburgh), or with anti-FUS/TLS polyclonal antibody (a gift from Moreau-Gachelin, from Institut Curie, Paris).
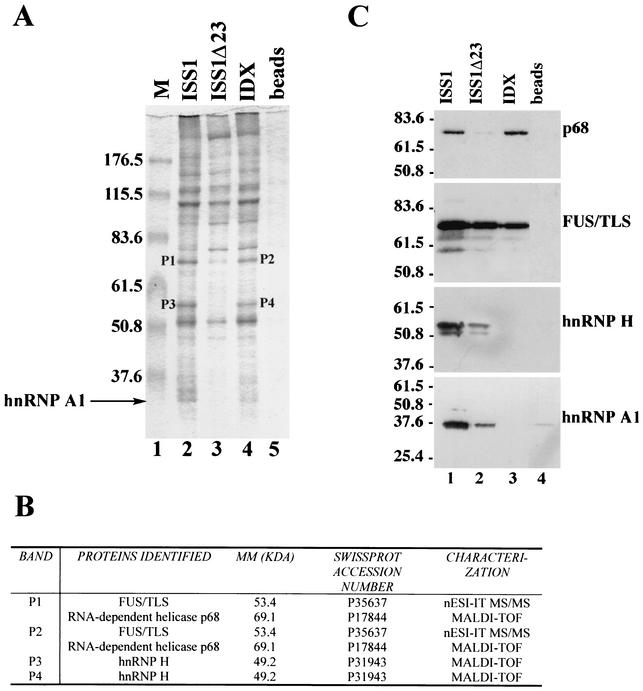
FIG. 6.
FUS/TLS, hnRNP H, and the ATP-dependent RNA helicase p68 are associated to rasISS1 and IDX. (A) Coomassie blue staining of RNA affinity-purified factors from HeLa nuclear extracts separated by 8% SDS-polyacrylamide gel electrophoresis. Agarose beads coupled to rasISS1 RNA (lane 2), ISS1Δ23 (lane 3), or IDX (lane 4) or alone (lane 5) were incubated with nuclear extract. Lane 1, size markers (M). hnRNP A1 band is indicated (lane 2). Four bands were excised from the gel and microsequenced (P1 to P4). (B) Identification of the four protein bands. MM, molecular mass. Protein identity was assessed either by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF) or by nanospray ion trap tandem mass spectrometry (nESI-ITMS/MS). (C) Western blot of the gel in panel A assayed with each of the specific antibodies depicted on the right. The values on the left indicate the molecular mass in kilodaltons.
Total HeLa SR purification and recombinant proteins.
Total SR proteins from HeLa cells were prepared as previously described (65). Recombinant GST-hnRNP A1 and GST-UP1 were expressed in Escherichia coli BL21 strain and purified (15). Recombinant baculovirus expressed and purified SRs were obtained as described elsewhere (16). At the end of the purification steps, all proteins were dialyzed against buffer Roeder D. pCG-SR (p40, 9G8, and SC35) protein expression vectors were a gift from J. F. Caceres (Western General Hospital, Edinburgh). We prepared full-length pCG-ASF protein expression vector by PCR with two suitable oligodeoxynucleotides.
Peptide microsequencing of protein bands excised from denaturing gels.
SDS-polyacrylamide gels were stained with Coomassie blue, and protein bands were excised. Prior to in-gel digestion, protein bands were reduced with 10 mM dithiothreitol and alkylated with 55 mM iodoacetamide. Enzymatic digestion was performed with trypsin (Promega, Madison, Wis.). Protein identification by mass spectrometry (matrix-assisted laser desorption time-of-flight mass spectrometry and nanospray ion trap tandem mass spectrometry) and database search are described elsewhere (13).
RESULTS
The region of c-H-ras gene encompassing the terminal coding exons is depicted in Fig. 1A. The alternatively spliced exon IDX (82 nucleotides [nt]), located between exons 3 (E3) and 4a (E4a), is flanked by a short, upstream intron (94 nt) (D1) and a comparatively long, downstream one (521 nt) (D2) (21). Our first aim was to analyze which cis-acting sequence elements control the inclusion or skipping of IDX in the mature mRNA transcript and therefore the production of protein p19H-RasIDX. First, we prepared different plasmid constructions encompassing the ∼1-kb region between the NcoI-NotI restriction fragment. Then, we assessed the transcribed pre-mRNAs in splicing assays.
A sequence element located within intron D2 represses splicing of the upstream intron D1.
Exon IDX is known to be poorly included in the mature transcript in in vitro splicing of a two-intron pre-mRNA (19) and substrate N in this work. Hence, we focused on the possible roles of intronic sequences in regulating this alternative splicing. We therefore constructed two sets of single-intron pre-mRNA substrates (Fig. 1B) to test the ability of IDX to splice to E3 (as an exon acceptor; i.e., construct 1N and derivatives) or to E4a (as an exon donor; i.e., construct 2N and derivatives).
First, when IDX was coupled to exon 3, we compared two substrates in which IDX was either flanked by D2 intronic sequences (construct 1N) or joined at its 3′ end to E4a (identical to when intron D2 has already been spliced; i.e., construct NΔD2). Surprisingly, although 1N was very inefficient in in vitro splicing assays, the efficiency of excision of intron D1 was activated to a high level in NΔD2 substrate (Fig. 1B, left, compare lanes 2 and 4). This dramatic increase of splicing efficiency could be due either to the presence of enhancing sequences in E4a which helped in the exonic definition of IDX or to the removal of inhibitory sequences present in the 5′ end of D2 intron. To test these possibilities, we designed the substrate 1NΔD2, in which exon IDX is not followed by downstream D2 intronic sequences. 1NΔD2 was also efficiently spliced (Fig. 1B, left, compare lanes 6 and 2; note the approximately eightfold increase in the splicing rate), pointing to a negative effect of some element(s) within intron D2 on the upstream intron D1 splicing. In addition, the presence of the first 11 nt of the 5′SS of D2 further increased splicing efficiency compared to that of 1NΔD2 (see construct 1NΔD2-5′SS and compare lanes 8 and 6, Fig. 1B). Therefore, the 5′SS of IDX does not account for the inhibition of D1 splicing but instead improves IDX definition and stimulates D1 splicing. Splicing efficiency, in terms of product accumulation, was also correlated with formation of splicing complexes; 1NΔD2 formed A, B, and C complexes, which are typical of spliceosome assembly, much more effectively than 1N (Fig. 1B, right, compare lanes 3 and 4 with lanes 1 and 2).
We then checked whether intron D1 might also contain similar negative regulators. In substrate 2N and derivatives (NΔD1 and 2NΔD1; see the upper diagram in Fig. 1B), IDX is coupled to E4a and acts as a donor exon. The deletion of D1 sequence does not significantly improve the splicing of the downstream intron, suggesting that it does not contain an inhibitory element (Fig. 1B, left, compare lanes 14 and 10; the splicing efficiencies are similar [∼10%]). In contrast, the joining of E3 to IDX has a positive effect on D2 splicing, possibly because E3 contains enhancer sequences or the 3′ part of D1 impairs the splicing reaction (Fig. 1B, left, lane 12).
From these data, we can conclude that the intronic sequence between nucleotides 2721 and 2771 (the PstI site) in the intron D2 of c-H-ras gene displays properties of a splicing suppressor, and we will call it rasISS1 (ras intronic splicing suppressor 1).
To further analyze rasISS1, we removed this sequence from the two-intron substrate N, generating NΔISS1 pre-mRNA (Fig. 1C). As assessed by in vitro splicing, this resulted in a 2.5-fold increase of IDX inclusion in the mature transcript, confirming the role of rasISS1 as a splicing suppressor (Fig. 1C, left, lanes 3 and 5). However, as the increase in IDX splicing is not as dramatic as in the single-intron substrates, we cannot rule out the possibility that other cis-acting sequences in introns D1 and D2 may be involved in the regulation of IDX inclusion in construct N pre-mRNA. Besides, when IDX sequence in construct N was substituted by other exonic sequences of similar size from E3 or E4a of the c-H-ras gene, yielding constructs N(IDX→E3) and N(IDX→E4a), the splicing balance shifted to almost 100% inclusion of the substituted exon in the mRNA. This indicates that rasISS1 might act in concert with IDX sequences to repress recognition of the alternative exon (Fig. 1C, right, compare lanes 4 and 5 to lane 3).
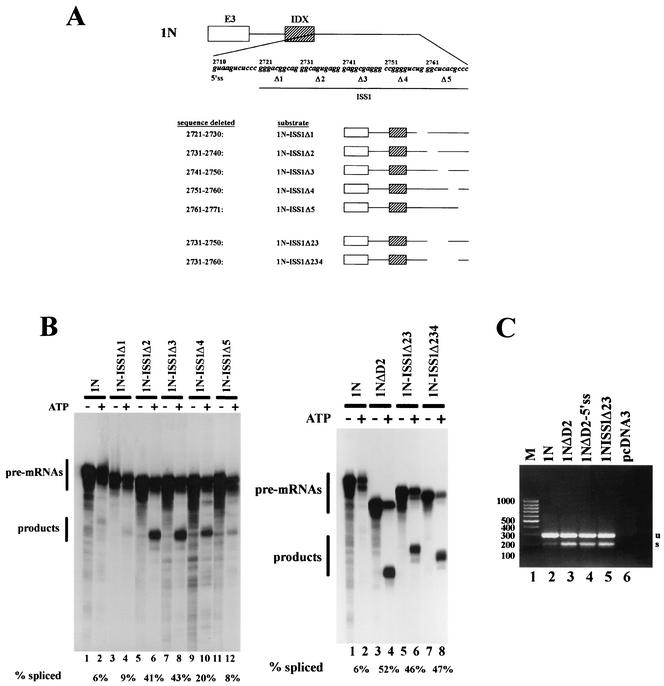
The rasISS1 region was then analyzed in more detail. We divided its 51 nt in five regions and then deleted each to form five constructs (Fig. 2A). The first and the last deletion (1N-ISS1Δ1 and 1N-ISS1Δ5) produced no significant improvement in the splicing reaction (Fig. 2B, left, compare lanes 4 and 12 with results for 1N in lane 2), indicating that the removed sequences are not relevant for splicing inhibition. In contrast, deletions Δ2, Δ3, and Δ4 produced a sharp increase in splicing efficiency (Fig. 2B, left, see lanes 6, 8, and 10 and the quantification below). To determine the smaller deletion in rasISS1 sufficient to restore D1 splicing, we designed constructs 1N-ISS1Δ23 and 1N-ISS1Δ234. Figure 2B, right, shows that the double deletion 1N-ISS1Δ23 is sufficient to almost completely restore splicing, in contrast to 1N-ΔD2 and 1N-ISS1Δ234 (compare lane 6 to lanes 4 and 8). Hence, 1N-ISS1Δ23 was used in further experiments.
FIG. 2.
The inhibitory sequence within rasISS1 maps to nt 2731 to 2750. (A) rasISS1 covers the region 2721 to 2772 nt downstream of exon IDX. The upper diagram shows the five regions (1 to 5) into which rasISS1 was divided and the corresponding deleted substrates assayed in in vitro splicing assays. The five sequences deleted and the five substrates are depicted. In addition, pre-mRNAs 1N-ISS1Δ23 (lacking nt 2731 to 2750) and 1N-ISS1Δ234 (lacking nt 2731 to 2760) are also shown. (B) Left, in vitro splicing assays of the five consecutive substrates deleted and the undeleted substrate 1N. Odd lanes, negative controls without ATP. Quantification of each splicing reaction (% spliced) is shown below each lane. Right, splicing of pre-mRNAs 1N-ISS1Δ23 (lanes 5 and 6) and 1N-ISS1Δ234 (lanes 7 and 8), in comparison to that of the undeleted pre-mRNA 1N (lanes 1 and 2) and the substrate lacking the whole rasISS1 sequence (pre-mRNA 1NΔD2) (lanes 3 and 4). Odd lanes, controls without ATP. (C) RT-PCR analysis of total RNA extracted from HeLa cells transiently transfected with pcDNA3 carrying the different minigenes shown (lanes 2 to 5) or with the empty vector (lane 6). Lane 1, molecular size markers (M) with sizes indicated at left. u, unspliced; s, spliced.
To confirm the role of rasISS1 intronic sequence in vivo, we transiently transfected HeLa cells with minigenes carrying the different sequence variants analyzed in vitro. Figure 2C shows a reverse transcription-PCR (RT-PCR) analysis of the RNAs obtained from the transfected cells; a pattern of splicing activation similar to that in the in vitro assays was observed.
RasISS1 forms a specific complex in HeLa nuclear extracts.
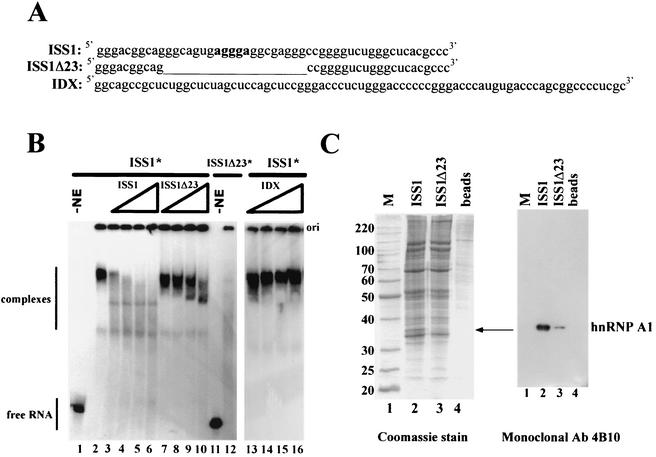
The rasISS1 regulatory sequence could, among other things, exert its function by binding to trans-acting factors or by promoting a secondary structure in the c-H-ras pre-mRNA, preventing IDX splicing. To check if rasISS1 could form stable complexes with nuclear factors in splicing conditions, we performed gel shift assays with the in vitro-labeled rasISS1 RNA and HeLa nuclear extracts. rasISS1 was seen to form a stable and specific complex (Fig. 3B, lane 2), as demonstrated in the competition assay with an excess molar amount of the same cold rasISS1 sequence or with ISS1Δ23 and IDX as control RNAs (Fig. 3B, compare lanes 3 to 6 with lanes 7 to 10 and lanes 13 to 16). Note that only at the highest titration point could the cold RNA ISS1Δ23 partially dissociate the rasISS1* complex (Fig. 3B, lane 10). Besides, ISS1Δ23 RNA is not capable of forming a complex comparable to that of rasISS1 (Fig. 3B, lane 12), as it appears as a faint smear in the presence of nuclear extract. These gel shift data agree with the ability of 1N-ISS1Δ23 pre-mRNA to splice efficiently compared to 1N, suggesting that the high-ordered structured rasISS1 complex is likely to be responsible for splicing inhibition.
FIG. 3.
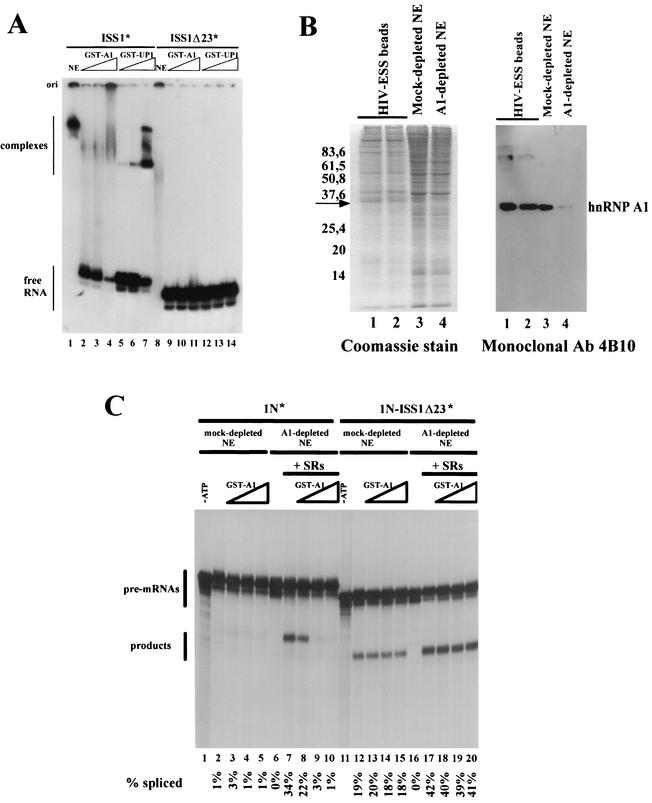
rasISS1 forms a specific complex in HeLa nuclear extracts and binds hnRNP A1. (A) Sequences corresponding to ISS1, ISS1Δ23, and IDX RNA. The sequence deleted in ISS1Δ23 RNA with respect to ISS1 is depicted as a thin line. Nucleotides in bold type in ISS1 represent one putative A1-binding motif. (B) Electrophoretic mobility shift assay performed with homogeneously labeled (*) ISS1 or ISS1Δ23 RNA in nuclear extract (NE), either alone or in competition conditions with increasing amounts of cold competitor RNAs. Lanes 1 and 11, free ISS1 and ISS1Δ23 RNA without extract; lanes 2 and 12, each RNA incubated with NE; lanes 3 to 6, ISS1 reactions with increasing amounts (2, 4, 8, and 16 pmol, respectively) of cold-transcribed ISS1 RNA added; lanes 7 to 10, ISS1 reactions with equivalent amounts of cold ISS1Δ23 added; lanes 13 to 16, ISS1 reactions with equivalent amounts of cold IDX RNA added. Competitor RNAs were preincubated for 10 min in the extract before addition of the labeled ISS1. (C) Left, Coomassie staining of RNA affinity-purified factors from HeLa nuclear extracts separated by 12.5% SDS-polyacrylamide gel electrophoresis. Agarose beads, either coupled to ISS1 RNA (lane 2) or ISS1Δ23 (lane 3) or alone (lane 4), were incubated with nuclear extract. The arrow indicates a specific ISS1-binding protein. The numbers on the left indicate protein molecular mass standards in kilodaltons. Right, Western blot analysis of the protein gel shown on the left with the anti-hnRNPA1 monoclonal antibody (Ab) 4B10.
hnRNP A1 is a component of the rasISS1 complex.
Using an RNA affinity purification approach, we then aimed to detect factors that selectively bind to rasISS1 RNA compared to ISS1Δ23 (negative control). The in vitro-transcribed RNAs were covalently coupled to agarose beads and incubated with HeLa nuclear extract (Fig. 3C). A very low background of proteins from nuclear extracts remained bound to beads alone (Fig. 3C, lane 4), but several bands were eluted from the beads linked to the RNAs. One band of ∼35 kDa was found to bind to rasISS1 RNA but not to ISS1Δ23 RNA (Fig. 3C, compare lanes 2 and 3), and because of its molecular weight and the consensus RNA-binding site present in rasISS1, it was thought to be hnRNP A1. Binding of hnRNP A1 to specific RNA sequences has previously been investigated by SELEX, and a high-affinity binding motif UAGGG(A/U) has been defined (8). A search for putative hnRNP A1 binding sites within rasISS1 of intron D2 in c-H-ras pre-mRNA revealed at least one motif which resembles the consensus sequence with five out of six nucleotides (see Fig. 3A and the sequence in bold type in rasISS1 RNA) as well as others to a lesser extent. Western blotting of the gel with monoclonal antibody 4B10 (a generous gift from G. Dreyfuss), identified the protein as hnRNP A1 (Fig. 3C). To confirm the ability of hnRNP A1 to bind to rasISS1, the GST-A1 fusion protein (a generous gift from B. Chabot) as well as its truncated version (UP1, lacking the C-terminal Gly-rich domain) were expressed and purified in E. coli and assayed in RNA-binding experiments. Both full-length and truncated versions of hnRNP A1 could bind to the rasISS1 RNA fragment (Fig. 4A, lanes 2 to 4 and 5 to 7) but failed to bind to the ISS1Δ23 RNA, where the central GAGGGA motif was absent (Fig. 4A, lanes 9 to 11 and 12 to 14). This confirms that rasISS1 contains hnRNP A1 binding sites.
FIG. 4.
Reconstitution of 1N splicing inhibition by addition of hnRNP A1 recombinant protein. (A) Gel mobility shift assay with labeled ISS1 or ISS1Δ23 RNAs and nuclear extract (NE) or recombinant GST-A1 or GST-UP1. RNA probes, indicated above the autoradiogram, were incubated either with nuclear extract (lanes 1 and 8), with 2, 4, or 8 pmol of recombinant protein GST-A1 (lanes 2 to 4 and 9 to 11), or with the same increasing amounts of recombinant GST-UP1 (lanes 5 to 7 and 12 to 14). Migration of the free RNA probes and the complexes is indicated on the left. (B) Depletion of hnRNP A1 from the HeLa nuclear extract. Left, Coomassie staining of HIV-ESS affinity-purified factors. Lanes 1 and2, eluted factors of the first and second rounds of depletion from nuclear extract incubated with HIV-ESS-coupled beads; lanes 3 and 4, 5 μl of mock-depleted extract (incubated with uncoupled agarose beads) or of the A1-depleted nuclear extract. The band corresponding to hnRNP A1 is indicated. Right, Western blot, performed with monoclonal antibody (Ab) 4B10, of the SDS-polyacrylamide gel on the left. The lane numbers correspond to the same samples on both panels. (C) Reconstitution of splicing inhibition by addition of recombinant (GST-fused) hnRNP A1. In vitro splicing reactions were performed with the HeLa NE depleted twice with HIV-ESS-bound beads (A1-depleted NE) or mock-depleted NE as shown in panel B. Lanes 7 to 10 and 17 to 20, splicing reactions performed in A1-depleted NE supplemented with 200 ng of total HeLa cell SR proteins in the absence or presence of 4, 8, or 16 pmol of purified recombinant hnRNP A1; lanes 2 to 5 and 12 to 15, identical reactions with mock-depleted NE without additional SR proteins; lanes 1 and 11, negative controls without ATP; lanes 6 and 16, control reaction for the A1-depleted NE without additional SR proteins. hnRNP A1 was added to the reaction mixtures prior to the addition of the splicing substrates.
Recombinant hnRNP A1 protein reconstitutes splicing inhibition of intron D1 in A1-depleted nuclear extracts.
To determine whether hnRNP A1 was required for specific inhibition of 1N pre-mRNA, we depleted it from the HeLa nuclear extract used in our in vitro assays by coupling agarose beads to a previously reported A1-binding RNA sequence, the ESS from HIV type 1 (HIV-1) tat exon 2 (10). After two rounds of incubation, more than 90% of hnRNP A1 had been bound by the bead-immobilized RNA (Fig. 4B, lanes 3 and 4). This somewhat aggressive treatment of nuclear extract results in a marked reduction in splicing activity, even for the mock-depleted nuclear extracts (presumably because of a nonspecific depletion of SR proteins) (11). As a consequence, we observed that our control substrate, 1N-ISS1Δ23, was less efficiently spliced in this new mock-depleted nuclear extracts (compare Fig. 4C, lane 12, with Fig. 2) and that in A1-depleted nuclear extracts, it was not processed at all (Fig. 4C, lane 16); this may be due to the removal of other essential factors by the HIV-ESS RNA. Since SR proteins are required for constitutive splicing activity, a preparation of total native SR proteins from HeLa cells was added to the depleted extracts to improve splicing efficiency (Fig. 4C, lane 17). In the A1-depleted SR protein-supplemented nuclear extract, when GST-A1 recombinant protein was added to the reaction, there was no inhibition of the 1N-ISS1Δ23 control substrate (Fig. 4C, compare lanes 18 to 20 with lane 17). There was also no inhibition in the mock-depleted nuclear extract (Fig. 4C, lanes 13 to 15). In contrast, for 1N substrate, which contains the entire rasISS1 sequence, the depletion of hnRNP A1 and supplementation by native SR proteins could restore the splicing reaction (Fig. 4C, lane 7); however, further addition of GST-A1 protein resulted in the splicing inhibition of intron D1 (Fig. 4C, lanes 8 to 10). Thus, this result demonstrates that the addition of hnRNP A1 is sufficient for the complete suppression of intron D1 splicing and that this suppression is dependent on the sequence present in nucleotides 2731 to 2750 in intron D2 of c-H-ras pre-mRNA.
SC35 and SRp40 antagonize the inhibitory effect of hnRNP A1 on intron D1 splicing.
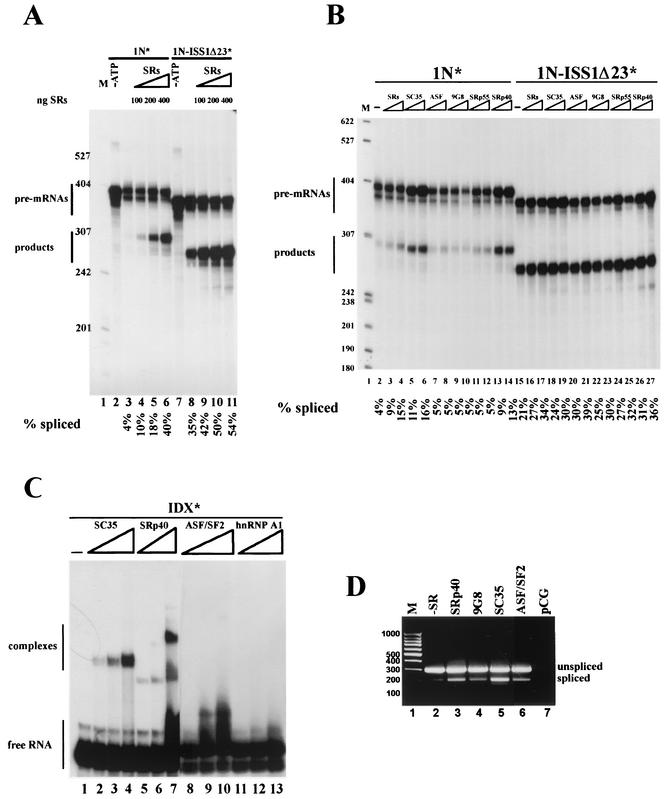
SR proteins were demonstrated to promote the splicing of 1N pre-mRNA in an effective way in the A1-depleted nuclear extract (Fig. 4C, lane 7), but we also wanted to test whether this effect could be seen in a nondepleted extract. When increasing amounts of native purified SR proteins were added to the splicing mix, the reaction was considerably enhanced, as can be determined by the amount of product rendered (Fig. 5A, compare lanes 4 to 6 with lane 3). Native SR proteins were also capable of increasing the splicing of 1N-ISS1Δ23 pre-mRNA (Fig. 5A, compare lanes 9 to 11 with lane 8).
FIG. 5.
SC35 and SRp40 activate the splicing of 1N pre-mRNA in vitro and in vivo. (A) In vitro splicing of 1N and 1N-ISS1Δ23 pre-mRNAs with increasing amounts of total HeLa native SR proteins. Lanes 4 to 6 and 9 to 11, SRs (100, 200, and 400 ng) were assayed; lanes 2 and 7, control reactions without ATP; lanes 3 and 8, control reactions without additional SRs; lane 1, size markers (M). Quantitation of each reaction is shown below each lane. (B) In vitro splicing assays of 1N and 1N-ISS1Δ23 pre-mRNAs with total or individual baculovirus-expressed SR proteins. Lanes 3 and 16, 100 ng of total native SRs added to the splicing reaction; lanes 4 and 17, 200 ng of total native SRs added to the splicing reaction; all other lanes, 8 and 16 pmol of each recombinant baculovirus-expressed SR protein assayed in each couple of reactions, with the identity of theproteins indicated above each lane; lanes 2 and 15, no extra SR proteins added; lane 1, size markers (M). The quantitation of each reaction is shown below each lane. (C) Gel shift assay performed with homogeneously labeled IDX RNA and increasing amounts (3, 6, and 12 pmol) of recombinant baculovirus expressed SC35 (lanes 2 to 4), SRp40 (lanes 5 to 7), ASF/SF2 (lanes 8 to 10), and hnRNP A1 (lanes 11 to 13). Lane 1, labeled RNA only. (D) RT-PCR analysis of total RNA extracted from HeLa cells transiently cotransfected with pCG carrying the different SR proteins shown (lanes 3 to 6) and pcDNA3 carrying 1N minigene; lane 1, molecular weight markers (M); lane 2, control cotransfection with the empty vector pCG; lane 7, control transfection with vector pCG alone. We have checked that all constructs express significant amounts of each SR protein by Western blot analysis, using an anti-T7 tag antibody, of the HeLa cell lysates transiently transfected with the T7-tagged SR proteins (not shown).
To check if all individual SR proteins had the same ability to overcome the suppressing effect of rasISS1 on intron D1, we tested separately the baculovirus-expressed recombinant proteins SC35, ASF/SF2, 9G8, SRp55, and SRp40 on the in vitro splicing of 1N and 1N-ISS1Δ23 substrates (Fig. 5B). Surprisingly, although all of the SR proteins could enhance the splicing of 1N-ISS1Δ23 to various degrees, only SC35 and SRp40 could efficiently improve the splicing of 1N (Fig. 5B, see lanes 5 and 6 and lanes 13 and 14). This indicates a sequence specificity requirement for the action of SR proteins in 1N substrate. In addition, as all the SR proteins tested activated intron D1 splicing when nucleotides 2731 to 2750 were deleted (ISS1Δ23), this indicates that the ability of SC35 and SRp40 to activate splicing of 1N is not mediated at a general level (i.e., a global enhancement of splicing) but most likely is mediated by a specific counteraction of rasISS1 present in intron D2. Presumably, and because it has been shown above that hnRNP A1 acts as a D1 splicing repressor through binding to rasISS1, SC35 and SRp40 might enhance 1N splicing due to their ability to antagonize hnRNP A1 action.
Since SR proteins have been shown to bind mainly to exonic sequences (61), we next checked whether SC35 and SRp40 could bind to exon IDX in gel shift assays. Using increasing amounts of SC35 or SRp40 resulted in a mobility shift of the RNA and the formation of a complex (Fig. 5C, compare lanes 2 to 4 and 5 to 7 with lane 1). In contrast, no detectable complex was formed with ASF/SF2 or GST-hnRNP A1 (Fig. 5C, lanes 8 to 10 and 11 to 13, respectively), indicating that none of these proteins can bind to IDX RNA, at least under the same conditions as SC35 and SRp40 can. These data agree with the above demonstration that SC35 and SRp40, but not ASF/SF2, are able to counteract the splicing inhibition of hnRNP A1. In addition, overexpression in HeLa cells of the T7-tagged SRp40, 9G8, SC35, or ASF/SF2 proteins, together with the 1N minigene, resulted in a similar pattern of action, as SRp40 and SC35 were the most efficient SR species in the inclusion of IDX exon, as assessed by RT-PCR assays (Fig. 5D, compare lanes 3 and 5 to lane 2). As the results described above indicate that IDX and rasISS1 may interact with each other, we searched for other factors binding to IDX and/or rasISS1, which could mediate a bridge bringing these sequences together. Using the RNA affinity purification approach, we identified two pairs of protein bands that are both present in IDX and rasISS1 RNAs but not in ISS1Δ23 RNA (Fig. 6A, compare lane 3 with lanes 2 and 4). After microsequencing, three proteins were identified (Fig. 6B), namely, FUS/TLS, the ATP-dependent RNA helicase p68, and hnRNP H. We confirmed the presence of these three proteins and also of hnRNP A1 by Western blotting (Fig. 6C). While FUS/TLS appeared with all three RNAs, indicating a nonspecific interaction, p68 helicase was present only with ISS1 and IDX samples, pointing to a possible role for this protein in D1 splicing regulation (see the upper panels in Fig. 6C). hnRNP H showed a pattern similar to that of hnRNP A1, being much more abundant in the ISS1 sample than in the control ISS1Δ23 and being absent with IDX RNA (see the lower panels in Fig. 6C).
In vivo suppression of RNA helicase p68 promotes the alternative splicing of c-H-ras pre-mRNA.
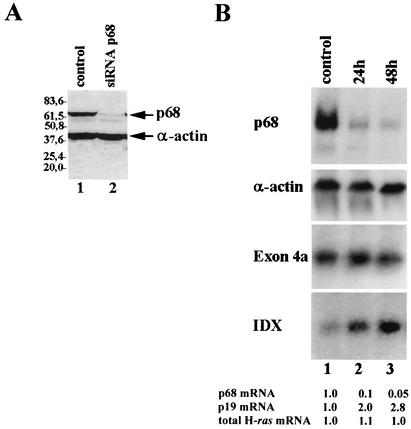
Since the binding of p68 to ISS1 but not to ISS1Δ23 (which lacks the core of the intronic inhibitory region) suggested a negative function in splicing regulation, we performed RNA interference experiments on HeLa cells to specifically suppress expression of p68 helicase and checked for its effect on H-ras alternative splicing. We used a plasmid-based system to express short interfering RNA targeted against p68 mRNA (7). Forty-eight hours posttransfection, the p68 level was reduced to less than 10% of the original amount, as assessed by Western blotting (Fig. 7A, compare lane 2 to the control lysate shown in lane 1) or Northern blotting (Fig. 7B, upper panel), and in comparison with α-actin. Interestingly, while the total amount of endogenous H-ras transcripts was unaffected (as seen with a probe directed against exon 4a, which detects both p21 and p19H-ras mRNAs), p19H-ras mRNA suffers a nearly threefold increase (as seen with the specific probe against IDX exon), indicating that p68 helicase has a negative role in IDX selection. This finding, together with the previous observations that IDX and ISS1 might act in concert, prompted us to perform a prediction of the secondary structure of IDX linked to rasISS1. As shown in Fig. 8B, the Mfold program (51) predicts a long interaction between IDX and rasISS1 that might indicate that rasISS1 traps the IDX structure in a long stem-loop. Furthermore, this trapping structure might be conserved from mice to humans; however, the mouse structure is less stable than that of other species (Fig. 8B, see the legend). Finally, we wanted to test whether the whole RNA sequence IDXISS1 could compete with any inhibitory factor(s) regulating D1 splicing in our in vitro system. As shown in Fig. 8C, only the joined IDXISS1 sequence could promote an increase in the splicing efficiency when added in excess to the reaction, whereas IDX or ISS1 sequences added separately could not (Fig. 8C, compare lanes 10 to 13 with lanes 2 to 5 or lanes 6 to 9), strengthening the role of a possible secondary structure in alternative splicing regulation.
FIG. 7.
RNA interference of p68 RNA helicase in HeLa cells promotes an increase in p19H-ras mRNA. (A) Western blot analysis of HeLa cell lysates transiently expressing the plasmid pSUPER-p68 (lane 2) or the empty vector (lane 1). Cells were harvested 48 h posttransfection, and the blot was probed with the monoclonal antibodies PAb204 (anti-p68) and Sigma (mouse anti-α-actin) as a loading control. (B) Northern blot analysis of total RNA (5 μg) from HeLa cells transiently transfected with construct pSUPER-p68 (lanes 2 and 3) or vector alone (lane 1). RNA was extracted 24 h (lane 2) or 48 h (lanes 1 and 3) posttransfection. The identity of each probe used is described on the right. mRNA levels shown below were determined by normalizing against the levels of α-actin mRNA, which was assessed in the same blot. The level of each mRNA in the control sample was arbitrarily given a value of 1.
FIG.8.
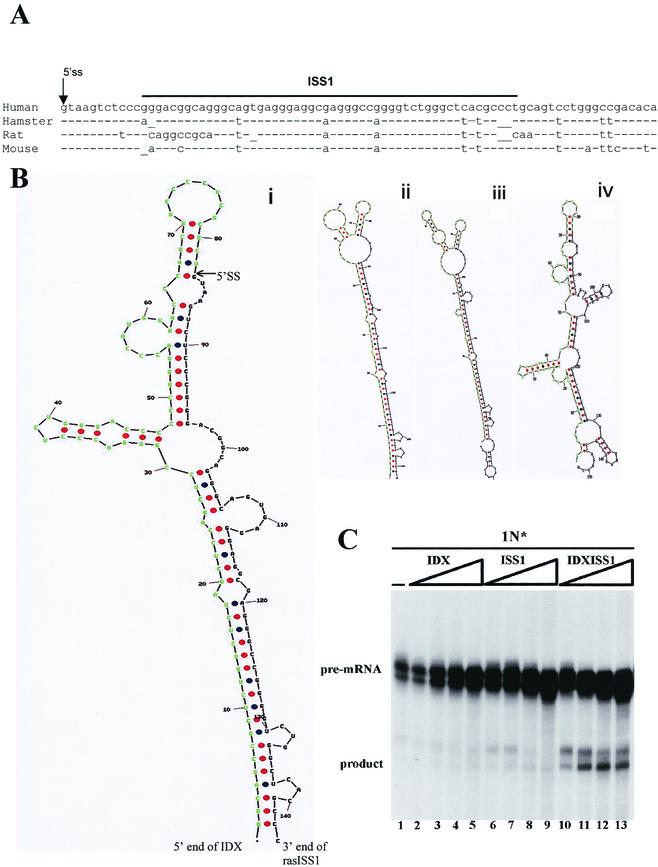
IDX and ISS1 act in concert to repress 1N splicing. (A) Conservation of rasISS1 sequences across species. The alignment and comparison of rasISS1 sequences from human (ACN V00574), hamster (ACN M84166.1), rat (ACN M13011), and mouse (AF081118) sources was performed with MacMolly Tetra version 3.10. (B) Prediction of the secondary structure of IDX-rasISS1 obtained from the same species and folded by the Mfold program, version 3.1. It is hypothesized that an IDX sequence is seized by a long interaction with the intronic rasISS1. Shown are the IDX sequence (green) and contiguous and downstream 5′ SS and rasISS1 (black). Panels show human (i) (ΔG = −63.7 kcal/mol), hamster (ii) (ΔG = −61.4 kcal/mol), rat (iii) (ΔG = −62.4 kcal/mol), and mouse (iv) (ΔG = −56.5 kcal/mol) structures. (C) Competing IDXISS1 RNA can restore splicing activity of 1N substrate. In vitro splicing assay of 1N pre-mRNA was performed in the presence of 5, 10, 20, or 40 pmol of cold IDX RNA (lanes 2 to 5), ISS1 RNA (lanes 6 to 9), or IDXISS1 RNA (lanes 10 to 13).
DISCUSSION
Here, we demonstrate that an intron silencer sequence, ras ISS1, acts as a negative regulator of IDX inclusion, mainly by preventing its 3′SS from being properly recognized, an effect that is clearly seen in the single-intron substrates, where ras ISS1 inhibits splicing of intron D1 completely (Fig. 1) but not the splicing of intron D2. Interestingly, rasISS1 is conserved from mice to humans (Fig. 8A). By means of nuclear extract depletion and add-back experiments, hnRNP A1 has been identified as one factor mediating the rasISS1 negative effect. The amino-terminal form of hnRNP A1, UP1, containing the RNA-binding domain, formed two defined bands when interacting with rasISS1 (Fig. 4), suggesting that two binding sites for A1 might be present in this RNA. Some RNA-binding sequences for hnRNP A1 have been previously described, namely, CUAGACUAGA in the ESS and AUAGAAGAAGAA in the Janus regulator of HIV tat exon 2 (10, 49); UACCUUUAGAGUAGG in the ISS of human hnRNP A1 pre-mRNA, which modulates 5′SS selection (17); UUAGAUUAGA in the mouse hepatitis virus RNA transcription regulatory region (44); and UAGGGCAGGC in an ESS in K-SAM exon of human FGF receptor 2 (23). It is remarkable that together with the SELEX winner sequence identified previously (8), all of these hnRNP A1-binding sequences contain unique or tandem repeats of the sequence UAG(G/A) at their core, unlike the rasISS1 core sequence reported in this work (GGCAGUGAGGGAGGCGAGGG). The whole rasISS1 sequence, however, rendered a gel shift interaction with recombinant hnRNP A1 (Fig. 4), most likely through the binding to the three AGG motifs, which match three residues of the core A1 binding sequence. It is therefore clear that the spectrum of RNA sequences binding to A1 is broad, and only functional studies like these are capable of identifying new hnRNP A1 interactive elements.
To date, there have been three proposed mechanisms for inhibition by hnRNPs: (i) a direct competition for overlapping enhancer-binding sites that are recognized by stimulatory factors, which prevent SS recognition (48); (ii) the binding of hnRNPs to several sites around or within a silenced exon, followed by dimerization of the bound proteins, which might cause relevant portions of the pre-mRNA to loop out, which would make them unavailable for splicing, as happens in hnRNP A1 pre-mRNA (3) and the FGF receptor 2 pre-mRNA (14); and (iii) initial binding of hnRNP A1 to a high-affinity site on an exon and then nucleating the cooperative assembly of inhibitory hnRNP complexes that coat the pre-mRNA. In this final case, the cooperative binding of hnRNP A1 molecules interferes directly with the initial steps of spliceosome assembly or antagonizes the action of nearby enhancers (49, 66). In our c-H-ras system, although deletion of the whole rasISS1 sequence results in efficient splicing of the 1N substrate, we cannot exclude the possibility that in the complete pre-mRNA, some overlapping between enhancer and inhibitory sites might be present. In fact, hnRNP H has two putative binding sequences within rasISS1 (GGGA), indicating possible competition between hnRNPs H and A1 for this site. In addition, and although we have not detected binding of hnRNP A1 to other sites surrounding or within exon IDX, the formation of a stable stem-loop trapping IDX by rasISS1 cannot be completely ruled out.
We have shown that SC35 and SRp40 can antagonize the inhibitory action of hnRNP A1 on IDX inclusion, presumably through binding to ESEs within IDX. The consensus heptamer GUUC(G/C)AG described previously (16, 59) as putative binding sites for SC35 is observed in IDX (as GCUCCAG). Further, the functional consensus GRYY(C/A)CYR described by Liu et al. (45) is also observed at three different IDX positions (GACCCCCC, GACCCAUG, and GGCCCCUC). The heptamer consensus UGGGAGC described by Tacke et al. (57) as a putative binding site for SRp40 is also observed at three different IDX positions (UGGGACC, CGGGACC, and UGUGACC). Two distinct models have been proposed for the stimulation of splicing by ESE elements. While early studies supported a U2AF recruitment model according to which ESE-bound SR proteins facilitate U2AF recruitment to the polypyrimidine tract and 3′SS by RS domain-mediated interactions (31), another study suggests that the primary function of ESE elements is to somehow antagonize the effect of a juxtaposed splicing silencer (4). From our study and others (18), both functions could be present. SR proteins could stimulate the assembly of splicing factors and thus increase the splicing efficiency (in a RNA binding-dependent or -independent manner, therefore affecting 1N-ISS1Δ23 pre-mRNA), and as seen for the 1N substrate, they could bind to an ESE and counteract a nearby splicing repressor complex consisting of at least hnRNP A1. Our results, then, reveal the first mammalian gene which can be submitted to specific action of the antagonistic regulation of A1 and SR proteins.
We have also isolated three more proteins, namely, FUS/TLS, hnRNP H, and the ATP-dependent RNA helicase p68, that are linked to rasISS1 and/or IDX RNAs and which may be bridging the intronic and exonic sequences. FUS/TLS (also known as hnRNP P2) has been connected to RNA maturation (64), among other nuclear processes. Its consensus RNA-binding sequence found by SELEX contains a conserved GGUG motif (43), which is not present in either IDX or rasISS1. This points to a degeneration in its RNA binding sequences (as is common for other hnRNPs) or, alternatively, to an interaction with IDX mediated by other proteins. Interestingly, FUS/TLS has been found to bind to SC35 (64) and also hnRNP A1 (67), which we have demonstrated to be implicated in the regulation of IDX inclusion. hnRNP H was recently shown to regulate the inclusion of a regulated exon in the env gene of HIV-1 in combination with SC35 and other SR proteins, through its binding to a GGGA motif (11). In addition to the GGGA sequences in rasISS1, three other GGGA motifs are present in the IDX, suggesting that hnRNP H might play an important role in the alternative splicing of the IDX. Finally, p68 is a well-characterized ATP-dependent ATPase and RNA helicase, but its function and substrates are not well understood. A recent work has identified p68 as the unwinding factor necessary to destabilize U1snRNA-5′SS interaction prior to the first catalytic step of splicing (46), but until now, no involvement of p68 helicase in alternative splicing regulation has been reported. Therefore, as far as we know, we have for the first time described a function for p68 in splicing control. Accordingly, it has also been shown that the closely related p72 RNA helicase plays a role in alternative splicing regulation of CD44 pre-mRNA (34). The mechanism by which p68 helicase can inhibit IDX recognition and splicing remains obscure. One important observation is that rasISS1 exerts its function in the presence of IDX; when another exonic sequence like c-H-ras exon 3 or exon 4a replaces IDX, rasISS1 is not active. In addition, we have demonstrated that the sequence IDXISS1, but not IDX or ISS1 separately, is capable of restoring splicing efficiency in vitro when present in competing amounts, reinforcing the plausibility of a secondary structure encompassing both the exonic and intronic regions. In this context, it is tempting to hypothesize the existence of a strong binding site for a helicase which could stabilize and destabilize the structure and thereby exert its function in splicing regulation. One attractive hypothesis is that IDX and rasISS1 might form several secondary structures that regulate this alternative splicing, with p68 helicase then being a factor that unwinds a more active secondary structure to a less active secondary structure (in terms of IDX inclusion).
Further analysis of the action and interplay between all of these regulatory factors and secondary structure elements will be necessary to elucidate the fine regulation of IDX inclusion in mature transcripts and, consequently, the expression of the alternative protein p19H-Ras.
Acknowledgments
This work was supported by Fundación Ramón Areces and MCyT grants PB98-0642 and BMC2002-03282, but it was started with a grant from the Asociación Española contra el Cáncer and La Marató de TV3. This work was supported by funds from the CNRS, the INSERM, and the Association pour la Recherche sur le Cancer. S.G. was a recipient of a BEFI fellowship.
We thank the staff of the IGBMC facilities for their assistance. We thank Ruth Willmott for revising the manuscript.
REFERENCES
- 1.Achsel, T., and Y. Shimura. 1996. Factors involved in the activation of pre-mRNA splicing from downstream splicing enhancers. J. Biochem. 120:53-60. [DOI] [PubMed] [Google Scholar]
- 2.Bai, Y. D., D. Lee, T. D. Yu, and L. A. Chasin. 1999. Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 27:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 5.Blencowe, B. J., R. Issner, J. A. Nickerson, and P. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgeois, C. F., M. Popielarz, G. Hildwein, and J. Stevenin. 1999. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol. 19:7347-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 10.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlo, T., R. Sierra, and S. M. Berget. 2000. A 5′ splice site-proximal enhancer binds SF1 and activates exon bridging of a microexon. Mol. Cell. Biol. 20:3988-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrascal, M., S. Carujo, O. Bachs, and J. Abian. 2002. Identification of p21Cip1 binding proteins by gel electrophoresis and capillary liquid chromatography microelectrospray tandem mass spectrometry. Proteomics 2:455-468. [DOI] [PubMed] [Google Scholar]
- 14.Carstens, R. P., E. J. Wagner, and M. A. Garcia-Blanco. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7388-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartegni, L., M. Maconi, E. Morandi, F. Cobianchi, S. Riva, and G. Biamonti. 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259:337-348. [DOI] [PubMed] [Google Scholar]
- 16.Cavaloc, Y., C. F. Bourgeois, L. Kister, and J. Stevenin. 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5:468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chabot, B., M. Blanchette, I. Lapierre, and H. LaBranche. 1997. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 17:1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chew, S. L., H. X. Liu, A. Mayeda, and A. R. Krainer. 1999. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96:10655-10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codony, C., S. Guil, C. Caudevilla, D. Serra, G. Asins, A. Graessmann, F. G. Hegardt, and M. Bach-Elias. 2001. Modulation in vitro of H-ras oncogene expression by trans-splicing. Oncogene 20:3683-3694. [DOI] [PubMed] [Google Scholar]
- 20.Cohen, J. B., S. D. Broz, and A. D. Levinson. 1989. Expression of the H-ras proto-oncogene is controlled by alternative splicing. Cell 58:461-472. [DOI] [PubMed] [Google Scholar]
- 21.Cohen, J. B., S. D. Broz, and A. D. Levinson. 1993. U1 small nuclear RNAs with altered specificity can be stably expressed in mammalian cells and promote permanent changes in pre-mRNA splicing. Mol. Cell. Biol. 13:2666-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Gatto, F., and R. Breathnach. 1995. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 15:4825-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dirksen, W. P., R. K. Hampson, Q. Sun, and F. M. Rottman. 1994. A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J. Biol. Chem. 269:6431-6436. [PubMed] [Google Scholar]
- 25.Dreyfuss, G., M. J. Matunis, S. Pinolroma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of messenger RNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 26.Dye, D. T., M. Buvoli, S. A. Mayer, C. H. Lin, and J. G. Patton. 1998. Enhancer elements activate the weak 3′ splice site of α-tropomyosin exon 2. RNA 4:1523-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eperon, I. C., D. C. Ireland, R. A. Smith, A. Mayeda, and A. R. Krainer. 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 12:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eperon, I. C., O. V. Makarova, A. Mayeda, S. H. Munroe, J. F. Caceres, D. G. Hayward, and A. R. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 30.Gallego, M. E., R. Gattoni, J. Stevenin, J. Marie, and A. Expert-Bezancon. 1997. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the β-tropomyosin alternative exon 6A. EMBO J. 16:1772-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo, W., G. J. Mulligan, S. Wormsley, and D. M. Helfman. 1991. Alternative splicing of β-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 5:2096-2107. [DOI] [PubMed] [Google Scholar]
- 33.Hertel, K. J., and T. Maniatis. 1998. The function of multisite splicing enhancers. Mol. Cell 1:449-455. [DOI] [PubMed] [Google Scholar]
- 34.Honig, A., D. Auboeuf, M. M. Parker, B. W. O'Malley, and S. M. Berget. 2002. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 22:5698-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huh, G. S., and R. O. Hynes. 1993. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol. Cell. Biol. 13:5301-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey, M. B., J. Bryan, T. A. Cooper, and S. M. Berget. 1995. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol. Cell. Biol. 15:3979-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen, K. B., K. Musunuru, H. A. Lewis, S. K. Burley, and R. B. Darnell. 2000. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA 97:5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin, W., E. S. Huang, W. Bi, and G. J. Cote. 1998. Exon sequence is required for regulated RNA splicing of the human fibroblast growth factor receptor-1 α-exon. J. Biol. Chem. 273:16170-16176. [DOI] [PubMed] [Google Scholar]
- 39.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Luhrmann, M. A. Garciablanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 40.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 41.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 42.Lavigueur, A., H. Labranche, A. R. Kornblihtt, and B. Chabot. 1993. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with Sr proteins and stimulates U2-snRNP binding. Genes Dev. 7:2405-2417. [DOI] [PubMed] [Google Scholar]
- 43.Lerga, A., M. Hallier, L. Delva, C. Orvain, I. Gallais, J. Marie, and F. Moreau-Gachelin. 2001. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 276:6807-6816. [DOI] [PubMed] [Google Scholar]
- 44.Li, H. P., X. Zhang, R. Duncan, L. Comai, and M. M. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. USA 94:9544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, H. X., S. L. Chew, L. Cartegni, M. Q. Zhang, and A. R. Krainer. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, Z. R. 2002. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 48.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchand, V., A. Mereau, S. Jacquenet, D. Thomas, A. Mougin, R. Gattoni, J. Stevenin, and C. Branlant. 2002. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323:629-652. [DOI] [PubMed] [Google Scholar]
- 50.Marshall, C. J. 1996. Ras effectors. Curr. Opin. Cell Biol. 8:197-204. [DOI] [PubMed] [Google Scholar]
- 51.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 52.Mayeda, A., D. M. Helfman, and A. R. Krainer. 1993. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol. 13:2993-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-messenger-RNA splicing by hnRNP-A1 and splicing factor-SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 54.Min, H. S., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 55.Stoss, O., M. Olbrich, A. M. Hartmann, H. Konig, J. Memmott, A. Andreadis, and S. Stamm. 2001. The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J. Biol. Chem. 276:8665-8673. [DOI] [PubMed] [Google Scholar]
- 56.Sun, Q., A. Mayeda, R. K. Hampson, A. R. Krainer, and F. M. Rottman. 1993. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 7:2598-2608. [DOI] [PubMed] [Google Scholar]
- 57.Tacke, R., Y. Chen, and J. L. Manley. 1997. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl. Acad. Sci. USA 94:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tacke, R., and J. L. Manley. 1999. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11:358-362. [DOI] [PubMed] [Google Scholar]
- 59.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, Z., H. M. Hoffmann, and P. J. Grabowski. 1995. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA 1:21-35. [PMC free article] [PubMed] [Google Scholar]
- 61.Watakabe, A., K. Tanaka, and Y. Shimura. 1993. The role of exon sequences in splice site selection. Genes Dev. 7:407-418. [DOI] [PubMed] [Google Scholar]
- 62.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 63.Wu, J. I., R. B. Reed, P. J. Grabowski, and K. Artzt. 2002. Function of quaking in myelination: regulation of alternative splicing. Proc. Natl. Acad. Sci. USA 99:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, L., L. J. Embree, S. Tsai, and D. D. Hickstein. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 273:27761-27764. [DOI] [PubMed] [Google Scholar]
- 65.Zahler, A. M. 1999. Purification of SR protein splicing factors. Methods Mol. Biol. 118:419-432. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 67.Zinszner, H., R. Albalat, and D. Ron. 1994. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 8:2513-2526. [DOI] [PubMed] [Google Scholar]