Abstract
The septins are a family of cytoskeletal proteins present in animal and fungal cells. They were first identified for their essential role in cytokinesis, but more recently, they have been found to play an important role in many cellular processes, including bud site selection, chitin deposition, cell compartmentalization, and exocytosis. Septin proteins self-associate into filamentous structures that, in yeast cells, form a cortical ring at the mother bud neck. Members of the septin family share common structural domains: a GTPase domain in the central region of the protein, a stretch of basic residues at the amino terminus, and a predicted coiled-coil domain at the carboxy terminus. We have studied the role of each domain in the Saccharomyces cerevisiae septin Cdc11 and found that the three domains are responsible for distinct and sometimes overlapping functions. All three domains are important for proper localization and function in cytokinesis and morphogenesis. The basic region was found to bind the phosphoinositides phosphatidylinositol 4-phosphate and phosphatidylinositol 5-phosphate. The coiled-coil domain is important for interaction with Cdc3 and Bem4. The GTPase domain is involved in Cdc11-septin interaction and targeting to the mother bud neck. Surprisingly, GTP binding appears to be dispensable for Cdc11 function, localization, and lipid binding. Thus, we find that septins are multifunctional proteins with specific domains involved in distinct molecular interactions required for assembly, localization, and function within the cell.
Septins are highly conserved cytoskeletal elements found in fungi, mammals, and all eukaryotes examined thus far, with the exception of plants. The septin proteins assemble into filaments that lie underneath the plasma membrane (5, 16, 20). In Saccharomyces cerevisiae, where they were first identified, septins are visible as electron-dense cortical rings at the mother bud neck (5). In multicellular organisms, they are found at the cleavage furrow and other cortical locations (14, 34, 48, 63). Consistent with their localization, septins have been shown to be required for cytokinesis in yeast, Drosophila melanogaster, and mammalian cells (14, 26, 35).
Recent evidence in yeast has demonstrated that septins participate in a variety of other cellular processes, including cell morphogenesis (19, 25, 32), bud site selection (6, 18, 56), chitin deposition (9), cell cycle regulation (4, 40), cell compartmentalization (3, 36a), and spore wall formation (13, 61). In mammalian cells, septins are involved in exocytosis (31). Since septins participate in many cellular processes, it is not surprising that a diverse set of proteins have been found associated with the yeast septin cytoskeleton (12, 21, 28). The precise manner in which these proteins interact with septins at a molecular level has not been investigated previously.
The septins have a highly conserved structure. They contain a central GTP-binding domain flanked by a basic region at the amino terminus, and most septins contain a coiled-coil domain at the carboxy terminus (15). In mammalian cells, the function of the basic region is to bind phosphoinositides. In vitro experiments have shown that the basic region of the H5 septin protein was able to specifically bind phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] and phosphatidylinositol (3,4,5)-triphosphate [PI(3,4,5)P3]; mutation of the basic residues abolished this binding (66). The role of this domain has not been analyzed in vivo, and the functions of the GTP-binding and coiled-coil domains have not been studied previously.
We have investigated the in vivo functions and biochemical activities of the different domains of the yeast septin Cdc11, with the goal of understanding how septins assemble, localize, and function within the cell. In yeast, five septins, Cdc3, Cdc10, Cdc11, Cdc12, and Shs1, localize to the mother bud neck in vegetatively growing cells (24, 46). Cdc3 and Cdc12 are essential for growth at all temperatures (38), whereas Cdc10 and Cdc11 are required only at elevated temperatures (18, 20). Shs1 is a nonessential septin (46). Cells containing temperature-sensitive mutations in either CDC3, CDC10, CDC11, or CDC12 delay at a G2 checkpoint (4) and arrest at the restrictive temperature, forming extensive chains of highly elongated cells (25).
In this study, we have made mutations in each of the different domains of Cdc11 (15, 31) and have investigated their effects on Cdc11 function, localization, and activity. We find that the different domains have distinct roles in vivo and in vitro. The basic region is required for binding to phosphatidylinositol 4-phosphate [PI(4)P] and phosphatidylinositol 5-phosphate [PI(5)P] and for localization. The GTP-binding domain is required for septin-septin interactions, localization, and neck targeting, and the carboxy terminus of Cdc11 is important for localization and interactions with Cdc3 and the polarity protein Bem4. Our results provide insight into septin assembly and targeting within the cell and how septins interact with other proteins to carry out their diverse functions.
MATERIALS AND METHODS
Yeast strains.
For morphological and localization studies, we used a haploid cdc11-Δ::HIS3 strain obtained from the diploid strain YAC662 (CDC11/cdc11Δ::HIS3 SWE1/swe1Δ::LEU2 in an S288c background) expressing CDC11 constructs from the CDC11 promoter in the centromeric pRS414 plasmid (7). A pep4-3 ura3-52 yeast strain (47) was used for protein expression. The SEY6210 background (49) was used for the study of stt4-4 and pik1-83 mutants, and the PJ69-4A background (29) was used for the two-hybrid experiments. Yeast media and standard techniques were described previously (51).
Plasmids and recombinant DNA techniques.
Constructs used for morphology studies were prepared in the pRS414 plasmid (7); each construct has 654 bp upstream and 297 bp downstream of the CDC11 open reading frame. The mutations cloned are indicated in Table 1; details of their construction are available upon request. For localization studies, C-terminal fusion constructs of Cdc11 with the green fluorescent protein (GFP) GFP(S35T) (39) in pRS414 were used; these constructs also express the Cdc11 proteins from the endogenous promoter. Expression constructs were cloned into the pEGH plasmid (68), which overexpresses the proteins as six-His double-tagged amino-terminal glutathione S-transferase (GST) fusions from a galactose-inducible promoter. A complete list of those constructs with their relevant features is provided in Table 1. The sequences of the primers used to generate those constructs are available upon request. The different plasmids were transformed into yeast strain YAC662 by the lithium acetate method (23).
TABLE 1.
Plasmids used in this study
| Plasmid type | Plasmid designation | Plasmid construction | Reference or source |
|---|---|---|---|
| 2μm | pAC100 | pEGH-GST-6xHIS | 68 |
| pAC101 | pEHG-GST-6xHIS-CDC3 | 68 | |
| pAC102 | pEGH-GST-6xHIS-CDC10 | 68 | |
| pAC103 | pEGH-GST-6xHIS-CDC11 | 68 | |
| pAC104 | pEGH-GST-6xHIS-CDC12 | This study | |
| pAC105 | pEGH-GST-6xHIS-CDC11-1-45 | This study | |
| pAC106 | pEGH-GST-6xHIS-CDC11-1-258 | This study | |
| pAC107 | pEGH-GST-6xHIS-CDC11-Δ347-415 | This study | |
| pAC108 | pEGH-GST-6xHIS-cdc11-G29A, G32A, G34A | This study | |
| pAC109 | pEGH-GST-6xHIS-cdc11-R12Q, K13Q, R14Q, K15Q, H16Q | This study | |
| pAC110 | pEGH-GST-6xHIS-cdc11-R12Q, K13Q, R14E, K15Q, H16Q | This study | |
| Centromeric | pAC111 | pRS414-CDC11 | This study |
| pAC112 | pRS414-cdc11-Δ12-16 | This study | |
| pAC113 | pRS414-cdc11-Δ18-20 | This study | |
| pAC114 | pRS414-cdc11-Δ347-415 | This study | |
| pAC115 | pRS414-CDC11-GFP | This study | |
| pAC116 | pRS414-cdc11-Δ12-16-GFP | This study | |
| pAC117 | pRS414-cdc11-Δ18-20-GFP | This study | |
| pAC118 | pRS414-cdc11-R12Q, K13Q, R14Q, K15Q, H16Q-GFP | This study | |
| pAC119 | pRS414-cdc11-R12Q, K13Q, R14E, K15Q, H16Q-GFP | This study | |
| pAC120 | pRS414-cdc11-G29A, G32A, G34A-GFP | This study | |
| pAC121 | pRS414-cdc11-R35A-GFP | This study | |
| pAC122 | pRS414-cdc11-Δ347-415-GFP | This study | |
| pAC123 | pRS414-cdc11-G230E-GFP | This study | |
| pAC124 | pRS414-cdc11-N40E-GFP | This study |
Standard recombinant DNA techniques were performed as described previously (52). Yeast genes and gene fragments were amplified by PCR with a 4:1 mix of Taq (Qiagen) and Pfu (Stratagene) polymerases using genomic or cloned DNA. PCR amplification products were gel purified with the GeneClean kit (Bio101 Systems) before use. Sequences of all amplified and cloned DNA products, as well as DNA sequences of the mutated clones, were verified by automatic DNA sequencing (W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University).
In order to delete CDC11, the entire CDC11 open reading frame was replaced with the PCR-generated selectable marker HIS3 by the method described previously (41) (details upon request). The swe1-Δ::LEU2 deletion has been described previously (43).
Determination of the sequence of cdc12-1.
To determine the DNA sequence of cdc12-1, the gene was amplified by PCR from genomic DNA and cloned into the pCR2.1-TOPO plasmid (Invitrogen). Six independent clones were sequenced. All clones differed in only one nucleotide from the corresponding CDC12 annotated sequence, causing a change in one amino acid. The Gly227 present in Cdc12 (encoded by a GGG codon) was replaced by Glu (GAG codon in cdc12-1). This is an important change, since it introduces a negative charge in a conserved region of the septin protein.
Preparation of the yeast GST-septin fusions.
To purify the yeast proteins Cdc3, Cdc10, Cdc11, and Cdc12, 50-ml cultures of the pep4 strain carrying the described constructs in the pEGH vector were grown in synthetic complete medium (SI) without uracil with raffinose as the primary carbon source for 7 h at 30°C. Protein expression was induced at an optical density at 600 nm of 1.0 by adding 5.5 ml of a filtered aqueous solution containing 40% (wt/vol) galactose. After 4 h of growth at 30°C, yeast cells were collected by centrifugation and washed two times in cold water, and yeast cell pellets were kept frozen at −70°C.
Fusion proteins were purified at 4°C as follows. Five hundred microliters of cold yeast protein purification lysis buffer (50 mM Tris-Cl [pH 7.4] 1 mM EGTA, 0.1% [vol/vol] Triton X-100, 0.1% [vol/vol] β-mercaptoethanol, complete protease inhibitor cocktail [Roche]) was added to a frozen yeast cell pellet together with 300 μl of cold zircon beads (0.5-mm diameter). Cells were lysed by 2 min of vigorous shaking at 4°C. After lysis, samples were centrifuged for 1 min at 13,000 × g, and the protein extract was kept on ice. Five hundred microliters of fresh lysis buffer was added to the cell pellet, and the lysis and spin process was repeated four more times. Extracts obtained were pooled and cleared by centrifugation at 13,000 × g for 10 min at 4°C. The supernatant was mixed with 400 μl of Sepharose-glutathione beads (Pharmacia) equilibrated in lysis buffer, and the volume of the suspension was increased to 10 ml by adding fresh cold lysis buffer. The binding incubation was performed for 1 h at 4°C with rocking. Proteins bound to the Sepharose-glutathione beads were washed 3 times with lysis buffer containing 0.5 M NaCl, 10 times with wash buffer (50 mM Tris-Cl [pH 7.4], 0.1 mM EGTA, 0.1% [vol/vol] β-mercaptoethanol), and 3 times with wash buffer containing 0.6 M sucrose and kept in aliquots at −70°C. Fusion proteins were eluted from Sepharose-glutathione beads immediately before each experiment by the addition of 1/10 volume of 0.2 M glutathione to the Sepharose-glutathione suspension equilibrated in the appropriate buffer. After incubation for 5 min on ice, purified protein was separated from the Sepharose-glutathione beads by centrifugation for 1 min at 13,000 × g. The elution process was repeated one more time. To verify that endogenous yeast septins were not present in the purified GST-septin products, we tested the absence of endogenous Cdc11 in all the purified GST-septins by Western blot analysis, using commercial anti-Cdc11 antibody (Santa Cruz Biotech).
GTP-binding assay.
To test the binding of GTP to GST-Cdc11 and GST-Cdc11G29A, G32A, G34A, we used the method described by Field et al. (16). Briefly, Sepharose-glutathione beads containing 2 μg of each protein were washed in reaction buffer (50 mM KCl, 20 mM HEPES [pH 7.0], 1 mM EGTA, 3 mM MgCl2, 1 mM dithiothreitol), and the protein eluted in 320 μl of the same buffer containing 20 mM glutathione. Three 80-μl aliquots were incubated in the presence of 5 μCi of [α-32P]GTP along with 2 μM cold GTP at 23°C for 15 h. The protein present in each reaction mixture was filtered through a nitrocellulose membrane (Hybond-C Extra; Amersham) using a manifold filtration device, and the membrane was washed several times with cold reaction buffer before detection of the [α-32P]GTP bound by autoradiography. A fourth aliquot was used to quantify, by immunoblot analysis, the GST fusion protein present in the reaction mixtures using polyclonal anti-GST antibody (Upstate Biotechnology). As positive and negative controls, the GTP binding of Cdc42 and GST, respectively, were also analyzed (Cdc42 data are not shown).
Phosphoinositide-binding assays.
The binding of recombinant proteins to lipids was detected by protein lipid overlay assays (8, 11). We used commercial strips of nitrocellulose (phosphatidylinositol phosphate [PIP] strips) purchased from Echelon (Salt Lake City, Utah), in which 100 pmol of each phosphoinositide were immobilized. Membranes were blocked for 60 min at room temperature in binding buffer containing 10 mM Tris (pH 7.4), 150 mM NaCl, 3% bovine serum albumin (fatty acid-free), and 0.1% (vol/vol) Tween 20. GST fusion proteins were eluted from glutathione-Sepharose beads immediately before each experiment by incubation with a final concentration of 20 mM glutathione in binding buffer. About 2 μg of each recombinant GST fusion protein was incubated separately overnight at 4°C with a PIP strip in 3 ml of buffer. After the strip was washed, GST was visualized by conventional Western blotting using a polyclonal rabbit anti-GST antibody (Upstate Biotechnology).
For the protein pull-down assays, we used PI(4)P-bound agarose beads, purchased from Echelon, equilibrated in binding buffer (10 mM HEPES [pH 7.0], 150 mM NaCl, 0.5% [vol/vol] Nonidet P-40). Purified recombinant GST proteins were eluted from the gluthathione-Sepharose beads that had been equilibrated in binding buffer as described above. About 2 μg of each GST fusion protein was incubated with the PI(4)P-agarose beads overnight at 4°C in the binding buffer. After extensive washes with the binding buffer, 1 volume of 2× Laemmli sample buffer was added to the beads, and the bound protein was eluted by heating the samples to 95°C for 5 min. Eluted proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blotting using the polyclonal rabbit anti-GST antibody.
To test the effects of GDP and γ-S-labeled GTP on binding of GST-Cdc11 to phosphoinositides, we used 10 μg of purified protein in each experiment and the methods described previously (58, 67). Briefly, GST-Cdc11 bound to glutathione-Sepharose beads was washed three times and resuspended in 100 μl of Tris-buffered saline buffer containing 1 mM EDTA. γ-S-labeled GTP or GDP was added to a final concentration of 100 μM or 1 mM, respectively. The bead suspension was incubated at 30°C for 15 min, and the loading reaction was stopped by addition of MgCl2 to a final concentration of 60 mM. Protein was eluted from the glutathione-Sepharose beads in Tris-buffered saline buffer containing 60 mM MgCl2 and 20 mM glutathione and incubated with blocked PIP strips in the blocking buffer described above containing 60 mM MgCl2.
Two-hybrid analyses.
Yeast two-hybrid analyses were performed using the Gal4-based system (29). The different DNA fragments used for two-hybrid analyses were inserted between the EcoRI and PstI restriction sites of the vectors pGAD-C1 and pGBDU-C1 (29). PCR-amplified DNA fragments containing the region to be expressed flanked by sequence present in the destination vectors were used to cotransform yeast cells together with the EcoRI/PstI-digested destination vectors. The collection of plasmids used for the two-hybrid assays are listed in Table 2. Primers used for the construction of the plasmids are available upon request. The yeast reporter strain PJ69-4α was transformed with the GAL4 activation domain-containing pGAD-C1 vectors (LEU2 marked), whereas the PJ69-4a strain was transformed with the GAL4 DNA-binding domain pGBDU-C1 plasmids (URA3 marked). To test for interactions, diploid cells, resulting from mating of appropriate haploid strains, were isolated and tested for the reporter genes of this system (HIS3, ADE2 and lacZ). In order to test the GAL4-driven transcription activity of the HIS3 reporter, cells from log-phase cultures in synthetic complete (SC) medium minus leucine and uracil were spotted or streaked onto plates of SC medium minus leucine, uracil, and histidine (containing 1 mM 3-aminotriazole where indicated). The spots contained similar cell densities.
TABLE 2.
Plasmids used for the two-hybrid analysesa
| Plasmid designation | Plasmid construction | Reference or source |
|---|---|---|
| pAC200 | pGBDU-C1 | 29 |
| pAC201 | pGBDU-C1-CDC11 | This study |
| pAC202 | pGBDU-C1-CDC3 | This study |
| pAC203 | pGBDU-C1-CDC12 | This study |
| pAC204 | pGBDU-C1-cdc11-347-415 | This study |
| pAC205 | pGBDU-C1-cdc11-Δ347-415 | This study |
| pAC206 | pGBDU-C1-cdc11-1-100 | This study |
| pAC207 | pGBDU-C1-cdc11-101-217 | This study |
| pAC208 | pGBDU-C1-cdc11-218-415 | This study |
| pAC209 | pGBDU-C1-cdc11-Δ347-415, N40E | This study |
| pAC210 | pGBDU-C1-BEM4 | This study |
| pAC250 | pGAD-C1 | 29 |
| pAC251 | pGAD-C1-CDC11 | This study |
| pAC252 | pGAD-C1-cdc11-347-415 | This study |
| pAC253 | pGAD-C1-cdc11-Δ347-415 | This study |
All plasmids were the 2μm type.
Examination of cell morphology and subcellular localization.
Mid-log-phase cells grown for 7 h at the indicated temperatures were fixed for 10 min with 2% formaldehyde. Fixed cells were then washed and resuspended in phosphate-buffered saline solution. Cells were examined by phase-contrast and differential interference microscopy using a Leitz Aristoplan microscope, and the images were captured using a Princeton Micromax charge-coupled device camera and Scanalytics IPLab software. The same cultures were used to examine the localization of the GFP-tagged proteins. Random fields of cells for each sample were visualized under the fluorescence microscope, and the localization of the GFP was recorded and quantified as described previously (53).
RESULTS
Different domains of Cdc11 are important for function.
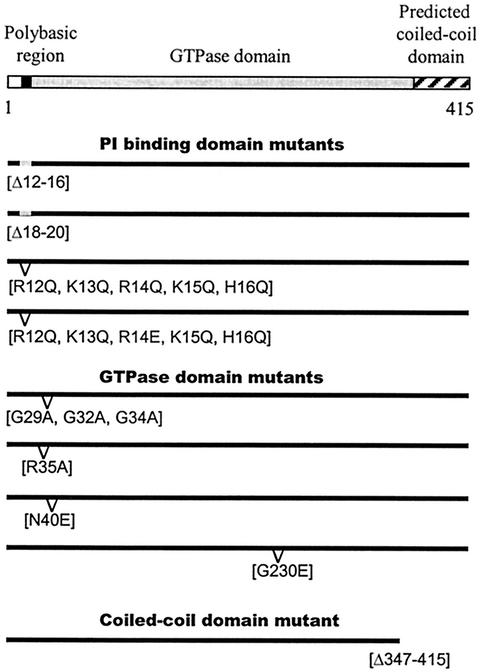
Cdc11 has three domains (Fig. 1): a short region comprised of basic amino acids near the amino terminus (residues 12 to 16 and residues 18 and19), a central domain which is homologous to small GTPases (residues 23 to approximately 259), and a potential coiled-coil domain at the carboxy terminus (residues 360 to 415).
FIG. 1.
Schematic representations of the cdc11 alleles used in this study. cdc11-Δ12-16 (Δ12-16) and cdc11-Δ18-20 (Δ18-20) contain deletions of five and three residues, respectively, located in the polybasic region. The cdc11-R12Q, K13Q, R14Q, K15Q, H16Q mutant (R12Q, K13Q, R14Q, K15Q, H16Q) has the positively charged residues deleted in cdc11-Δ12-16 replaced by the neutral glutamine (Q). cdc11-R12Q, K13Q, R14E, K15Q, H16Q (R12Q, K13Q, R14E, K15Q, H16Q) is similar to the previous mutant but has a glutamic acid (E) in position 14. cdc11-G29A, G32A, G34A (G29A, G32A, G34A) has been mutated in its nucleotide-binding P loop by replacing three glycine residues with alanine. The cdc11-R35A (R35A), cdc1-G230E (G230E), and cdc11-N40E (N40E) alleles have the indicated point mutations, and the coiled-coil domain of Cdc11 has been removed in the cdc11-Δ347-415 (Δ347-415) mutant.
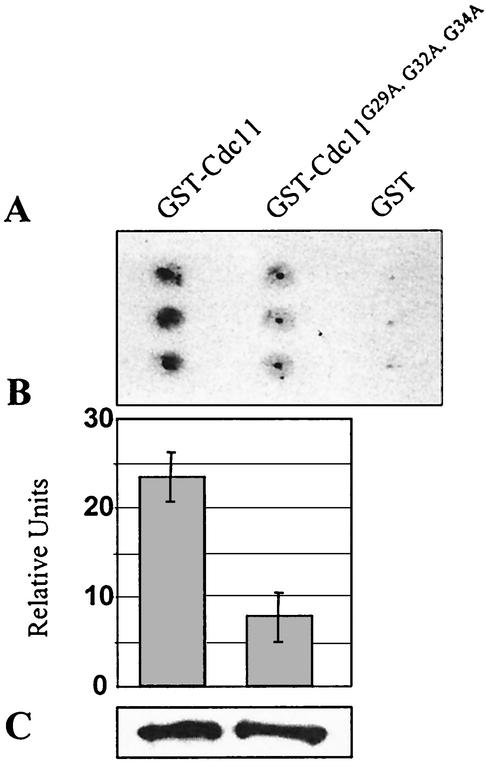
To determine the functions of these different regions, we first prepared a series of mutations that specifically affected each domain (Fig. 1). Two deletion mutations, cdc11-Δ12-16 and cdc11-Δ18-20, which remove different segments of the basic domain were constructed. We also constructed two replacement mutations, cdc11-R12Q, K13Q, R14Q, K15Q, H16Q and cdc11-R12Q, K13Q, R14E, K15Q, H16Q, that substitute the basic residues with either uncharged glutamine residues or glutamine residues plus one acidic residue, respectively. Two strains containing mutations in the conserved residues of the putative P-loop motif, cdc11-G29A, G32A, G34A and cdc11-R35A were also prepared. Analogous mutations in other GTPases prevent GTP binding and nucleotide hydrolysis (22, 36, 54, 64). We tested whether wild-type Cdc11 binds GTP and whether the cdc11-G29A, G32A, G34A allele is defective in binding using in vitro assays. As shown in Fig. 2, the GST-Cdc11 fusion protein binds GTP, but the GST-Cdc11G29A, G32A, G34A exhibited a severe defect in GTP binding. Additional mutations in the GTPase domain were also constructed. A mutant allele, cdc11-G230E, was generated in the GTP-binding domain; the mutation is analogous in position and nature to one that causes a temperature-sensitive growth defect in cdc12 (cdc12-1). An interesting PCR-induced mutation in the GTPase-binding domain (N40E) that appeared in the course of this study was also constructed and analyzed. Finally, we prepared an allele lacking the carboxy-terminal coiled-coil domain, cdc11-Δ347-415.
FIG. 2.
Effect of the mutation Cdc11G29A, G32A, G34A on GTP binding. Equal amounts of purified GST-Cdc11 and GST-Cdc11G29A, G32A, G34A proteins were incubated in triplicate in the presence of [α-32P]GTP before being filtered onto a nitrocellulose membrane. The [α-32P]GTP present on the membrane after washes was detected by autoradiography (A), and the intensity of the spots was quantified (B). The quantity of protein present in each sample was determined by immunodetection, using an anti-GST antibody (C).
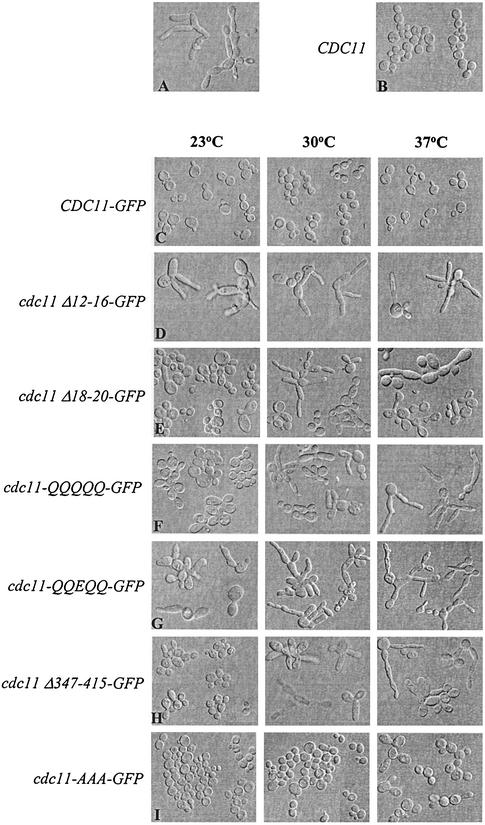
To determine the roles of each region of Cdc11 in vivo, we analyzed the abilities of centromeric plasmids containing the different alleles to complement a cdc11-Δ strain (see Materials and Methods). In order to determine the subcellular locations of the different mutant proteins (described below), the constructs were fused to the GFP coding region and analyzed as Cdc11-GFP fusion proteins. The Cdc11-GFP protein is fully functional. At 23°C, cdc11-Δ strains exhibit severe growth and morphological defects; the cells are highly elongated and are defective in cytokinesis, resulting in chains of elongated cells. cdc11-Δ cells containing the CDC11-GFP allele are indistinguishable from wild-type cells in growth and morphology at all temperatures tested, 23, 30, and 37°C (compare Fig. 3A, B, and C).
FIG. 3.
Effect of expression of different cdc11 alleles in cdc11-Δ cells. Strains were cultured overnight at 23°C, diluted, and then shifted to 23, 30, or 37°C. After 7 h of growth at each temperature, log-phase cells were fixed and the cellular morphology was observed. A representative composite is shown in each case. (A) cdc11-Δ cells (strain YAC660) were cultured for 3 days at 23°C due to its low growth rate. (B) Strain cdc11-Δ expressing Cdc11 from the centromeric vector pRS414 under control of its own promoter (YAC685) was grown at 30°C. (C) cdc11-Δ cells expressing Cdc11-GFP from pRS414 under the control of its own promoter (YAC731); (D) cdc11-Δ cells carrying pRS414-cdc11 Δ12-16-GFP (YAC780); (E) cdc11-Δ cells expressing cdc11-Δ18-20-GFP (YAC728); (F) cdc11-Δ cells carrying pRS414-cdc11-R12Q, K13Q, R14Q, K15Q, H16Q-GFP (YAC804); (G) cdc11-Δ cells expressing cdc11-R12Q, K13Q, R14E, K15Q, H16Q-GFP (YAC808); (H) cdc11-Δ cells expressing cdc11Δ-347-415-GFP (YAC793); (I) cdc11-Δ cells carrying pRS414-cdc11-G29A, G32A, G34A-GFP (YAC715); (J) cdc11-Δ cells carrying pRS414-cdc11-G230E-GFP (YAC704); (K) cdc11-Δ cells expressing cdc11-N40E-GFP (YAC881).
Mutations in the basic region cause loss of CDC11 function. The most severe defect is observed in the cdc11-Δ12-16 strain; this strain fails to complement the morphological defects of the cdc11-Δ strain at all temperatures (Fig. 3D) and grows almost as slowly as the cdc11-Δstrain. cdc11-R12Q, K13Q, R14E, K15Q, H16Q, cdc11Δ18-20 and cdc11-R12Q, K13Q, R14Q, K15Q, H16Q strains exhibit temperature-sensitive defects. At 23°C, cdc11-R12Q, K13Q, R14E, K15Q, H16Q displays moderate defects in morphology and cytokinesis, whereas the defects of cdc11Δ18-20 and cdc11-R12Q, K13Q, R14Q, K15Q, H16Q strains are mild (Fig. 3E, F, and G). At 30°C, the defects are more severe, and at 37°C, the strains are similar to the cdc11-Δ strains. Thus, the basic region is important for Cdc11 function.
Deletion of the coiled-coil domain revealed that it is required for Cdc11 function at elevated temperatures. The cdc11-Δ347-415 strain exhibits only a modest morphological defect at 23°C, with a subset of cells (<15%) exhibiting slightly elongated buds and cytokinesis defects. At 30 and 37°C, the defect is more severe, and all cells are highly elongated and exhibit cytokinesis defects (Fig. 3H). Therefore, the coiled-coil domain is also required for proper Cdc11 function, particularly at higher temperatures.
Mutations in the GTPase domain yielded different effects depending upon the nature of the mutation. Surprisingly, cdc11-G29A, G32A, G34A cells, which are defective in nucleotide binding (Fig. 2), had morphologies and growth rates similar to those of wild-type cells (Fig. 3I). Only modest morphological defects were observed at 37°C. Similar results were obtained when the cdc11-R35A mutation was analyzed (data not shown). The cdc11-G230E mutation produced a temperature-sensitive defect similar to the defect caused by the cdc12-1 allele (25). cdc11-G230E cells were similar to wild-type cells at 23°C but exhibited more-pronounced defects in growth and morphology at both 30 and 37°C; at 37°C, they were similar to cdc11-Δ cells (Fig. 3J). cdc11-N40E cells also exhibit temperature-sensitive morphological defects similar to those of cdc11-G230E cells, except that the defect was not as severe at 37°C (Fig. 3K). These results indicate that sequences in the GTPase domain are important for Cdc11 function, especially at elevated temperatures, but that GTP binding is not required at moderate and low temperatures.
To ensure that these results were not affected by the presence of GFP at the carboxy terminus, we also analyzed the cdc11-Δ12-16, cdc11-Δ18-20, and cdc11-Δ347-415 alleles on a centromeric plasmid without the GFP fusion. Results identical to those found for the GFP fusion alleles were observed (data not shown). Taken together, these results indicate that the basic region of Cdc11 is required for function, the coiled-coil domain and other parts of the GTPase domain are required at the elevated temperatures, and GTP binding is not required for Cdc11 function.
Yeast septin proteins bind PI(4)P and PI(5)P independent of guanine nucleotide.
The yeast septins are associated with the cell periphery, and the mammalian H5 septin has been shown to bind PI(4,5)P2 and phosphatidylinositol (3,4,5)-triphosphate [PI(3,4,5)P3] (66). Therefore, it was reasonable to test whether yeast septins might bind phospholipids, although we expected that septins would not bind PI(3,4,5)P3, since yeast cells lack this phospholipid (10).
We first tested the abilities of GST-Cdc3, GST-Cdc10, GST-Cdc11, and GST-Cdc12 to bind a collection of phosphoinositide lipids spotted onto a membrane. As shown in Fig. 4, all four yeast septins tested bind PI(4)P and PI(5)P and have much lower affinities for PI(3,4)P2, PI(3,5)P2, PI(3,4,5)P3, and PI(3)P. Interestingly, the septins do not significantly bind phosphatidic acid, phosphatidylserine, phosphatidylethanolamine, phosphatidylinositol (PI), lysophosphatidic acid, lysophosphocholine, or sphingosine 1-phosphate. Thus, the four septins specifically bind a subset of phosphoinositides.
FIG. 4.
Analysis of the phosphoinositide-binding properties of yeast septins. The ability of the indicated yeast GST-septins to bind to different lipids was analyzed using a protein lipid overlay assay (11). Abbreviations: S 1-P, sphingosine 1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine; LPA, lysophosphatidic acid; LPC, lysophosphocholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine.
The GDP-bound form of the mammalian H5 septin is required for binding to phospholipids (66). To determine whether guanine nucleotide binding is required for the interaction of Cdc11 with PI(4)P and PI(5)P, we tested the different mutants for their abilities to bind to the different phosphoinositides. The mutant Cdc11 lacking the C-terminal coiled-coil domain, GST-Cdc11Δ347-415, and the GTP-binding site mutant, GST-Cdc11G29A, G32A, G34A each bound PI(4)P or PI(5)P as well as wild-type Cdc11, indicating that neither guanine nucleotide binding nor the coiled-coil region is important for binding (Fig. 5A).
FIG. 5.
Lipid-binding properties of GST-Cdc11. (A) Wild-type and mutant GST-Cdc11 fusion proteins were tested for their lipid-binding properties as described in the legend to Fig. 4. (B) GST-Cdc11 fusion protein was loaded with GDP and γ-S-labeled GTP before incubation with the lipid-containing membrane. (C) Alignment of the polybasic region located 10 residues amino terminal with respect to the P loop in the mammalian H5 protein and the seven septins encoded in the yeast genome. The one-letter code for amino acids is used. Basic residues are shown in bold type. Asterisks mark the five positively charged amino acids replaced in the GST-Cdc11R12Q, K13Q, R14Q, K15Q, H16Q and GST-Cdc11R12Q, K13Q, R14E, K15Q, H16Q mutants. Note that the location of the basic residues in Spr3 is further from the P loop than for the other septins. (D) Protein lipid overlay assay using the Cdc11 fusion mutants where the five basic residues in positions 12 to 16 have been replaced as indicated. (E) Pull-down assay. The Western blots were treated with an anti-GST antibody. The bands in lanes 1 and 2 show the input protein, and the bands in lanes 3 and 4 show protein associated with the PI(4)P agarose beads. Lanes 1 and 3 contain wild-type GST-Cdc11, and lanes 2 and 4 contain GST-Cdc11R12Q, K13Q, R14E, K15Q, H16Q.
We also studied the roles of nucleotide binding in septin interaction with PI(4)P and PI(5)P using a different assay. GST-Cdc11 was first loaded with GDP and γ-S-labeled GTP, and the samples were then incubated with the membrane containing the different phosphoinositides. As shown in Fig. 5B, GDP-bound GST-Cdc11 and γ-S-labeled GTP-bound GST-Cdc11 proteins bound equally well to PI(4)P and PI(5)P. These results confirm that binding of Cdc11 to the different phosphoinositides is independent of the presence and type of guanine nucleotide.
Phosphoinositide binding requires the basic region of Cdc11.
The mammalian H5 septin requires the highly conserved basic region (Fig. 5C) in order to bind phosphoinositides (66). To identify the region of Cdc11 required for phosphoinositide binding, we tested whether truncated Cdc11 proteins containing only the amino-terminal sequences, GST-Cdc111-45 and GST-Cdc111-258, bound to different phospholipids. Both of these constructs bound to PI(4)P and PI(5)P, like the full-length protein, indicating that a phospholipid-binding domain lies at the amino terminus (data not shown). To determine if the basic region of Cdc11 is required for binding, we tested whether the GST-Cdc11R12Q, K13Q, R14Q, K15Q, H16Q and GST-Cdc11R12Q, K13Q, R14E, K15Q, H16Q proteins would bind lipids. As shown in Fig. 5D, GST-Cdc11R12Q, K13Q, R14Q, K15Q, H16Q bound extremely poorly and GST-Cdc11R12Q, K13Q, R14E, K15Q, H16Q failed to bind the lipids.
To confirm that the basic region at the amino terminus is important for lipid binding, we used a different approach consisting of a lipid pull-down assay. Commercial PI(4)P-agarose beads were incubated with GST-Cdc11 and GST-Cdc11R12Q, K13Q, R14E, K15Q, H16Q proteins. The beads were pelleted and washed, and the bound protein was eluted with sodium dodecyl sulfate and analyzed by immunoblot analysis. As shown in Fig. 5E, GST-Cdc11 associates with the PI(4)P-agarose beads (lane 3), but GST-Cdc11R12Q, K13Q, R14E, K15Q, H16Q does not (lane 4). Thus, the basic region at the amino terminus of Cdc11 is required for binding to phosphoinositides.
PI(4)P is necessary for proper in vivo septin organization.
To determine the role of PI(4)P in septin function in vivo, we investigated how the septin localization is affected in yeast cells defective for PI 4-kinase activity. In yeast, most of the PI 4-kinase activity is due to the products of the essential PI 4-kinase genes STT4 and PIK1 (2).
In order to analyze the effect of low levels of PI(4)P on septin organization, we examined the localization of Cdc11-GFP in strains containing a temperature-sensitive allele (stt4-4 or pik1-83 or both) (2). Mid-log-phase cells growing at 23°C were either incubated at the same temperature or shifted to 37°C for 3 h. As shown in Fig. 6, approximately 20% of the budded wild-type cells did not display a visible septin signal from the Cdc11-GFP fusion protein at permissive or restrictive temperatures. Similarly, approximately 20% of the budded stt4-4, pik1-83, and stt4-4 pik1-83 double mutant cells lacked Cdc11-GFP staining when cells were grown at 23°C. In contrast, the Cdc11-GFP staining was absent in 40% of the exponentially growing stt4-4 or pik1-83 budded cells at the restrictive temperature. Interestingly, 80% of the cells lacked Cdc11-GFP staining in stt4-4 pik1-83 double mutant strains (Fig. 6). The correlation between the quantity of PI(4)P present in yeast cells and the number of cells with normal septin structures suggests that PI(4)P plays a role in either transport, formation, or stabilization of the septin cytoskeleton components.
FIG. 6.
Quantification of budded yeast cells with undetectable Cdc11-GFP in mutants lacking PI(4)P. Strains SEY6210 (wild type [wt]), AAY102 (stt4-4), AAY104 (pik1-83), and AAY105 (stt4-4 pik1-83) (2) were transformed with a centromeric plasmid expressing Cdc11-GFP under the CDC11 promoter. Log-phase cultures of the corresponding strains were grown at the indicated temperature for 3 h before fixation. The number of budded cells displaying Cdc11-GFP signal was quantified in at least 250 cells. The percentage of budded cells in which Cdc11-GFP staining was not observed is plotted on the y axis.
Localization of Cdc11 requires the basic, GTP-binding, and coiled-coil domains, but not GTP binding.
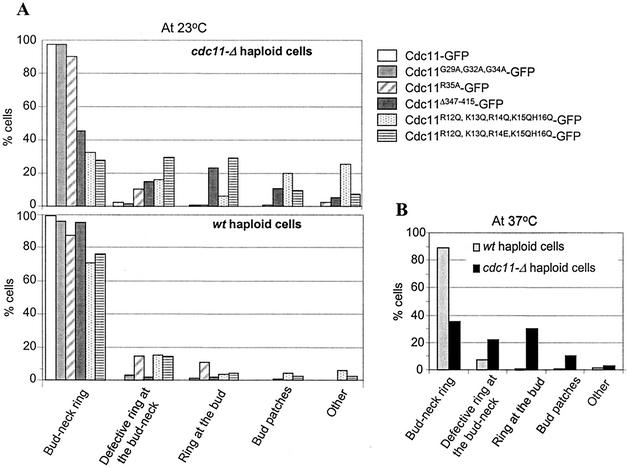
To determine the regions of Cdc11 necessary for localization to the bud neck, we examined the localization of the different mutant proteins tagged with GFP at their carboxy termini. These alleles were introduced into a cdc11-Δ strain, and cells were grown at 23°C. Full-length Cdc11 fused to GFP localized as a ring at the incipient bud site in unbudded cells and at the bud neck in budded cells, as described previously (Fig. 7A and 8A) (19). Examination of the different Cdc11-GFP mutants revealed that several regions of the protein are important for localization. Deletion of the coiled-coil domain (Cdc11Δ347-415-GFP) or mutations in the basic region (Cdc11R12Q, K13Q, R14E, K15Q, H16Q-GFP) resulted in significant defects in neck structures (Fig. 7B and C); 60 to 70% of the cells contained either defective rings or rings misplaced in the bud (Fig. 8A). Similar results were observed for the Cdc11R12Q, K13Q, R14Q, K15Q, H16Q-GFP construct, although many of the cells contained patches in the bud instead of rings (Fig. 7D and 8A). Surprisingly, Cdc11G29A, G32A, G34A-GFP and Cdc11R35A-GFP proteins still localized normally at 23°C (Fig. 7E and 8A and data not shown), indicating that GTP binding is not important for localization. However, for both strains at 37°C, only 35% of the cells displayed normal localization (Cdc11R35A-GFP data shown in Fig. 8B). These results indicate that at 23°C, the basic region and the coiled-coil domain are required in for proper localization of Cdc11 cdc11-Δ cells, but GTP binding is not; at 37°C, all three domains are required.
FIG. 7.
Subcellular localization of GFP-tagged cdc11 alleles. cdc11Δ (A to E) and CDC11 (wild type) (F to J) cells were transformed with centromeric plasmids expressing Cdc11-GFP (A and F), Cdc11Δ347-415-GFP (B and G), Cdc11-R12Q, K13Q, R14E, K15Q, H16Q-GFP (C and H), Cdc11R12Q, K13Q, R14Q, K15Q, H16Q-GFP (D and I), or Cdc11G29A, G32A, G34A-GFP (E and J). Cells were grown overnight at 23°C, diluted into fresh SC medium lacking Trp, grown until mid-exponential phase at the same temperature, and fixed. Different localization patterns are indicated by the arrows.
FIG. 8.
Quantification of the subcellular localization of different alleles of Cdc11-GFP in cdc11-Δ and CDC11 (wild-type [wt]) cells. (A) The localization of different alleles of Cdc11-GFP cells, treated as indicated in the legend to Fig. 7, was examined. More than 200 budded cells were observed in each category, and the results are presented as a percentage of the total number of budded cells. (B) Localization of the Cdc11R35A-GFP allele at 37°C in both wild-type and cdc11-Δ mutant cells.
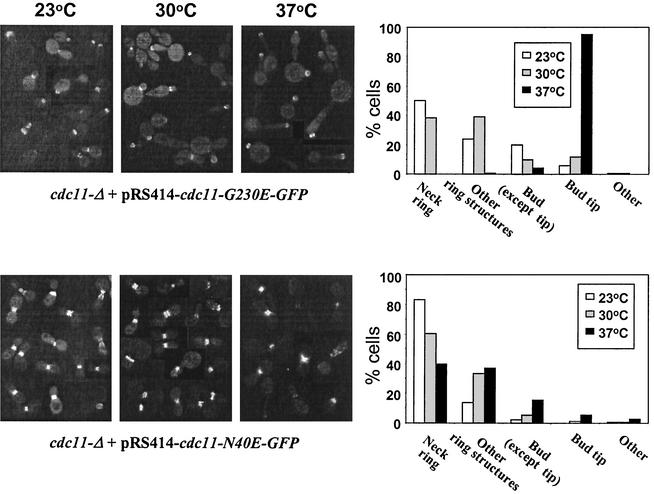
We also examined the effects of mutating other parts of the GTP-binding domain on Cdc11 localization. cdc11-Δ cells containing Cdc11G230E-GFP exhibited a temperature-dependent effect on subcellular localization. When cells were grown at 23 and 30°C, approximately 50 and 40% of the cells exhibited their proper localization at the bud neck, respectively. In contrast, at 37°C, the mutant protein was always mislocalized to the bud tip and usually appeared as rings (Fig. 9). In cdc11-Δ cells expressing Cdc11N40E, a similar but less severe temperature-sensitive defect was observed. Most of the cells exhibited normal Cdc11 localization at 23°C, but at 37°C, the percentage decreased to 40%; many of the defective cells formed rings, but they had aberrant shapes (Fig. 9). Thus, although GTP binding is not required for Cdc11 localization, sequences in this domain are required for proper localization; the residue at position 40 is important for formation of normal rings, whereas the residue at position 230 is needed for targeting of the ring to the neck.
FIG. 9.
Subcellular distribution of alleles of GFP-tagged Cdc11 with point mutations in a cdc11-Δ background at different temperatures. cdc11-Δ cells expressing either Cdc11G230E-GFP or Cdc11N40E-GFP from a centromeric plasmid were grown overnight, diluted in fresh SC medium lacking Trp, and grown until mid-exponential phase for 7 h at the indicated temperatures. The cellular localization of Cdc11G230E-GFP and Cdc11N40E-GFP was examined in more than 200 budded cells. Examples of localization of both alleles are shown in the micrographs on the left, while quantification of the localization of alleles for the indicated categories at each temperature are shown in the graphs on the right.
The aberrant localization of many Cdc11 mutant proteins is rescued by an extra copy of wild-type Cdc11.
In addition to localizing the different Cdc11 variants in a cdc11-Δ strain, we also analyzed a number of them in a wild-type cell. Interestingly, many of the proteins now localized normally. Cdc11 protein lacking its coiled-coil domain (Cdc11Δ347-415-GFP) localized normally in more than 90% of the budded cells at 23°C (Fig. 7G and 8A). Similarly, the presence of the native Cdc11 dramatically reduced the severe mislocalization of Cdc11R12Q, K13Q, R14E, K15Q, H16Q-GFP and Cdc11R12Q, K13Q, R14Q, K15Q, H16Q-GFP; more than 70% of these mutants localized normally (Fig. 7H and I and 8A). In addition, native Cdc11 also rescues the defective localization of Cdc11R35A-GFP at 37°C (Fig. 8B). Thus, the presence of a wild-type Cdc11 protein can help rescue the localization defect of many Cdc11 mutant constructs.
The coiled-coil domain is not required for Cdc11 interaction with itself or Cdc12 but facilitates interaction with Cdc3 and Bem4.
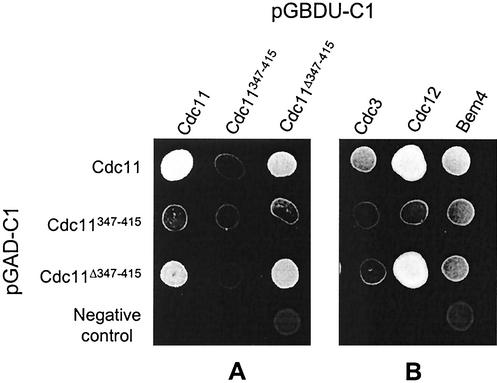
The observation that mutant Cdc11 protein defects can be rescued by the presence of a wild-type protein suggests that Cdc11 directly or indirectly interacts with itself; the presence of the wild-type protein presumably helps target the defective protein to the mother bud neck. To determine whether Cdc11 can interact with itself or other septins, two-hybrid experiments were performed. It has been reported that Cdc11 interacts with Cdc12 and Bem4 by two-hybrid analyses and that Cdc11 and Cdc3 coimmunoprecipitate (12, 21, 28). To determine whether Cdc11 can interact with itself, the coding region of CDC11 and fragments containing and lacking the coiled-coil domain were fused to the coding sequence of the GAL4 activation domain. Similarly, the same fragments were fused to the sequence of the GAL4 DNA-binding domain. Diploid cells coexpressing both constructs were examined for their ability to activate a Gal4-responsive HIS3 construct by conferring growth in the absence of histidine. They were also examined for their ability to activate Gal4-responsive lacZ and ADE2 genes. As shown in Fig. 10A, using the HIS3 assay, full-length Cdc11 protein interacts with itself and with a construct lacking the coiled-coil domain. Full-length Cdc11 does not interact with the coiled-coil domain, and a construct containing the coiled-coil domain does not interact with either full-length Cdc11 protein or any of the truncated forms, even though the fusion proteins were expressed and detected by immunoblot analyses (data not shown). Similar results were observed using β-galactosidase and adenine growth assays (not shown). Thus, the coiled-coil domain is not important for the interaction of Cdc11 with itself.
FIG. 10.
Two-hybrid interactions between Cdc11 and other yeast proteins. Six-microliter samples of log-phase cultures at similar cell densities were spotted onto plates containing SC medium minus Ura, Leu, and His. The yeast cells carry two plasmids: (i) the pGAD-C1 plasmid (marked with LEU2) harboring fragments of DNA encoding either the complete coding region of CDC11 (Cdc11), the coiled-coil domain of Cdc11 (Cdc11347-415), Cdc11 lacking its coiled-coil domain (Cdc11-Δ347-415), or the plasmid without any insert as a negative control, and (ii) the pGBDU-C1 plasmid (marked with URA3), carrying the already described fragments of CDC11 (A) or the full coding region of CDC3, CDC12, or BEM4. An interaction between different fragments leads to expression of the reporter gene HIS3 that enables growth on the plate.
To further determine the region required for the association of Cdc11 with itself, a set of additional constructs were prepared and tested. We found that a central region of Cdc11 (residues 101 to 217) is sufficient for interaction with either full-length Cdc11 or the protein lacking the coiled-coil domain (Fig. 11A). Thus, the interacting region can be delineated to a 117-amino-acid region in the center of the GTPase domain. Additional sequences likely influence interaction of Cdc11 with itself, because the Cdc11N40E protein, which has a mutation not far from the septin interacting domain, fails to interact with Cdc11 constructs (Fig. 11B). In summary, the entire GTP-binding domain of Cdc11 is not required for self-association, but an interacting region in the center of this domain is sufficient. The activity of the interacting region may be influenced by nearby residues (Fig. 11C).
FIG. 11.
Mapping the region of Cdc11 required for interaction with itself. (A) Yeast strains harboring the pGAD-C1 plasmid (marked with LEU2) expressing wild-type Cdc11 or Cdc11Δ347-415 or lacking an insert and the pGBDU-C1 plasmid (marked with URA3) carrying DNA encoding the indicated fragments of CDC11 are shown. Yeast cells were grown on SC medium minus Ura, Leu, and His; only cells carrying fragments of Cdc11 capable of interacting with Cdc11 can grow on this medium. (B) Yeast strains harboring the pGBDU-C1 plasmid expressing Cdc11Δ347-415 or Cdc11Δ347-415, N40E and pGAD-C1 plasmid expressing Cdc11, Cdc11347-415, or Cdc11Δ347-415 were grown on SC medium minus Ura, Leu, and His. Six microliters of each yeast liquid culture carrying the indicated constructs were spotted onto a plate containing the same medium. (C) Schematic representation of the truncated Cdc11 proteins used in the two-hybrid analyses. The complete coiled-coil domain of Cdc11 is included in the fragment Cdc11347-415. The fragments capable of interacting with Cdc11 (as detected by two-hybrid analysis) are shown in black boxes.
We also tested the interaction of Cdc11 with Cdc3 and Cdc12 using two-hybrid assays. Full-length Cdc11 and Cdc11 lacking the coiled-coil region interact with Cdc12, but only the full-length Cdc11 interacts well with Cdc3 (Fig. 10B), suggesting that (i) the noncoiled-coil region is important for binding Cdc12 and (ii) the carboxy terminus facilitates interaction between Cdc11 and Cdc3. The carboxy terminus of Cdc11 alone does not interact with Cdc3, suggesting that other parts of the molecule are also important for this interaction.
We also explored the interaction of Cdc11 with the polarity protein Bem4. Interestingly, full-length Cdc11, the amino terminus, and the coiled-coil domain each interact with Bem4. However, the truncated constructs of Cdc11 do not interact as strongly as the full-length protein does (Fig. 10B). These results suggest that both the coiled-coil region and the sequence that comprises the basic region plus the GTPase domain interact with Bem4.
DISCUSSION
Although septins have now been found in a variety of organisms, few details as to their assembly and function have been elucidated. In this report, we have mapped several functional domains of the yeast septin Cdc11 and ascertained the functions of these domains in vivo. A basic domain which is required for binding to PI(4)P and PI(5)P was found; this domain is also important for proper localization and function (i.e., normal cell morphology, cytokinesis, and growth). A coiled-coil domain is also important for proper localization and function, especially at higher temperatures. This region is not required for self-assembly but facilities interaction with Cdc3 and can interact with Bem4. Last, the GTP-binding domain is important for interaction of Cdc11 with itself, other septins, and Bem4. This region is also required for proper localization and function. Unexpectedly, GTP binding is not essential for phospholipid binding, localization, or function at lower temperatures but is important for proper localization at higher temperatures. These observations provide a model for the function and assembly of septins.
The phosphoinositide-binding domain is important for phosphoinositide binding and localization.
Previous studies have found that the mammalian H5 septin binds PI(4,5)P2 and PI(3,4,5)P3 (66). Although PI(4,5)P2 is present in yeast, it binds very poorly to Cdc3, Cdc10, Cdc11, and Cdc12. Instead, yeast septins preferentially bind to PI(4)P and PI(5)P (Fig. 4). Two different pools of PI(4)P have been found in yeast cells which are synthesized by the PI 4-kinases Stt4 and Pik1. Stt4 localizes to the plasma membrane, and its activity is required for the maintenance of vacuole morphology, cell wall integrity, and actin cytoskeleton organization (1). In contrast, Pik1 localizes to the nucleus and trans-Golgi membranes, and its PI(4)P product is a regulator of transport from the Golgi body membranes to plasma membrane (2, 57, 62). Due to its very low basal levels, PI(5)P has only recently been identified in a few types of mammalian cells under particular activation states, and it has not yet been identified in yeast cells (59). Our results using the stt4-4 mutant strain (Fig. 6) are consistent with the possibility that PI(4)P is likely to be present at the plasma membrane where it contributes to the septin organization, localization, or stability. Since pik1-83 cells also exhibit a decrease in the number of visible septin structures, we speculate that the septin-lipid interaction may initiate in the Golgi complex.
It is likely that the phosphoinositide-binding region of the yeast septins may help to anchor the septin complex to the membrane. Presumably, the multitude of binding sites present in a septin polymer would lead to a high-affinity interaction with the membrane. Phosphoinositide binding is important for proper localization of the Cdc11 septin complex; mutation of this site often results in mislocalized rings, many of which exhibit an aberrant structure (Fig. 7C, D, H, and I). However, some cells with mutations in the region of basic residues exhibit normal rings. This residual normal localization may be due to the fact that most, but not all, of the basic residues have been eliminated or to the presence of other domains of Cdc11 that interact with plasma membrane components.
The results of our studies are consistent with those of other cytoskeletal elements. Although well-defined domains (e.g., PH domains) have been shown to specifically bind phospholipids, small stretches of basic amino acids have also been shown to bind these molecules (44). Most of the cytoskeletal proteins that show phosphoinositide-binding properties (cofilin, profiling, gelsolin, α-actinin, and vinculin) have a lysine/arginine-rich region involved in this activity, similar to the septins, indicating that this is a general feature of these proteins (27, 30, 42, 44, 60, 65).
The GTPase domain is important for self-association, binding other proteins, and targeting to the mother bud neck.
The GTPase domain was found to be important for association of Cdc11 with itself, with Cdc12, and with Bem4 by two-hybrid analysis (Fig. 10). These results do not prove that these interactions are direct; they might be mediated through other members of the septin complex. Nevertheless, it is clear that these interactions occur through a region upstream of the coiled-coil domain. We found that a central region in this domain is required for interaction of Cdc11 with itself (Fig. 11A), indicating that only a portion of this domain mediates the interaction. Since a mutation in a residue upstream of the interacting domain (N40E) can disrupt this interaction (Fig. 11B), it is likely that flanking sequences can influence Cdc11 self-association. This mutation and others in the region do not result in complete loss of protein levels or function, because the mutant proteins are detected at wild-type levels by indirect immunofluorescence, and many of the cells exhibit normal Cdc11 localization.
Mutations in the GTPase domain yield a variety of interesting results. The N40E and G230E mutations cause a temperature-sensitive localization and morphological defects, indicating that this region is important for septin function (Fig. 9). Interestingly, the G230E mutation generally still allows septin ring formation at the elevated temperature, but the rings are usually mislocalized to the bud tip. This indicates that this residue does not affect polarized targeting to the bud tip and might interact with a protein involved in targeting or maintenance of Cdc11 to the neck. A similar result has been observed previously for the bud neck proteins Cdc12, Myo1 and Chs2: all of these proteins mislocalize to the bud tip in a temperature-sensitive cdc3-1 strain grown at the restrictive temperature (50). Presumably, these proteins are still targeted by the polarized secretory machinery but are not properly directed or anchored at the neck.
Mutation of the GTP-binding site so that Cdc11 can no longer bind GTP revealed a number of surprising results. This mutation did not affect the binding of Cdc11 to phospholipids or its cellular localization and had only a small effect on Cdc11 function. This result is unexpected, since this site is highly conserved in all septins. Perhaps the GTP binding of other septins can substitute for the lack of activity in Cdc11, thereby accounting for the lack of a detectable phenotype at 23°C and a modest defect at 37°C.
The mammalian H5 septin has been shown to require GDP for efficient phospholipid binding (66). Therefore, it was unexpected that GST-Cdc11G29A, G32A, G34A can still bind phosphoinositides even though it has reduced its ability to bind guanine nucleotides (Fig. 5A). This finding is supported by the fact that incubation with either γ-S-labeled GTP or GDP had no effect on the phospholipid-binding activity of Cdc11 (Fig. 5B). Thus, the yeast Cdc11 protein appears to differ from the mammalian H5 protein. The difference between yeast and mammalian septin proteins may be due to the different roles septin play in yeast and mammalian cells. Mammalian septins are implicated in some cellular events that are absent in yeast cells, and likewise, yeast septins have roles not present in mammalian cells (reviewed in reference 35). In addition, although septins play a role in cytokinesis in both mammalian and yeast cells, the mechanisms involved in this process are not identical.
What is the role of the GTP-binding domain if it does not affect lipid binding or localization? Perhaps it plays a role in the dynamics of assembly or disassembly of septins. In this respect, septins may be similar to tubulin, where nucleotide binding controls microtubule assembly (45, 55). Other similarities between septins and tubulin have been proposed previously (17, 18). However, a recent study demonstrated that in vitro polymerization of different purified mammalian septins is not affected by the nucleotide-binding state of the septins (33). Thus, if GTP binding is important for the kinetics of assembly or disassembly of septins in vivo, it must involve other factors.
The coiled-coil domain is not important for self-association but is important for association with other proteins.
The carboxy-terminal coiled-coil domain is not required for Cdc11 interaction with itself or Cdc12. This result is not completely surprising, since there are septins that lack a carboxy-terminal coiled-coil domain (S. cerevisiae Cdc10) (18) yet still associate with the septin complex. However, the coiled-coil domain may help Cdc11 interact with Cdc3, since loss of the coiled-coil domain reduced the strength of this interaction in a two-hybrid assay (Fig. 10B). Thus, presumably both coiled-coil interactions and noncoiled-coil interactions are important for septin filament assembly. Presumably multivalent interactions involving both domains lead to a strong, and stable cytoskeletal complex.
The coiled-coil domain may also help mediate interactions with other proteins, since loss of this region reduced binding to Bem4 and the coiled-coil domain by itself can directly interact with Bem4. Since the construct lacking the coiled-coil domain also bound Bem4 (Fig. 10B), we suggest that the coiled-coil domain, together with other parts of the protein, contributes to optimal Bem4 binding. This coiled-coil domain may mediate functionally important protein interactions, since loss of this region resulted in a reduction of proper protein localization and in the formation of elongated and connected cells; particularly at higher temperatures in a cdc11-Δ strain.
After we submitted this manuscript, Lee et al. (37) published a study analyzing the role of the coiled-coil domain with different results and conclusions. As in our study, they found that the coiled-coil domain is not essential for interactions with other septins, since cells lacking this domain still interact with other septins in the two-hybrid assay. However, they did not observe any effect on protein localization or function upon deleting this region. The reason for this is not entirely clear but may stem from the fact that a wild-type construct was still present in these experiments. They used a cdc11-Δ strain containing a pGAL1-GST-CDC11 plasmid and examined effects after growth on glucose for 1 h. The presence of the wild-type GST-Cdc11 that remained after growth in glucose or which is still expressed from the GAL1 promoter may have suppressed any observable defects. In our strains in which Cdc11Δ347-415-GFP or Cdc11Δ347-415 were expressed and wild-type Cdc11 protein was not present, aberrant cell morphologies and cytokinesis defects were readily apparent at 30°C and were even more severe at higher temperatures (Fig. 3H and 7B). These defects were significantly reduced in strains containing wild-type Cdc11 (Fig. 7G and 8A). Fifty percent of cdc11-Δ budded cells presented normal localization of Cdc11Δ347-415-GFP; in contrast, 90% of CDC11 budded cells localized Cdc11Δ347-415-GFP properly at 23°C (Fig. 8A). Thus, the presence of the wild-type protein suppresses the defects of the mutant protein. In summary, the coiled-coil domain of Cdc11 is necessary for interaction with one or more proteins important for its localization at the bud neck; this domain may also contribute to the stabilization of the septin complex at the neck.
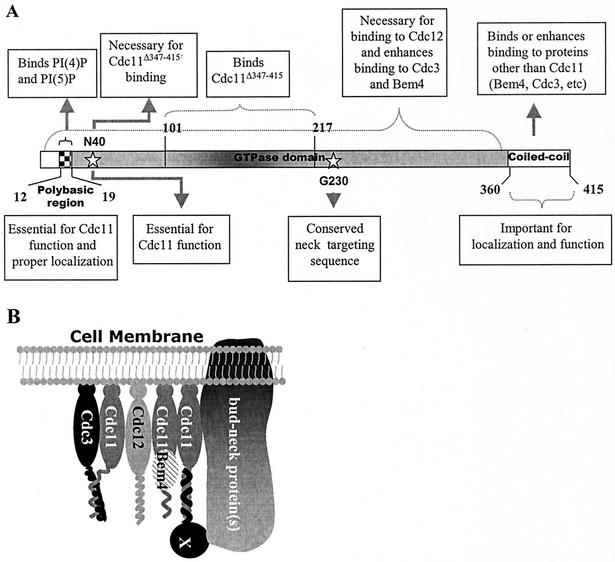
A model for septin function.
On the basis of the observations of this study, we propose the following model to describe the manner in which septins carry out their functions (Fig. 12A). We propose that the septin basic region is required for general interaction with membranes. Interaction with other proteins may also be important for cortical association, because when the basic region is mutated, thereby destroying the phosphoinositide-binding activity, the protein is still found at the cortex. We also propose that Cdc11 homodimerizes through the central part of its GTPase domain. In fact, perhaps all septins function as rudimentary homodimers or heterodimers that assemble into higher-order structures (Fig. 12B). The GTP-binding domain also has a role in neck targeting. Last, the coiled-coil domain may help promote specific interactions between several of the septins and mediate associations with other proteins. Cdc11Δ347-415, which lacks the coiled-coil domain, does not bind well to Cdc3, nor does it bind strongly to Bem4. Furthermore, the coiled-coil domain of Cdc11 can interact directly with Bem4. We propose that septin coiled-coil domain constitutes a protein interaction region and that distinct molecular interactions between this domain and that of many other proteins may enable the numerous and diverse protein associations underlying septin cytoskeletal function.
FIG. 12.
A model for Cdc11 function. (A) Summary of the different regions of Cdc11 required for its interactions and activity. (B) A model for how Cdc11 associates with other proteins at the bud neck. A putative protein(s) that interacts with Cdc11 through the coiled-coil domain and with other proteins anchored to the bud neck is indicated by the X.
Acknowledgments
We thank Scott D. Emr and Anjon Audhya for generously providing yeast strains. We also thank Daniel Gelperin, Jessie Hanrahan, Anuj Kumar, and Susana Vidan for critically reading the manuscript.
This work was supported in part by grant GM36494 from the National Institutes of Health (to M.S.). A.C. received a postdoctoral fellowship from the Spanish Ministerio de Educación y Ciencia.
REFERENCES
- 1.Audhya, A., and S. D. Emr. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2:593-605. [DOI] [PubMed] [Google Scholar]
- 2.Audhya, A., M. Foti, and S. D. Emr. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11:2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. [DOI] [PubMed] [Google Scholar]
- 4.Barral, Y., M. Parra, S. Bidlingmaier, and M. Snyder. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byers, B., and L. Goetsch. 1976. A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casamayor, A., and M. Snyder. 2002. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5:179-186. [DOI] [PubMed] [Google Scholar]
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Deak, M., A. Casamayor, R. A. Currie, C. P. Downes, and D. R. Alessi. 1999. Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 451:220-226. [DOI] [PubMed] [Google Scholar]
- 9.DeMarini, D. J., A. E. M. Adams, H. Fares, C. D. Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dove, S. K., F. T. Cooke, M. R. Douglas, L. G. Sayers, P. J. Parker, and R. H. Michell. 1996. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390:187-192. [DOI] [PubMed] [Google Scholar]
- 11.Dowler, S., G. Kular, and D. R. Alessi. 2002. Protein lipid overlay assay. Sci. STKE http://stke.sciencemag.org/cgi/content/full/sigtrans;2002/129/p16. [DOI] [PubMed]
- 12.Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen, W. Guo, K. G. Kozminski, M. W. Lau, J. J. Moskow, A. Tong, L. R. Schenkman, A. McKenzie III, P. Brennwald, M. Longtine, E. Bi, C. Chan, P. Novick, C. Boone, J. R. Pringle, T. N. Davis, S. Fields, and D. G. Drubin. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fares, H., L. Goetsch, and J. R. Pringle. 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132:399-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fares, H., M. Peifer, and J. R. Pringle. 1995. Localization and possible functions of Drosophila septins. Mol. Biol. Cell 6:1843-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faty, M., M. Fink, and Y. Barral. 2002. Septins: a ring to part mother and daughter. Curr. Genet. 41:123-131. [DOI] [PubMed] [Google Scholar]
- 16.Field, C. M., O. al-Awar, J. Rosenblatt, M. L. Wong, B. Alberts, and T. J. Mitchison. 1996. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Field, C. M., and D. Kellogg. 1999. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 9:387-394. [DOI] [PubMed] [Google Scholar]
- 18.Flescher, E. G., K. Madden, and M. Snyder. 1993. Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 122:373-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford, S. K., and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 12:281-292. [DOI] [PubMed] [Google Scholar]
- 20.Frazier, J. A., M. L. Wong, M. S. Longtine, J. R. Pringle, M. Mann, T. J. Mitchison, and C. Field. 1998. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 22.Gelperin, D., L. Horton, J. Beckman, J. Hensold, and S. K. Lemmon. 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7:1268-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haarer, B. K., and J. R. Pringle. 1987. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother bud neck. Mol. Cell. Biol. 7:3678-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwell, L. H. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265-276. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell, L. H., R. K. Mortimer, J. Culotti, and M. Culotti. 1974. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics 74:267-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartwig, J. H., S. Kung, T. Kovacsovics, P. A. Janmey, L. C. Cantley, T. P. Stossel, and A. Toker. 1996. D3 phosphoinositides and outside-in integrin signaling by glycoprotein IIb-IIIa mediate platelet actin assembly and filopodial extension induced by phorbol 12-myristate 13-acetate. J. Biol. Chem. 271:32986-32993. [DOI] [PubMed] [Google Scholar]
- 28.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 29.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janmey, P., J. Lamb, P. Allen, and P. Matsudaira. 1992. Phosphoinositide-binding peptides derived from the sequences of gelsolin and villin. J. Biol. Chem. 267:11818-11823. [PubMed] [Google Scholar]
- 31.Kartmann, B., and D. Roth. 2001. Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J. Cell Sci. 114:839-844. [DOI] [PubMed] [Google Scholar]
- 32.Kim, H. B., B. K. Haarer, and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112:535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinoshita, M., C. M. Field, M. L. Coughlin, A. F. Straight, and T. J. Mitchison. 2002. Self- and actin-templated assembly of mammalian septins. Dev. Cell 3:791-802. [DOI] [PubMed] [Google Scholar]
- 34.Kinoshita, M., S. Kumar, A. Mizoguchi, C. Ide, A. Kinoshita, T. Haraguchi, Y. Hiraoka, and M. Noda. 1997. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11:1535-1547. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita, M., and M. Noda. 2001. Roles of septins in the mammalian cytokinesis machinery. Cell Struct. Funct. 26:667-670. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu, K., W. J. Driscoll, Y. C. Koh, and C. A. Strott. 1994. A P-loop related motif (GxxGxxK) highly conserved in sulfotransferases is required for binding the activated sulfate donor. Biochem. Biophys. Res. Commun. 204:1178-1185. [DOI] [PubMed] [Google Scholar]
- 36a.Kusch, J., A. Meyer, M. P. Snyder, and Y. Barral. 2002. Microbubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16:1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, P. R., S. Song, H.-S. Ro, C. J. Park, J. Lippincott, R. Li, J. R. Pringle, C. De Virgilio, M. S. Longtine, and K. S. Lee. 2002. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6906-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine, M. S., D. J. DeMarini, M. L. Valencik, O. S. Al-Awar, H. Fares, C. De Virgilio, and J. R. Pringle. 1996. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8:106-119. [DOI] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 40.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. J. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenz, M. C., R. S. Muir, E. Lim, J. McElver, S. C. Weber, and J. Heitman. 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158:113-117. [DOI] [PubMed] [Google Scholar]
- 42.Lu, P.-J., W.-R. Shieh, S. G. Rhee, H. L. Yin, and C.-S. Chen. 1996. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry 35:14027-14034. [DOI] [PubMed] [Google Scholar]
- 43.Ma, X. J., Q. Lu, and M. Grunstein. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10:1327-1340. [DOI] [PubMed] [Google Scholar]
- 44.Martin, T. F. 1998. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14:231-264. [DOI] [PubMed] [Google Scholar]
- 45.McKean, P. G., S. Vaughan, and K. Gull. 2001. The extended tubulin superfamily. J. Cell Sci. 114:2723-2733. [DOI] [PubMed] [Google Scholar]
- 46.Mino, A., K. Tanaka, T. Kamei, M. Umikawa, T. Fujiwara, and Y. Takai. 1998. Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 251:732-736. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell, D. A., T. K. Marshall, and R. J. Deschenes. 1993. Vector for the inducible overexpression of gluthatione S-transferase fusion protein in yeast. Yeast 9:715-723. [DOI] [PubMed] [Google Scholar]
- 48.Neufeld, T. P., and G. M. Rubin. 1994. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77:371-379. [DOI] [PubMed] [Google Scholar]
- 49.Robinson, J. S., D. J. Klionsky, L. M. Banta, and S. D. Emr. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and sorting of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roh, D.-H., B. Bowers, M. Schmidt, and E. Cabib. 2002. The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell 13:2747-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Santos, B., and M. Snyder. 2000. Sbe2p and Sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol. Biol. Cell 11:435-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop. A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15:430-434. [DOI] [PubMed] [Google Scholar]
- 55.Scheffers, D.-J., and A. J. M. Driessen. 2001. The polymerization mechanism of the bacterial cell division protein FtsZ. FEBS Lett. 506:6-10. [DOI] [PubMed] [Google Scholar]
- 56.Schenkman, L. R., C. Caruso, N. Page, and J. R. Pringle. 2002. The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J. Cell Biol. 156:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schorr, M., A. Then, S. Tahirovic, N. Hug, and P. Mayinger. 2001. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11:1421-1426. [DOI] [PubMed] [Google Scholar]
- 58.Self, A. J., and A. Hall. 1995. Measurement of intrinsic nucleotide exchange and GTP hydrolysis rates. Methods Enzymol. 256:67-76. [DOI] [PubMed] [Google Scholar]
- 59.Shisheva, A. 2001. PIKfyve: the road to PtdIns 5-P and PtdIns(3,5)-P2. Cell Biol. Int. 25:1201-1206. [DOI] [PubMed] [Google Scholar]
- 60.Sohn, R. H., J. Chen, K. S. Koblan, P. F. Bray, and P. J. Goldschmidt-Clermont. 1995. Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J. Biol. Chem. 270:21114-21120. [DOI] [PubMed] [Google Scholar]
- 61.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walch-Solimena, C., and P. Novick. 1991. The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1:523-525. [DOI] [PubMed] [Google Scholar]
- 63.Xie, H., M. Surka, J. Howard, and W. S. Trimble. 1999. Characterization of the mammalian septin H5: distinct patterns of cytoskeletal and membrane association from other septin proteins. Cell Motil. Cytoskeleton 43:52-62. [DOI] [PubMed] [Google Scholar]
- 64.Yoneya, T., M. Tagaya, F. Kishi, A. Nakazawa, and T. Fukui. 1989. Site-directed mutagenesis of Gly-15 and Gly-20 in the glycine-rich region of adenylate kinase. J. Biochem. (Tokyo) 105:158-160. [DOI] [PubMed] [Google Scholar]
- 65.Yonezawa, N., Y. Homma, I. Yahara, H. Sakai, and E. Nishida. 1991. A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. J. Biol. Chem. 266:17218-17221. [PubMed] [Google Scholar]
- 66.Zhang, J., C. Kong, H. Xie, P. S. McPherson, S. Grinstein, and W. S. Trimble. 1999. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 9:1458-1467. [DOI] [PubMed] [Google Scholar]
- 67.Zheng, Y., M. J. Hart, and R. A. Cerione. 1995. Guanine nucleotide exchange catalyzed by dbl oncogene product. Methods Enzymol. 256:77-84. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 293:2101-2105. [DOI] [PubMed] [Google Scholar]