Abstract
Retinoic acid receptors (RARs) are ligand-regulated transcription factors that play multiple roles in vertebrate development and differentiation. RARs as a class are capable of both repressing and activating target gene expression. Transcriptional repression is mediated through the recruitment of corepressor proteins such as SMRT. Notably, vertebrates encode three major forms of RARs, α, β, and γ, and these distinct RAR isotypes differ in the ability to recruit a corepressor. RARα strongly interacts with SMRT and can repress target gene transcription, whereas RARβ and -γ interact with SMRT only weakly and fail to repress. We report here the use of a genetic suppressor approach, based on a yeast two-hybrid interaction assay using Saccharomyces cerevisiae, for the isolation of RARβ mutants that have gained the RARα-like corepressor phenotype, i.e., a strong interaction with SMRT and the ability to repress gene expression in vertebrate cells. Analysis of these gain-of-function mutants indicates that the different corepressor interaction properties of RARα, -β and -γ are determined by a gating mechanism through which amino acid differences in the helix 3 region of these receptors influence the position of the receptor C-terminal helix 12 domain. As a consequence, the RARβ and RARγ receptors appear to adopt a constitutively closed helix 12 conformation in the absence of hormone that may approximate the conformation of RARα when bound to hormone agonist. This closed helix 12 conformation in RARβ and RARγ blocks corepressor binding, prevents repression, and permits significant levels of target gene activation even in the absence of hormone. We refer to this phenomenon as a “gate-latch” model of corepressor regulation.
Retinoic acid receptors (RARs) are hormone-regulated transcription factors that regulate multiple aspects of vertebrate development and differentiation (9, 44, 45, 47, 59, 68). RARs exert these effects principally by binding to specific DNA sequences and regulating the expression of adjacent target genes (9, 44, 45, 47, 59, 68). In common with many other members of the nuclear receptor family, RARs possess bimodal transcriptional properties and can function either by repressing or by activating transcription of their target genes (19, 47, 57). This dual mode of transcriptional regulation is dependent on the ability of the receptors to associate with coregulator proteins denoted coactivators and corepressors (reviewed in references 13, 19, 30, 35, 39, 47, 48, 53, 57, 65, 71, and 80). Corepressors such as SMRT and its paralog, N-CoR, bind the nuclear receptor directly and can tether additional components of a larger corepressor complex, including TBL-1 and an array of histone deacetylases (HDACs) (13, 19, 30, 35, 39, 47, 48, 53, 57, 65, 71, 80). Conversely, the known coactivators that can function with the RARs include SRC-1, GRIP-1, ACTR, p300/CBP, pCAF, and the DRIP/TRAP/Mediator complexes (13, 30, 35, 39, 47, 48, 65, 71, 80). In turn, corepressors and coactivators modulate transcription by modifying the chromatin template and by interacting with the general transcriptional machinery to promote or inhibit formation of a preinitiation complex (2, 4, 12, 23, 34, 49, 55, 60, 67, 72, 75, 77, 78).
Nuclear receptors typically recruit the SMRT/N-CoR corepressor complex and repress transcription in the absence of hormone or when bound to hormone antagonists (19, 57). Alternatively, the binding of an agonist by the receptor induces dissociation of the corepressor complex, the binding of coactivators, and transcriptional activation (19, 57). Corepressors are tethered to the unliganded nuclear receptor through the interaction of L/V-X-X-I/V-I amino acid motifs within SMRT and N-CoR with a docking surface on the receptor composed primarily of portions of helix 3 and helix 4/5 of the hormone binding domain (31, 32, 46, 51, 56, 79). In the absence of hormone, the C-terminal receptor helix 12 is thought to assume an extended conformation that allows corepressor access to its docking site (6, 19, 31, 32, 51, 56, 79). Conversely, the binding of a hormone agonist appears to induce a conformational change in the receptor, leading to the assumption by helix 12 of a more sequestered position lying across the receptor surface (7, 58). This sequestered helix 12 position occludes the corepressor docking site, resulting in release of corepressor, and simultaneously contributes to the formation of a new interaction surface that allows recruitment of the L-X-X-L-L amino acid motifs found in many coactivators (19, 31, 32, 42, 51, 56, 57, 79). Hormone antagonists confer additional conformations of helix 12 that destabilize coactivator binding and/or further favor corepressor binding (for examples, see references 8, 33, 54, 64, 66, 73, 79, and 85). Helix 12 therefore represents a hormone-operated, multiposition toggle switch that plays a key role in gating access of both corepressors and coactivators to their docking sites on the nuclear receptors.
Three distinct genetic loci encode RARs in vertebrates, resulting in the synthesis of three major RAR isotypes: α, β, and γ (9, 45, 68). Alternative mRNA splicing and promoter utilization result in further diversification within each isotype. The three RAR loci are strongly conserved in evolution and mediate distinctive biological functions, yet they encode receptors that share substantial amino acid sequence identity and possess similar biochemical properties (9, 45, 68). One intriguing difference among the different RAR isoforms that might contribute to their distinct functions in vertebrate development and physiology has been previously reported: RARα recruits the SMRT corepressor and represses target gene transcription in the absence of hormone, whereas RARβ (and RARγ) does neither (76; H. Hauksdottir, B. Farboud, and M. Privalsky, submitted for publication). Instead, RARβ and RARγ mediate significant levels of transcriptional activation even in the absence of hormone, and this activation is further augmented in response to the all-trans retinoic acid (ATRA) ligand (Hauksdottir, Farboud, and Privalsky, submitted). Unexpectedly, prior studies by Wong and Privalsky indicated that these different corepressor interaction properties of RARα, -β, and -γ map outside of the known corepressor docking site, suggesting that the ability of different isoforms of RAR to recruit a corepressor (and to repress transcription) is mediated by a region of the receptor distinct from the actual site of corepressor contact (76).
To better understand this phenomenon, we have now employed a genetic suppression-mutagenesis scheme to identify alterations in RARβ that are able to confer the RARα-like corepressor interaction phenotype. Using this method, we have determined that RARβ (and RARγ) has a fully functional SMRT docking site but that access of SMRT to this docking site is blocked by helix 12, which in RARβ and RARγ assumes a sequestered position in the absence of hormone that approximates the conformation of the agonist-bound RARα. This constitutively closed helix 12 conformation of RARβ and RARγ appears to be stabilized by hydrophobic interactions between amino acids in receptor helix 3 and helix 12. Disruption of this helix 3-helix 12 interaction by the amino acid substitutions found naturally in helix 3 of RARα, or by artificial mutagenesis of RARβ and RARγ, confers a more open helix 12 conformation, enhanced corepressor binding, and transcriptional repression in the absence of hormone. Consistent with this “gate-latch” model for helix 12 of RARβ and RARγ, these two isotypes also display an enhanced ability to recruit p160 coactivators and to activate transcription in the absence of hormone compared to that of the unliganded RARα. We conclude that tertiary interactions confer inherently different conformations on helix 12 of the unliganded RARα, -β, and -γ and that this evolutionarily programmed, differential gating of helix 12 contributes to the distinct cofactor recruitment and transcriptional properties of these different receptor isotypes.
MATERIALS AND METHODS
Molecular clones.
The molecular clones of human RARα, RARβ, and RARγ cDNAs were previously described, as were the constructs for pGBT9-SMRT (representing codons 71 to 769 in the TRAC1/SMRT clone), pGEX-KG-SMRT (representing codons 406 to 769 in the TRAC1/SMRT clone), pGEX-KG-SRC1 (representing codons 560 to 1136 of SRC1), and pGEX-KG-ACTR (representing codons 621 to 821 of ACTR) (11, 24, 28, 62, 76, 81). The RARβ isolate, originally obtained from a human hepatocellular carcinoma and designated HAP, encodes a Met at position 407 whereas most other RARβ isolates have been reported to encode a Leu in this position (16). This possible genetic polymorphism did not result in a functional difference in our assays, and comparable results were obtained with constructs in which Met 407 was replaced by a Leu (data not shown). Chimeras between the different RAR isotypes were constructed by ligation of suitable restriction fragments or by overlap extension PCR methodology (1), whereas point mutations were generated by a QuikChange oligonucleotide-mediated mutagenesis procedure (Stratagene, La Jolla, Calif.). Using standard restriction enzyme, PCR, and ligation procedures (1), the constructs of pACT2-RARα (representing codons 86 to 462 for RARα), pACT2-RARβ (representing codons 79 to 448 for RARβ), and pSG5-GAL4DBD-SMRT (representing codons 71 to 769 in the TRAC1/SMRT clone) were created. The pGEX-KG-SMRT(RID1) construct was created by using a PCR procedure to flank the SMRT receptor interaction domain 1(RID-1) CoRNR box motif (RVVTLAQHISEVITQDYTR) with BamHI and SmaI restriction sites (1); the resulting PCR fragment was excised and then joined in frame to the glutathione S-transferase (GST) sequence within pGEX-KG (21). All clones were confirmed by DNA sequence analysis.
Genetic suppression-yeast two-hybrid strategy.
The RARβ hormone binding domain was subjected to random mutagenesis by propagating our pACT2-RARβ clone in XL1-Red cells according to the instructions of the manufacturer (Stratagene). The mutated plasmids were recovered as a mixed population, and the sequences corresponding to the RARβ hormone binding domain were excised and reintroduced into a nonmutated pACT2 background (to exclude mutations generated within the vector backbone itself). This reconstituted pACT2-mutated RARβ plasmid population was then amplified and cotransformed together with pGBT9-SMRT into the Saccharomyces cerevisiae strain SFY526, thus selecting for growth in the absence of Leu and Trp (62). Separate yeast transformants (10,000) were selected, replica patched on fresh, appropriately supplemented synthetic dextrose agar plates, and tested for the ability to express β-galactosidase by use of a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) overlay technique (37). Briefly, the patched yeast colonies were permitted to grow for 24 h at 30°C; the plates were then overlaid with a molten 0.5% agarose-0.5 M NaPO4 (pH 7.0)-0.1% sodium dodecyl sulfate (SDS)-0.02% N,N-dimethyl formamide-0.2% X-Gal solution and were incubated for an additional hour at 37°C for colorimetric development. Yeast colonies that displayed a more intense blue color in the X-Gal assay than did control transformants containing the wild-type pACT2-RARβ (i.e., indicative of an enhanced RARβ-SMRT interaction) were identified visually, and the corresponding pACT2-mutant RARβ plasmid DNAs were recovered by transformation of Escherichia coli DH5α. Once recovered, the pACT2-mutant RARβ plasmid DNAs were amplified and reintroduced into fresh yeast SFY526 cells together with the pGBT9-SMRT vector (62). pACT2-mutant RARβ plasmids that retained the ability to produce intensified β-galactosidase expression in these secondary yeast transformants (i.e., retained the enhanced RARβ-SMRT interaction) were analyzed further by sequence determination, by a liquid-based β-galactosidase assay, and by the procedures described below.
Mammalian two-hybrid assay.
Approximately 5 × 104 CV-1b cells were aliquoted per well in 24-well culture plates containing Dulbecco modified Eagle's medium (DMEM) supplemented with hormone-depleted 10% fetal bovine serum. The cells were permitted to attach overnight at 37°C. Immediately prior to transfection, the cells were rinsed with phosphate-buffered saline and placed in DMEM supplemented with 10% hormone-depleted fetal bovine serum. Transfections were performed using the Effectene protocol as recommended by the manufacturer (Qiagen, Valencia, Calif.); 12.5 ng of pSG5-GAL4DBD-SMRT, 50 ng of pSG5-GAL4AD receptor, 50 ng of pGL2-GAL-17mer luciferase reporter, 25 ng of pCMV-lacZ (used as an internal transfection control), and 112.5 ng of pUC18 were employed per well (29, 62, 76). The culture medium was replaced 24 h later with fresh medium containing either ATRA or an equivalent amount of ethanol carrier alone. After an additional 24 h at 37°C, the cells were harvested and lysed and the luciferase and β-galactosidase activities were determined as previously described (29, 76).
Transient transfection assay.
CV-1b cells were plated in DMEM supplemented with hormone-depleted 10% fetal bovine serum as described above. The cells were rinsed with phosphate-buffered saline, placed in DMEM supplemented with 10% hormone-depleted fetal bovine serum, and then transfected using the Effectene protocol (Qiagen) and 50 ng of the pTK-Luc retinoic acid response element (RARE) reporter vector, 10 ng of the appropriate pSG5-RAR expression vector, 5 ng of pCMV-lacZ, and 185 ng of pUC18 (24). The culture medium was replaced 24 h later with fresh medium containing either ATRA or an equivalent amount of ethanol carrier alone. The cells were harvested 24 h later, and the luciferase and β-galactosidase activities were determined as previously described (24).
Protein-protein interaction assay in vitro.
Glutathione S-transferase (GST) corepressor and GST coactivator proteins were synthesized in E. coli BL-21 cells containing the corresponding pGEX-KG vector (pGEX-KG-SMRT, pGEX-KG-SRC1, or pGEX-KG-ACTR), the bacteria were lysed, and the GST fusion proteins were bound to a glutathione-agarose matrix (21). 35S-radiolabeled RARs were synthesized in vitro by a coupled transcription-translation (TnT) system (Promega, Madison, Wis.). Each radiolabeled receptor (typically 2 to 5 μl of TnT product per reaction) was then incubated with the immobilized GST fusion protein of interest (approximately 50 ng of GST fusion protein immobilized to 10 μl of agarose matrix per reaction) in a total volume of 120 μl of HEMG buffer (4 mM HEPES [pH 7.8], 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 10% glycerol, 1.5 mM dithiothreitol) at 4°C (62). The binding reactions were performed with 96-well multiscreen filter plates (Millipore, Bedford, Mass.) placed on a roller drum to ensure constant mixing. After a 3-h incubation, the filter wells were washed by centrifugation four times with 200 μl of ice-cold HEMG buffer each, and any radiolabeled RAR proteins remaining bound to the immobilized GST fusion proteins were subsequently eluted with 50 μl of 10 mM glutathione in 50 mM Tris-HCl, pH 7.8. The eluted proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (62) and were visualized and quantified using a PhosphorImager/STORM system (Molecular Dynamics, Sunnyvale, Calif.).
Protease resistance assay.
Using the coupled TnT system (Promega), 35S-radiolabeled RAR proteins were synthesized in vitro. For each time point, 1 μl of the TnT reaction products was diluted to 16 μl in 50 mM Tris-HCl (pH 6.8) containing either 1 μM ATRA or an equivalent amount of ethanol carrier. After 10 min on ice, 4 μl of carboxypeptidase Y (CPY) (1 mg/ml; Sigma) was added to each sample and the tubes were transferred to 10°C to initiate the digestion (28, 42). At each time point indicated, the proteolysis was terminated by the addition of 20 μl of SDS-PAGE buffer and the samples were rapidly frozen and stored on dry ice. The samples were subsequently quickly denatured by heating to 95°C for 10 min and were resolved by SDS-PAGE, and the proteolytic degradation products were visualized and quantified by PhosphorImager analysis.
RESULTS
RARα interacts with SMRT corepressor strongly and represses transcription in the absence of hormone, whereas RARβ and γ interact with SMRT poorly and fail to repress.
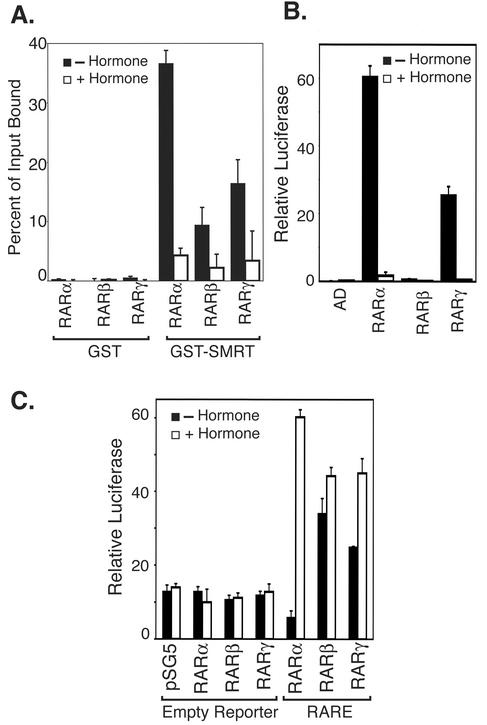
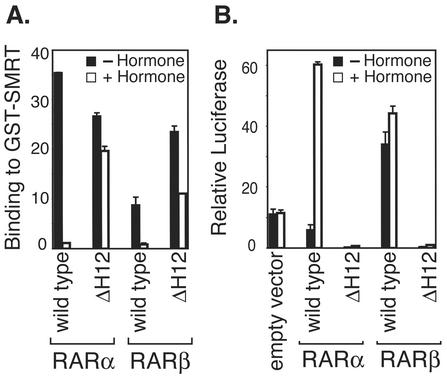
Consistent with prior reports (76), we determined that RARα bound to a GST-SMRT construct with high affinity in the absence of hormone, whereas RARβ and -γ displayed only a very weak interaction with GST-SMRT under the same conditions (Fig. 1A). Addition of ATRA disrupted both the strong binding seen for RARα and the residual binding observed for the RARβ and -γ isotypes, resulting in loss of all three receptor isotypes from the immobilized GST-SMRT (Fig. 1A). The impaired (relative to RARα) ability of RARβ and -γ to recruit SMRT in vitro was also observed in mammalian two-hybrid assays in vivo (Fig. 1B) when N-CoR was substituted for SMRT (76) and in the presence of a retinoid X receptor (RXR) heterodimer partner (data not shown). Paralleling these isotype-specific differences in corepressor binding, RARα repressed the expression of an RARE-thiamine kinase (TK) promoter luciferase reporter in transfected CV-1b cells in the absence of hormone, whereas RARβ and -γ failed to repress reporter gene expression under these conditions (Fig. 1C and 2B). Instead, both RARβ and -γ induced significant levels of reporter gene activation in the absence of hormone, and this activation was further increased by the addition of ATRA; RARα, in contrast, induced reporter gene activation only in the presence of the ATRA agonist (Fig. 1C and 2B). These differences in the abilities of RARα, -β, and -γ to induce repression versus activation of reporter expression in the absence of hormone were also observed in the presence of an RXRα expression vector, in L929 cells, and in the employment of GAL4-DNA binding domain (GAL4DBD) fusions of the corresponding RAR isotypes (Hauksdottir, Farboud, and Privalsky, submitted). Overexpression of SMRT in the transfected CV-1 cells did not convert either RARβ or RARγ into transcriptional repressors (data not shown).
FIG. 1.
RARα, but not RARβ or RARγ, interacts with SMRT corepressor in vitro and in vivo and represses reporter gene expression in transfected cells. (A) RARα efficiently binds to SMRT corepressor in a GST pulldown assay, but RARβ and RARγ do not. Radiolabeled RARα, -β, or -γ, as indicated below the panel, was synthesized in vitro by a coupled TnT reaction and incubated with a GST or a GST-SMRT construct previously immobilized to a glutathione-agarose matrix. Incubations were performed in the absence (filled bars) or presence (open bars) of 1 μM ATRA. The radiolabeled receptors remaining bound to the GST-SMRT matrix after repeated washing were eluted, resolved by SDS-PAGE, and quantified by PhosphorImager analysis. The percentage of each input protein bound to the GST or GST-SMRT matrix is shown; the averages and standard deviations from duplicate experiments are noted. (B) RARα, but not RARβ or RARγ, efficiently interacts with SMRT corepressor in a mammalian two-hybrid assay. pSG5 clones expressing GAL4AD alone or fused to the hormone binding domain of RARα, -β, or -γ (as noted below the panel) were cointroduced by transient transfection into CV-1 cells together with a GAL4DBD-SMRT construct and a GAL4-(17mer)-luciferase reporter vector. A pCMV-lacZ reporter was included as an internal normalization control. The cells were incubated in the absence or presence of 500 nM ATRA for 24 h prior to harvest and lysis. Relative luciferase activity, indicative of an interaction between receptor and corepressor, was determined. (C) The different RAR isoforms differ in their abilities to regulate reporter gene expression in the absence of hormone. An empty pTK-Luc reporter (Empty Reporter), or the same reporter containing three RAREs, was introduced into CV-1b cells by transfection together with a pSG5 vector expressing the native RARα, RARβ, or RARγ. A pCMV-lacZ reporter was included as an internal normalization control. The cells were incubated in the absence or presence of 500 nM ATRA for 24 h prior to lysis and analysis. Relative luciferase activity was calculated as described for panel B; two or more independent experiments, each in duplicate, were performed, and the averages and standard deviations are shown.
FIG. 2.
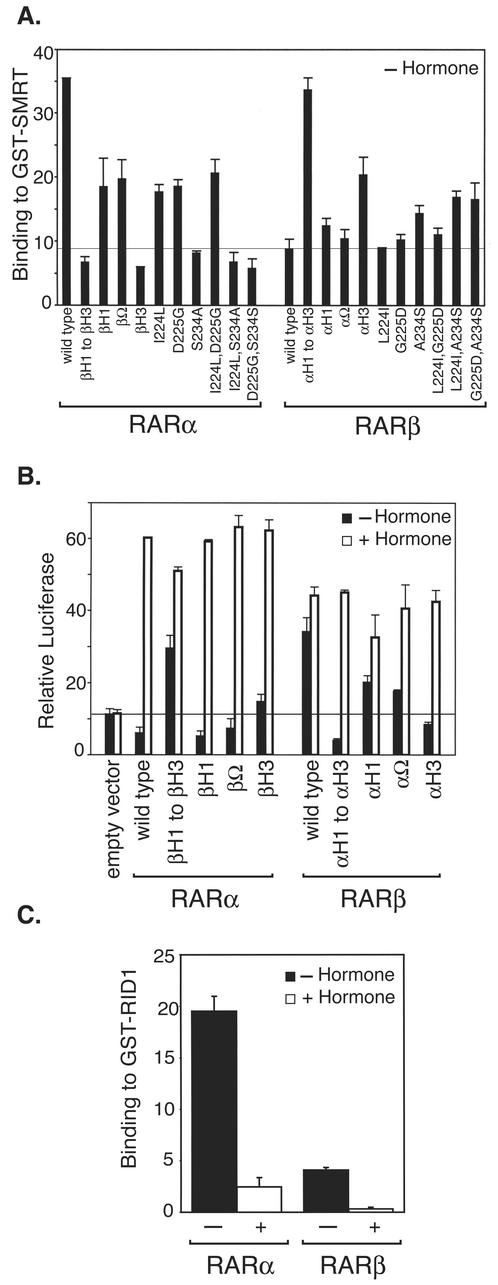
The differing corepressor and transcriptional regulatory properties of RARα and -β map primarily to helix 3 within the hormone binding domains of these receptors. (A) Chimeras of RARα that contain the helix 3 domain of RARβ display poor corepressor binding,whereas chimeras of RARβ that contain the helix 3 domain of RARα display efficient corepressor binding. Wild-type RARα, wild-type RARβ, or different chimeras and point mutants of the two isotypes, as indicated below the panel, were assayed as described for Fig. 1A for the ability to bind to a GST-SMRT construct in vitro. All assays were performed in the absence of hormone; binding of each RAR to a nonrecombinant GST construct was less than 1% of input. (B) Chimeras of RARα bearing the RARβ helix 3 domain activate reporter gene expression in the absence of hormone, whereas chimeras of RARβ possessing the RARα helix 3 domain repress expression. The different RAR chimeras and mutants described for panel A were tested for the ability to activate or repress the pTK-Luc-RARE reporter gene in transient transfections of CV-1b cells, as described for Fig. 1C. The cells were incubated in the absence (filled bars) or presence (open bars) of 500 nM ATRA and harvested 24 h later, and relative luciferase activity was determined. The average and standard deviation of two or more experiments are shown. (C) The different affinities of RARα and -β for SMRT are maintained when a corepressor domain restricted to the RID-1 region is employed. A GST protein interaction assay was performed as described for Fig. 1A but employed a recombinant GST-SMRT construct limited to the RID-1 domain (i.e., the more N terminal of the two RID domains in SMRT). The averages and standard deviations for two or more independent experiments, performed in duplicate, are shown.
The differences in corepressor recruitment by RARα and -β map primarily to differences in amino acid sequence in helix 3 of the ligand binding domain.
To map the receptor determinants responsible for the different corepressor recruitment properties of the different RAR isotypes, we created a series of chimeric constructs between RARα and -β and tested the chimeric receptors for the ability to bind to SMRT. Wong and Privalsky previously demonstrated that constructs of RARβ that contained the ligand binding domain derived from RARα displayed the RARα phenotype and interacted with SMRT with high affinity both in vitro and in vivo (76). Reciprocally, constructs of RARα that possessed the ligand binding domain from RARβ exhibited the RARβ phenotype and failed to efficiently interact with SMRT by either GST pulldown or by mammalian two-hybrid assay (76). Analysis of additional chimeras further mapped the relevant corepressor recruitment determinants to the N-terminal portion of the ligand binding domain, encompassing helices 1 through 3 (Fig. 2A and 3A). More detailed dissection demonstrated that the identity of the RAR helix 3 region played a dominant role in determining the SMRT binding properties of the two different RAR isoforms. For example, RAR chimeras that possessed an N-terminal half of helix 3 derived from the RARα sequence interacted with SMRT strongly, whereas RAR chimeras that possessed the corresponding N-terminal half of helix 3 derived from the RARβ sequence displayed little or no corepressor interaction (Fig. 2A). Nonetheless, it should be noted that the helix 1 and Ω loop subdomain of the RAR hormone binding domain did contribute, if more modestly, to the overall strength of the corepressor interaction (note that an RARβ construct containing the entire helix 1-Ω loop-helix 3 region of RARα interacted more strongly with SMRT than did an RARβ construct containing only the RARα helix 3; Fig. 2A).
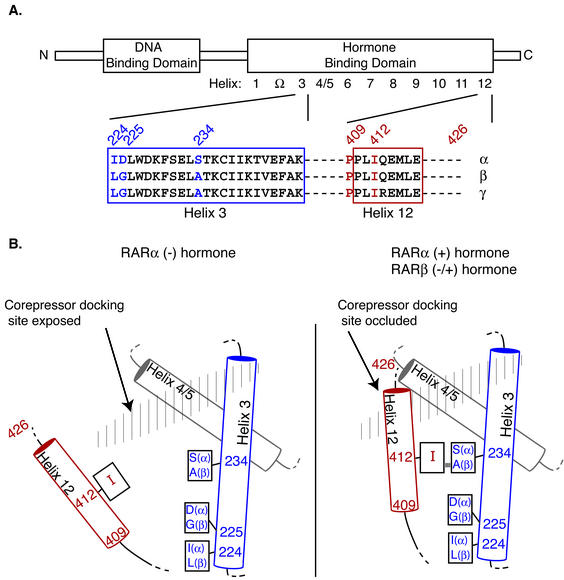
FIG. 3.
Schematics of the different RAR isotypes are presented. (A) A schematic representation of a generic RAR is depicted. The locations of the DNA and hormone binding domains, and the various structural features noted in the text, are indicated. The amino acid sequences of the different RAR isotypes are compared for helix 3 and helix 12. Amino acids described in the text are highlighted. (B) A model for the structures of the RARα and -β isotypes in the absence and presence of hormone is presented. The proposed orientations of helix 3, helix 4/5, and helix 12 in the absence or presence of a hormone agonist are shown for RARα and RARβ. The proposed docking surface for the corepressor (representing portions of helix 3 and helix 5/6) is depicted by hatched shading. The locations of amino acids 224, 225, 234, 409, 412, and 426 are indicated.
The ability to bind to SMRT in these interaction assays was also paralleled by transcriptional properties of these RAR constructs; RARβ chimeras that contained the relevant RARα helix 3 sequences lost the ability to activate reporter gene expression in the absence of hormone, whereas RARα chimeras that contained the equivalent helix 3 RARβ sequences failed to repress and displayed measurable hormone-independent activation instead (Fig. 2B). Here again, although the source of the helix 3 sequences played the dominant role in defining the transcriptional properties of the different isotypes in the absence of hormone, the identity of the helix 1 and the Ω loop also contributed (compare the effects of exchanging only helix 3 to the effects of exchanging the entire helix 1-Ω loop-helix 3 region; Fig. 2B).
There are three amino acid differences within the relevant regions of helix 3 of RARα and of RARβ, located at positions 224, 225, and 234 (to simplify discussion, we utilize a common numbering system for all three isotypes, as described in reference 58) (Fig. 3A). We therefore employed site-specific mutagenesis to exchange the RARα and -β sequences at these three helix 3 amino acids either individually or in sets of two. The majority of the isotype-specific corepressor interaction correlated with the identity of amino acid 234: an Ala 234-to-Ser substitution in RARβ significantly elevated the ability of this isotype to bind to SMRT, whereas the reciprocal Ser 234-to-Ala substitution in RARα resulted in a substantial decrease in SMRT binding (Fig. 2A). Nonetheless, individual or double mutations at codons 224 or 225 also affected, if more modestly, the RAR/SMRT interaction (Fig. 2A). We conclude that all three of these amino acid differences between helix 3 of RARα and helix 3 of RARβ contribute to the different corepressor interaction properties of these different isotypes but that codon 234 plays a dominant role in this regard.
The different corepressor interaction properties of RARα and -β reflect their abilities to interact with the previously identified (L/V)-X-X-I/V-I motif within the SMRT corepressor.
The SMRT and N-CoR corepressors are postulated to bind to RARα by the interaction of the nuclear receptor with L/V-X-X-I/V-I motifs within the corepressor (31, 32, 51, 56). Are the isotype-specific effects of helix 3 noted above manifested by altering the ability of RARα and -β to interact with these previously identified L/V-X-X-I/V-I corepressor motifs, or does helix 3 influence corepressor binding by contacting a previously unrecognized domain within the SMRT/N-CoR corepressor? To answer this question, we engineered a GST fusion protein that was restricted to the 19 amino acids immediately encompassing the L/I-X-X-I/V-I motif within the RID-1 of SMRT (i.e., the more N terminal of the two interaction domains in SMRT [76]). This abstracted SMRT subdomain retained the ability to bind to the radiolabeled RARα isoform but, in common with the native SMRT protein, exhibited little or no binding to the RARβ isoform (Fig. 2C). We conclude that the different helix 3 sequences of RARα and -β function by altering, directly or indirectly, the ability of these receptors to interact with the L/I-X-X-I/V-I motifs previously identified as the principal receptor contact surface on the SMRT/N-CoR corepressor.
Use of a yeast-based genetic suppression screen demonstrates that an intact receptor helix 12 is required for manifestation of the RARβ phenotype.
To better understand the molecular mechanism behind the different corepressor interaction properties displayed by RARα and -β, we next employed an unbiased genetic screen to identify any additional receptor domains involved in this phenomenon. We first devised a yeast two-hybrid assay in which a GAL4DBD-SMRT fusion construct and a GAL4 activation domain (GALAD)-RAR fusion construct were introduced together into S. cerevisiae cells bearing a GAL-17-mer-lacZ reporter (62). Consequently, an interaction between SMRT and RAR should manifest as enhanced β-galactosidase expression. The strong SMRT interaction properties of RARα, and the weak SMRT interaction properties of RARβ, were accurately mimicked in this system, with the RARα allele generating high levels of β-galactosidase expression and resulting in deep blue staining of the resulting recombinant yeast colonies in an X-Gal overlay assay, whereas the RARβ allele produced much lower levels of β-galactosidase expression and pale blue yeast colonies in the same assay (Fig. 4A and data not shown). Investigations with negative controls (devised by omitting either of the GAL4 fusion constructs, by using empty vectors, or by using irrelevant protein fusions in these assays) resulted in virtually undetectable levels of β-galactosidase expression and little or no X-Gal reactivity (Fig. 4 and data not shown).
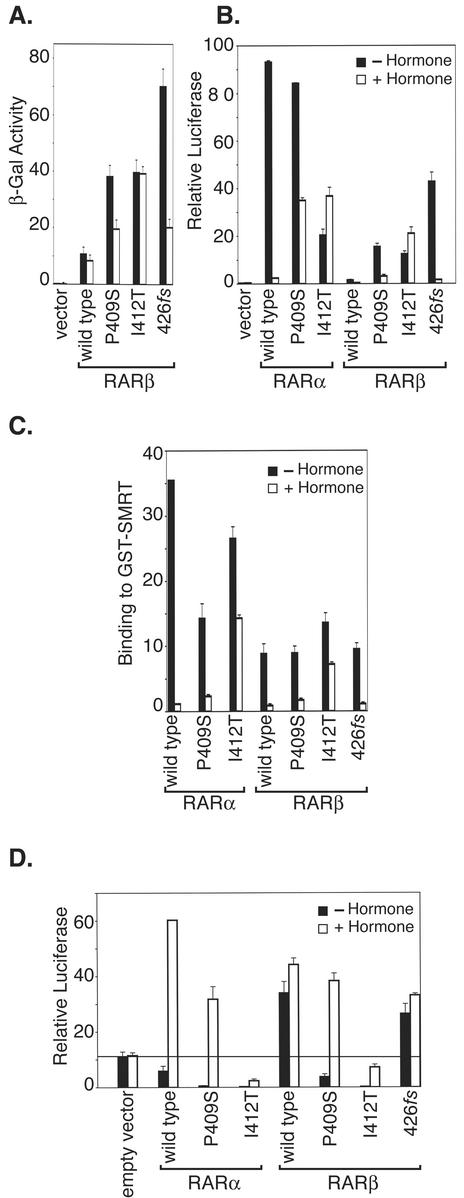
FIG. 4.
Gain-of-function mutations in the helix 12 domain of RARβ confer an elevated interaction with corepressor and the ability to repress reporter gene expression. (A) Mutations in helix 12 confer an elevated SMRT interaction on RARβ in a yeast two-hybrid analysis. Yeast cells were cotransformed with pACTII-GALAD-RARβ and pGBT9-GAL4DBD-SMRT constructs. Transformants were grown to stationary phase, diluted 1:5, and grown for an additional 12 h at 30°C in the absence (open bars) or presence (filled bars) of 1 μM ATRAprior to assay. Data are presented as β-galactosidase activity relative to the corresponding optical density of the culture at 600 nm. The results from four different transformants were combined for each mutant; the averages and standard deviations for three independent experiments are shown. (B) Mutations in helix 12 confer an elevated SMRT interaction on RARβ, but not RARα, in a mammalian two-hybrid analysis. The different helix 12 mutations assayed as described for panel A were tested for their effect on the ability of RARα and RARβ to interact with SMRT in a mammalian two-hybrid assay, as described for Fig. 1B; relative luciferase is plotted. (C) The I412T mutant enhances SMRT binding by RARβ, but not RARα, in a GST-protein interaction assay in vitro. The overall protocol was as described for Fig. 1A. (D) Mutations in helix 12 of RARβ confer the ability to repress target gene expression. The various helix 12 mutations were introduced into either the RARα or RARβ background, as indicated below the panel, and the resulting receptors were tested for the ability to repress or to activate the pTK-Luc-RARE reporter in the absence (filled bars) or presence (open bars) of 500 nM ATRA as described for Fig. 1C.
We next introduced random mutations into the RARβ molecular clone by its propagation in a mutSDT strain of E. coli, recovered the mutated RARβ cDNA population, excised the RARβ open reading frame, and introduced it back into a wild-type GAL4AD-plasmid vector backbone. The resulting GAL4AD-RARβ plasmid population was transformed into yeast possessing the wild-type GAL4DBD-SMRT, and the resulting yeast recombinants were screened visually for alterations in β-galactosidase expression. Of 10,000 colonies analyzed, 9 exhibited a significantly elevated level of β-galactosidase expression indicative of an enhanced interaction between RARβ and the SMRT corepressor (data not shown). The same increase in β-galactosidase expression was observed when the corresponding yeast recombinants were reisolated and reexamined in a liquid β-galactosidase assay and when the GAL4AD-RARβ mutant plasmids from these primary yeast transformants were recovered by transformation of E. coli and reintroduced into secondary yeast transformants bearing a naive GAL4DBD-SMRT construct (Fig. 4A and data not shown). These controls confirmed that the enhanced two-hybrid interaction phenomenon was inherent to the mutated RARβ alleles and was not due to either an epigenetic phenomenon or an unrecognized mutation in either the yeast host or in some other component of the two-hybrid system. An additional 287 mutant RARβ colonies displayed a nearly complete loss of β-galactosidase expression (i.e., white colonies instead of light blue); these “down” mutations, presumably receptor mutations that further disrupted the weak SMRT interaction characteristic of the wild-type RARβ, were not analyzed further.
The GAL4-RARβ plasmids that exhibited an enhanced SMRT interaction phenotype were recovered from the yeast and were subjected to DNA sequence analysis. The 9 plasmids represented three distinct mutations; all three mutant alleles retained the wild-type RARβ sequence throughout the helix 1 to helix 11 region but possessed substitution or frameshift mutations within or adjacent to receptor helix 12 (P409S, I412T, and a frameshift at 426 [426fs]; Fig. 3A). These RARβ “SMRT-up” mutants also displayed a detectably enhanced SMRT interaction in a mammalian two-hybrid analysis and (with the exception of 426fs) conferred strong transcriptional repression in transfected mammalian cells (Fig. 4B and D). The 426fs RARβ mutant paradoxically activates the reporter in our transfection experiments; although we do not understand the basis of this phenotype, it is possible that by appending novel amino acids onto the RAR C terminus, this frameshift has altered other properties of the receptor, such as coactivator recruitment, that might account for this effect.
The effects of these SMRT-up mutations were most pronounced when tested in the RARβ background; the same substitutions, when engineered into RARα, displayed much less of an effect on the already strong corepressor interaction properties of the latter isotype (Fig. 4B). The I412T RARβ mutant also displayed an enhanced SMRT interaction in a GST pulldown assay, whereas P409S and 426fs had relatively little or no effect in this in vitro assay (Fig. 4C); we presume that differences in the ionic environment or the presence of additional factors in intact cells stabilizes the interaction of the P409S and 426fs RARβ mutants with SMRT in two-hybrid and transcription assays in a manner that is not fully recapitulated in the GST pulldown. Overall, our results indicate that helix 12 interferes with the ability of the unliganded RARβ (but not RARα) to bind to SMRT.
RARβ possesses a corepressor binding site, but access to this site appears to be blocked by a constitutively closed position of helix 12.
In most unliganded nuclear receptors, helix 12 is thought to be in an extended position that permits access of a corepressor to a docking surface composed primarily of portions of receptor helix 3 and helix 4/5 (presented schematically in Fig. 3B). Binding of ligand by RARα is proposed to reorient helix 12 into a more sequestered position that occludes the corepressor binding site and generates a novel docking surface for coactivators (19, 57) (Fig. 3B). The location and nature of our intragenic suppressor mutations suggested that an isoform-specific interaction between RARβ helix 3 and helix 12 operates to gate helix 12 closed even in the absence of hormone, blocking access of SMRT to its docking site. Specifically, the Ala 234 in helix 3 of RARβ is in a position to interact with Ile 412 in helix 12 when helix 12 is gated closed (58); this hydrophobic pairing between helix 3 and helix 12 may be sufficient to favor the closed helix 12 conformation even in the absence of a ligand (Fig. 3B). Notably, this hydrophobic pairing is disrupted by the more-polar amino acids present in RARα (e.g., Ser at 234) or in our (I412T) RARβ mutant, both of which bind SMRT strongly (Fig. 3B). Our other RARβ mutants that confer an increase in the SMRT interaction are also likely to function by disrupting this helix 3-helix 12 interaction: the P409S substitution interrupts a hinge on which helix 12 rotates, and the 426 frameshift adds extraneous sequence to helix 12 that is likely to prevent proper packing of the folded helix 12 against the receptor surface (Fig. 3B). Significantly, simply deleting helix 12 also conferred strong corepressor binding and repression on RARβ (Fig. 5). Taken as a whole, our results indicate that the RARβ isoform possesses a fully functional corepressor docking site but that this site is occluded by a constitutively closed helix 12 position.
FIG. 5.
Deleting helix 12 greatly enhances the ability of RARβ to bind to SMRT corepressor in vitro and to repress target gene expression in transfected cells. (A) Deletion of helix 12 increases corepressor binding. Wild-type and C-terminal-deleted RARα (amino acids 1 to 403) and RARβ (amino acids 1 to 396) were tested for the ability to bind to GST-SMRT by the protocol described for Fig. 1A. (B) Deletion of helix 12 converts RARβ from a hormone-independent activator to a constitutive repressor of reporter gene expression in transiently transfected CV-1b cells. The protocol described for Fig. 1C was used.
A series of site-directed mutations that disrupt the hydrophobic interaction between amino acids 234 and 412 in RARβ convert RARβ into a transcriptional repressor in vivo and elevate corepressor binding in vitro.
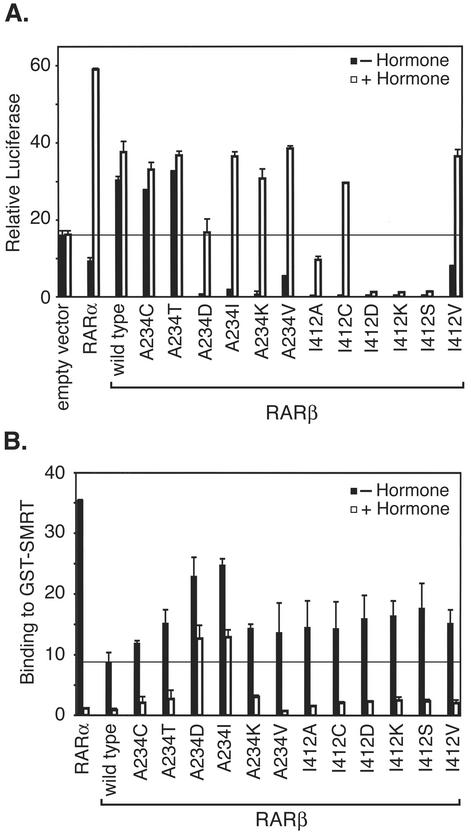
We next created and characterized a series of amino acid substitutions in helix 3 and helix 12 to further test our hypothesis that a hydrophobic interaction between these two domains was involved in gating access of the corepressor to the receptor. Replacement of Ala 234 in RARβ with Cys (approximately the same size and hydrophobicity as Ala) retained the RARβ phenotype of activating, rather than repressing, target gene expression in transfected cells (Fig. 6A). The A234T mutation in RARβ also retained the RARβ phenotype; by several criteria, threonine is considered to be more hydrophobic than serine, and this property may account for the RARβ-like ability of the A234T mutant to activate transcription in the absence of hormone.
FIG. 6.
Mutations in RARβ that disrupt the hydrophobic interaction surface between helix 3 and helix 12 increase corepressor binding and enhance repression of reporter expression. (A) Point mutations in helix 3 or 12 can convert RARβ into a repressor. The ability of wild-type RARα or wild-type RARβ or specific amino acid mutants of the latter to repress or activate the pTK-Luc-RARE reporter in transiently transfected CV-1b cells was determined as described for Fig. 1C. (B) Point mutations in helix 3 or helix 12 can confer enhanced corepressor binding on RARβ. The ability of wild-type RARα or RARβ or specific mutants of the latter to bind to SMRT in a GST protein interaction assay was determined as described for Fig. 1A.
In contrast, replacement of Ala 234 with amino acids expected to disrupt the interaction of helix 3 with helix 12 (replacements involving charged or bulky residues, such as A234D, A234K, or A234I), converted RARβ into the RARα-like phenotype, resulting in repression in the absence of hormone and activation in the presence of hormone (Fig. 6A). An A234V substitution, representing a hydrophobic amino acid of intermediate size, generated an intermediate phenotype (Fig. 6A). Replacing the helix 12 amino acid in RARβ with amino acids smaller or more polar than the wild-type Ile 412 (e.g., I412A, I412C, I412D, I412K, or I412S) also converted RARβ into an RARα-like repressor, whereas a more conservative replacement, I412V, generated an intermediate effect on reporter expression (Fig. 6A). Although the I412A and I412C mutants remained hormone responsive, the more highly polar or charged RARβ I412D, I412K, and I412S mutants failed to activate on addition of ATRA and behaved as constitutive repressors (Fig. 6A); possibly these latter mutations are sufficiently disruptive to the proper folding of helix 12, or to hormone binding, so as to prevent corepressor release-coactivator recruitment even in the presence of an agonist. In general, the amino acid substitutions that conferred repression on RARβ in transfected cells also elevated corepressor recruitment in vitro, although the effects were not always proportionate in both assays (Fig. 6B). In conclusion, a variety of (otherwise diverse) amino acid substitutions that shared the ability to disrupt the hydrophobic character of the helix 3-helix 12 interaction surface also shared the ability to confer enhanced corepressor binding and enhanced transcriptional repression on RARβ.
Consistent with our model, the RARα helix 12 is more accessible to protease in the absence than in the presence of ATRA, whereas the RARβ helix 12 is in a sequestered position (resistant to proteolytic degradation) both in the absence and the presence of hormone.
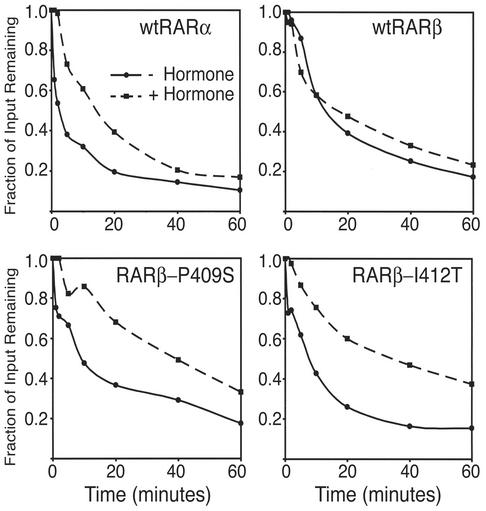
Unfortunately, no X-ray or nuclear magnetic resonance structure has been reported for the unliganded RARα, -β, or -γ. The position of helix 12 in these receptors, however, can be approximated by the use of CPY, an exopeptidase that serves as a probe of the solvent accessibility of the protein C terminus (28, 42, 83). Consistent with the prevailing model of hormone-driven helix 12 gating, the wild-type RARα was significantly more sensitive to CPY digestion in the absence of ATRA (indicative of an open helix 12 position) than in the presence of ATRA (suggestive of a more closed helix 12 position) (Fig. 7). In contrast, the wild-type RARβ was comparatively resistant to CPY digestion either in the absence or the presence of ATRA hormone, which is consistent with a constitutively sequestered helix 12 conformation (Fig. 7). Notably, our RARβ mutants that displayed an elevated (i.e., more RARα-like) SMRT interaction also displayed a more RARα-like pattern of CPY degradation: an increased sensitivity to carboxypeptidase digestion in the absence of hormone but protection in the presence of ATRA (Fig. 7 and data not shown). These results establish a correlation between the inability of the wild-type RARβ to recruit corepressor and the existence of a more sequestered helix 12 position in the unliganded conformation of this isotype.
FIG. 7.
RARβ is resistant to carboxypeptidase degradation in both the absence and presence of hormone, unlike RARα and the RARβ-P4009S or RARβ-I412T mutants. Radiolabeled receptor was synthesized in vitro by a coupled TnT reaction and incubated with CPY for the times indicated below the panel in the absence (solid line) or presence (dashed line) of 1 μM ATRA. The resulting proteolytic products were resolved by SDS-PAGE and were visualized and quantified by PhosphorImager analysis. The fraction of full-length receptor remaining at each indicated time point is shown. A representative experiment is provided; analogous results were obtained in three independent experiments.
RARβ is able to bind to p160 coactivators in the absence of hormone.
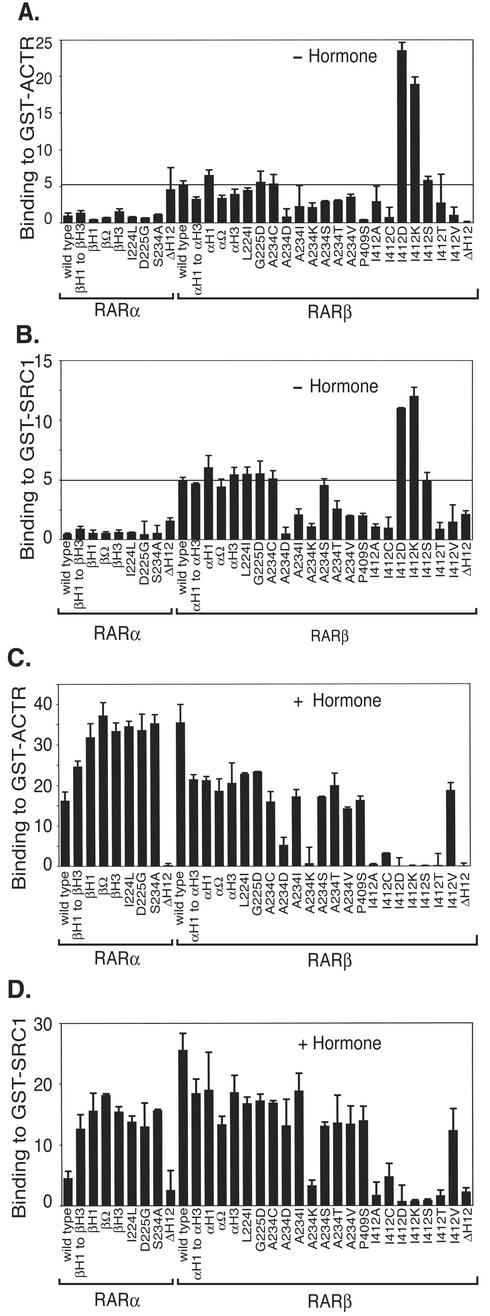
Our transfection studies established that RARβ not only fails to repress but also has a detectable ability to activate reporter gene expression even in the absence of ligand. To determine the basis for this property, we investigated whether RARβ possessed an elevated ability to bind to transcriptional coactivators in the absence of hormone compared to RARα. Indeed, the unliganded RARβ displayed a three- to ninefold-greater interaction with ACTR and SRC-1 in a GST pulldown assay than did unliganded RARα (Fig. 8A and B). This ability of RARβ to bind to coactivators in the absence of hormone was decreased by several of our mutations in helix 3 and helix 12 that increased corepressor binding; many of our substitutions at codon 234 and at codon 412, the P409S replacement, and the helix 12 deletion all disrupted coactivator binding by RARβ to a significant extent (Fig. 8B). Therefore, the enhanced coactivator binding by RARβ may be, in part, a consequence of the constitutively closed helix 12 conformation assumed by this isotype: the closed conformation in the unliganded RARβ may sufficiently mimic that of an agonist-bound RARα to form a docking site for coactivator recruitment. Nonetheless, note that coactivator binding by RARβ was further substantially increased by the addition of ATRA, suggesting that the conformation of helix 12 in the unliganded RARβ is not identical to the agonist-induced conformation (Fig. 8C and D). Furthermore, analysis of our chimeric RARα/β constructs indicated that regions of the RARβ receptor mapping outside of helix 3 were also critical contributors to the enhanced coactivator binding properties of this isotype relative to RARα. Two of our highly charged substitutions into RARβ helix 12, I412D and I412K, paradoxically increased coactivator binding (Fig. 8A and B). We conclude that RARβ possesses an inherent ability to interact with p160 coactivators in the absence of hormone and that this property overlaps, but is distinguishable from, the inability of this receptor to bind the corepressor.
FIG.8.
RARβ displays an elevated ability to interact with p160 coactivators in the absence of hormone relative to RARα. The ability of wild-type, chimeric, and point mutants of RARα and RARβ to bind in vitro to GST-ACTR (amino acids 621 to 821) (A and C) or to GST-SRC1 (amino acids 560 to 1136) (B and D) was analyzed by the same general protocol employed for SMRT as described for Fig. 1A. The percentage of binding (relative to input) of each receptor to the coactivator matrix is shown. Receptor binding to a GST-only control was 0.3 to 0.7% of input. The averages and standard deviations of at least two independent experiments, each performed in duplicate, are provided. The assay was performed both in the absence (A and B) and presence (C and D) of 1 μM ATRA.
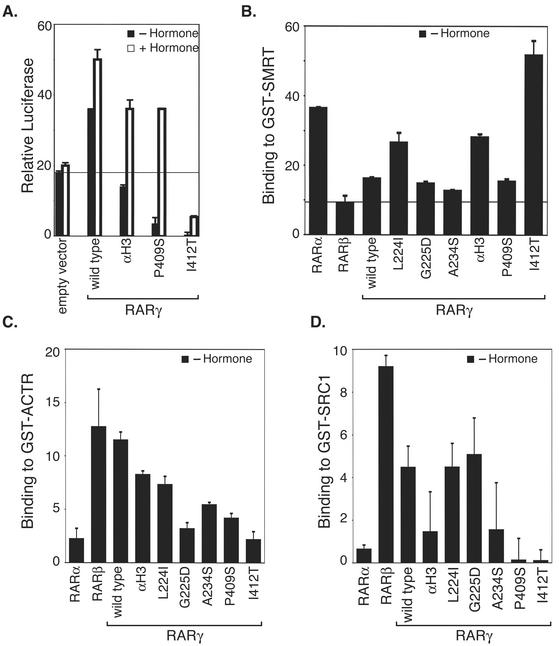
RARγ more closely resembles RARβ than RARα in its transcriptional and corepressor interaction properties.
We also investigated RARγ, the third major RAR isotype. RARγ possesses a helix 3 amino acid sequence identical to that of RARβ, as well as retaining the conserved Ile 412 in the helix 12 domain (Fig. 3A) (58). Consistent with these sequence identities, the transcriptional properties of RARγ were very similar to those of RARβ; RARγ activated, rather than repressed, reporter gene expression in the absence of hormone, and RARγ displayed a relatively weak interaction with the SMRT corepressor compared to RARα (Fig. 1). Molecular determinants similar to those in RARβ appeared to be responsible for these phenomena in RARγ: replacing helix 3 in RARγ with that of RARα, or introducing mutations in helix 12 that mimicked the enhanced SMRT mutations we isolated in RARβ, resulted in increased corepressor binding and transcriptional repression by RARγ (Fig. 9A and B). As with RARβ, the RARγ isotype displayed an elevated ability to interact in vitro with p160 coactivators in the absence of hormone (Fig. 9C and D). However, when examined in detail, RARγ did diverge from both RARβ and RARα in several of its properties. Most notably, the interaction RARγ displayed with SMRT in either the GST pulldown or mammalian two-hybrid assay, although weak, was nonetheless stronger that that observed with RARβ (Fig. 1). Also the relative contributions of amino acids 224, 225, and 234 to the corepressor binding phenotype differed in details for RARβ and RARγ, with the identity of amino acid 224 being more critical in the latter (Fig. 9B). It is possible that these differences in transcriptional regulation by RARγ versus that by RARα or RARβ reflect additional amino acid sequence differences either in helix 12 itself (most notably, amino acid 413 in RARγ is an Arg, whereas both RARβ and RARα have a Gln at this location; Fig. 3A) or in regions elsewhere in the coding domains of these different isotypes.
FIG. 9.
RARγ resembles RARβ in its corepressor binding, transcriptional regulation, and p160 coactivator binding properties. (A) RARγ activates reporter gene expression in the absence of hormone, and this property maps to helix 3. Using the procedure described for Fig. 1A, the ability of wild-type RARγ and the RARγ chimeras and mutants (as indicated below the panel) to regulate the expression of the pTK-Luc-RARE reporter was tested by transient transfection of CV-1b cells. (B) RARγ binds SMRT weakly, and this property maps primarily to helix 3 and helix 12. The ability of wild-type RARα, RARβ, and RARγ and of specific mutants of the last to bind to GST-SMRT in vitro was tested in the GST-protein interaction assay described for Fig. 1A. (C and D) RARγ interacts with p160 coactivators in the absence of hormone more strongly than does RARα. Using the GST protein interaction assay described for Fig. 7, the ability of wild-type RARα, RARβ, and RARγ to bind to ACTR (C) or to SRC-1 (D) was determined in vitro. The averages and standard deviations for at least two independent experiments are shown.
DISCUSSION
In the absence of hormone, RARα binds SMRT/N-CoR corepressor strongly and is a potent transcriptional repressor; RARβ and -γ, in contrast, bind SMRT/N-CoR only weakly and fail to repress and can activate transcription even in the absence of hormone.
In all vertebrates characterized to date, RARs are encoded by three distinct loci, denoted α, β, and γ. This coding scheme is evolutionarily tightly conserved, and the sequence conservation that a given isotype displays in comparison to its orthologs among different species is greater than that seen in comparison to its isotype paralogs within the same species. This evolutionary stability suggests that the different RAR isotypes perform distinct physiological functions. Consistent with this supposition, use of gene disruption techniques and isotype-specific ligands has confirmed that RARα, -β, and -γ play unique as well as overlapping roles in gene regulation (9, 68). In the work presented here, we have identified at least one molecular basis for the divergent biological capabilities of the different RAR isotypes: RARα can recruit the SMRT corepressor in the absence of a ligand and repress target gene expression, whereas the RARβ and RARγ isotypes are unable to efficiently bind the SMRT corepressor and instead possess the ability to activate transcription in both the absence and the presence of hormone. The different cofactor and transcriptional properties of RARα, -β, and -γ are likely to manifest in many aspects of retinoid signaling and help account for the incomplete biological redundancy previously noted for the different RAR isotypes. Note in this regard that the promoters for many of the RAR isoforms contain retinoid response elements and therefore respond to retinoic acid in a positive-feedback loop by which hormone induces additional receptor expression (26, 40, 41, 69); the constitutive activation properties of RARβ and RARγ are likely to influence the dynamics of these feedback loops quite differently than would the repressive properties of the unliganded RARα.
The different corepressor binding and transcriptional repression properties of RARα, -β, and -γ map to amino acid differences within helix 3 of the ligand binding domains of these receptors.
Use of chimeric constructs permitted us to demonstrate that the different corepressor interaction and transcriptional repression characteristics of RARα, -β, and -γ map predominantly to differences in the helix 3 sequences in the ligand binding domains of these different isotypes. RARs bearing the α isotype sequence within the N-terminal half of helix 3 bound to the SMRT corepressor strongly in vitro and repressed reporter gene expression in vivo, whereas RARs bearing the β or γ isotype sequence within the N-terminal half of helix 3 interacted only weakly with the corepressor and failed to repress target gene transcription. There are three amino acid differences, at positions 224, 225, and 234, between the relevant region of helix 3 of RARα and those of RARβ and -γ, and all three contribute to some extent to the differences in corepressor recruitment and repression noted for the different isotypes. Intriguingly, although the C-terminal half of helix 3 comprises a portion of the corepressor docking site in the nuclear hormone receptors, amino acids 224 and 225 are distal to the actual sites of contact between corepressor and receptor, and amino acid 234 lies on the boundary of the known corepressor docking surface (31, 32, 46, 51, 56, 79). In fact, both RARβ and -γ possess fully functional corepressor docking sites, but the accessibility of the corepressor to these docking sites appears to be restricted by the actions of helix 12, as discussed below.
A hydrophobic interaction between helix 3 and helix 12 stabilizes the closed helix 12 position in RARβ and RARγ in the absence of hormone, occluding corepressor binding and preventing repression.
To determine the molecular basis behind the inability of RARβ and RARγ to efficiently bind the SMRT corepressor, we utilized a yeast genetic screen to identity intragenic suppressors that can confer enhanced SMRT binding on the RARβ isoform. The enhanced SMRT-RARβ interaction mutations isolated by this approach all mapped within or proximal to the receptor helix 12 coding region. Simple deletion of helix 12 also conferred strong corepressor binding and strong transcriptional repression on both RARβ and RARγ. We conclude that all three RAR isotypes possess functional docking sites for the SMRT corepressor but that these sites in RARβ and -γ are rendered cryptic by the presence of the native helix 12 sequence. Notably, all three of the helix 3 amino acids that differ between RARα, -β, and -γ are in a position to influence the position of helix 12. Amino acid 234 (an Ala in RARβ and -γ but a Ser in RARα) is in a position to interact with Ile 412 in helix 12 when the latter helix is in the agonist-bound conformation (58). Similarly, amino acid 224 (a Leu in RARβ and -γ but an Ile in RARα) and amino acid 225 (a Gly in RARβ and -γ but an Asp in RARα) are in close proximity to helix 11 and to the base of helix 12 (58).
Allosteric changes centered on this and adjacent regions of the nuclear receptors have been shown to play critical roles in modulating the repositioning of helix 12 in response to antagonists and to receptor mutations (18, 38). We propose that a hydrophobic interaction between complementary portions of helix 3 (centered on Ala 234) and helix 12 (centered on Ile 412), perhaps further facilitated by the allosteric contributions of the amino acids at positions 224 and 225, helps gate helix 12 closed in RARβ and -γ in the absence of hormone and thus prevents corepressor binding by these isotypes. We further suggest that the helix 3 sequence of RARα disrupts this helix 3-helix 12 interaction sufficiently so that helix 12 assumes an open conformation, allowing access of the corepressor to its docking surface in the absence of hormone but also permitting helix 12 to gate shut and to release the corepressor upon the binding of hormone. A similar concept, although operating through a structurally distinct mechanism, has been proposed for the constitutive androstane receptor; this receptor, in the absence of a physiological ligand, appears to assume an agonist-like closed helix 12 conformation due, in part, to a stabilizing ionic interaction between the α-carboxylic acid of the receptor C terminus and a Lys located within helix 4 (17). Similarly, Ultraspiracle, the insect homolog of vertebrate RXR, also displays a closed helix 12 conformation in the absence of a ligand; this closed helix 12 conformation resembles that found in RXR and estrogen receptors when bound to certain mixed agonists-antagonists and is conferred on Ultraspiracle by an unusually positioned L1-3 loop that contacts and interlocks helix 3, helix 11, and helix 12 (5, 14).
Consistent with our hypothesis that RARβ and -γ assume a constitutively closed helix 12 conformation, disruption of the contact surface between helix 3 and helix 12 by the introduction of polar, charged, or bulky amino acid substitutions in either helix results in increased corepressor binding and increased transcriptional repression by RARβ and RARγ. The wide diversity of amino acid substitutions that confer corepressor binding on RARβ and on RARγ supports our argument that these mutations do not generate a corepressor docking site de novo but rather, by disrupting the interaction of helix 3 with helix 12, expose a existing, but previously cryptic docking site. Notably, our P409S substitution and 426fs mutations that also enhance SMRT binding by RARβ are similarly positioned so as to interfere with the ability of helix 12 to pivot correctly into the sequestered position, either by disrupting the pivot itself or by appending irrelevant protein sequence onto helix 12.
Naturally occurring mutations in these same receptor domains have been implicated in other contexts in which corepressor interaction is favored. For example, the PML-RARα oncoprotein plays a causal role in human acute promyelocytic leukemia, and the ability of this protein to recruit SMRT is closely associated with maintenance of the neoplastic phenotype (20, 22, 25, 27, 36, 43, 62). The native PML-RARα protein releases from the corepressor in response to high levels of ATRA, inducing differentiation of the leukemic cell and clinical remission (20, 22, 25, 27, 36, 43, 62). Two mutants of PML-RARα that are resistant to this ATRA-induced differentiation have been identified: one possesses a Leu-to-Pro substitution at position 411, whereas a second bears an Ile-to-Thr substitution equivalent to our RAR position 412 mutation (15, 63). Similarly, resistance to thyroid hormone syndrome, an inherited endocrine disease, is caused by the production of mutant thyroid hormone receptors (TRs) that fail to dissociate from the corepressor in response to physiological levels of hormone (10, 50, 61, 70, 84); the most abundant mutations identified in resistance to thyroid syndrome alter a Pro in the TRβ, corresponding to codon 409 in RAR, to Ala, Ser, Thr, or Asp (10, 82). Analogously, a Val-to-Met mutation in the peroxisome-proliferator-activated receptor γ at a site equivalent to RAR position 234, or a Pro-to-Leu substitution at a site equivalent to position 410, prevents corepressor release by peroxisome-proliferator-activated receptor γ in response to a ligand and is associated with severe inherited type II diabetes (3). Taken together, these results suggest that interactions between helix 3 and helix 12 play a critical role in the gating of helix 12 in many nuclear receptors and that defects in these interactions can lead to a variety of endocrine and neoplastic disorders.
Structural probes employing exopeptidase support the hypothesis that RARβ and -γ possess a constitutively closed helix 12 conformation.
A crystallographic or nuclear magnetic resonance analysis of the unliganded RARs has not been reported. We therefore employed CPY as an enzymatic probe of the solvent accessibility of helix 12 in the different receptor isotypes. Consistent with the accepted model of a hormone-induced alteration from an open to a closed helix 12 position, the C terminus of RARα is more resistant to CPY degradation in the presence, versus the absence, of the ATRA ligand. A similar ligand-induced alteration in the CPY susceptibility of helix 12 is also observed in the wild-type TR, which switches in a like manner from a transcriptional repressor to an activator in response to cognate hormone. In contrast, the C-terminal helix 12 of wild-type RARβ (and -γ) displays a hormone-independent resistance to CPY consistent with a constitutively sequestered helix 12 position. Furthermore, mutations in either helix 3 or 12 that confer corepressor binding on RARβ and -γ also confer CPY susceptibility on these isoforms, indicative of a close correlation between the exposed position of helix 12 and the ability to bind SMRT.
In addition to occluding corepressor binding, a closed conformation of helix 12 can also help form a docking surface for the L-X-X-L-L motifs that are found in many coactivators (19). In superficial agreement with this concept, both the RARβ and RARγ isotypes exhibit higher levels of hormone-independent p160 coactivator binding in vitro and hormone-independent transcriptional activation in vivo than does RARα. We note, however, that although both RARβ and -γ bind coactivators in the absence of hormone, addition of hormone induces still higher levels of p160 binding, indicating that the conformation of these receptors in the unliganded state is not identical to that assumed by the same receptors on the binding agonist. Additionally, receptor sequences outside of helix 3 and helix 12 also contribute to the elevated, hormone-independent coactivator binding properties of RARβ and -γ. Intriguingly, repression by unliganded RARβ generally proved to be dominant over activation; i.e., most of the RARβ mutants that recruit corepressor in vitro also repress reporter gene repression in transfected cells, even when these mutants retain the elevated SRC-1/ACTR binding characteristics of the unliganded wild-type RARβ. Although the specific coactivators responsible for the constitutive activation properties of RARβ and -γ in cells are not known, our results suggest that RARβ and -γ possess an innate ability to bind to p160 coactivators in the absence of hormone. However, for this innate coactivator binding to be manifested as target gene activation in vivo, it appears also necessary to prevent corepressor binding through the helix 3-helix 12 interaction elucidated in this work.
Helix 12 is a critical, multiposition toggle switch that controls corepressor and coactivator binding by nuclear hormone receptors and is programmed to operate differently in different receptor isoforms.
Helix 12 in the ligand binding domain of the nuclear receptors is surprisingly malleable and can assume any of a spectrum of different conformations; these conformations differ in the unliganded receptor, in the presence of different agonists and antagonists, in response to posttranslational modifications of the receptor, and under the influence of the receptor dimer partner (for examples, see references 5-8, 14, 52, 58, 64, 74, and 79). The different helix 12 conformations, in turn, play a key role in mediating the interactions of the receptor with auxiliary factors such as corepressors and coactivators (19, 57). Genetic defects that disrupt the ability of helix 12 to position properly in response to hormone lead to defects in cofactor recruitment and release that manifest as endocrine and neoplastic diseases (for examples, see references 3, 36, and 82). In the work reported here, we establish that different isotypes of the same receptor are evolutionarily programmed to display inherently different helix 12 conformations in the absence of hormone and therefore to manifest different cofactor recruitment and transcriptional regulatory properties. This observation helps explain the different roles that the three different RAR isotypes play in development and physiology and provides a rationale for the evolutionary conservation of these three distinct isotypes during the vertebrate divergence (9, 68). The closed position of helix 12 that we propose to be the null state of the unliganded RARβ and -γ need not be immutable; however, posttranslational modifications, antagonists, or interactions with other proteins may, in theory, be able to open the helix 12 conformation in RARβ and -γ and thereby confer corepressor binding and transcriptional repression. We are currently exploring this possibility.
Acknowledgments
This work was supported by Public Health Service grant DK53528 from the National Institute of Diabetes, Digestive, and Kidney Diseases. B.F. was also funded, in part, by a Public Health Service predoctoral training award, T32GM07377, from the National Institute of General Medical Sciences.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. J. Wiley, New York, N.Y.
- 2.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 3.Barroso, I., M. Gurnell, V. E. Crowley, M. Agostini, J. W. Schwabe, M. A. Soos, G. L. Maslen, T. D. Williams, H. Lewis, A. J. Schafer, V. K. Chatterjee, and S. O'Rahilly. 1999. Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880-883. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2001. Molecular biology. The histone modification circus. Science 292:64-65. [PubMed] [Google Scholar]
- 5.Billas, I. M., L. Moulinier, N. Rochel, and D. Moras. 2001. Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J. Biol. Chem. 276:7465-7474. [DOI] [PubMed] [Google Scholar]
- 6.Bourguet, W., M. Ruff, P. Chambon, H. Gronemeyer, and D. Moras. 1995. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature 375:377-382. [DOI] [PubMed] [Google Scholar]
- 7.Bourguet, W., V. Vivat, J. M. Wurtz, P. Chambon, H. Gronemeyer, and D. Moras. 2000. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5:289-298. [DOI] [PubMed] [Google Scholar]
- 8.Brzozowski, A. M., A. C. W. Pike, Z. Dauter, R. E. Hubbard, T. Bonn, O. Engstrom, L. Ohman, G. L. Greene, J. A. Gustafsson, and M. Carlquist. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753-758. [DOI] [PubMed] [Google Scholar]
- 9.Chambon, P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940-954. [PubMed] [Google Scholar]
- 10.Chatterjee, V. K. 1997. Resistance to thyroid hormone. Horm. Res. 48(Suppl. 4): 43-46. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 13.Chen, J. D., and H. Li. 1998. Coactivation and corepression in transcriptional regulation by steroid/nuclear hormone receptors. Crit. Rev. Eukaryot. Gene Expr. 8:169-190. [DOI] [PubMed] [Google Scholar]
- 14.Clayton, G. M., S. Y. Peak-Chew, R. M. Evans, and J. W. Schwabe. 2001. The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation. Proc. Natl. Acad. Sci. USA 98:1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cote, S., A. Rosenauer, A. Bianchini, K. Seiter, J. Vandewiele, C. Nervi, and W. H. Miller, Jr. 2002. Response to histone deacetylase inhibition of novel PML/RARα mutants detected in retinoic acid-resistant APL cells. Blood 100:2586-2596. [DOI] [PubMed] [Google Scholar]
- 16.de The, H., A. Marchio, P. Tiollais, and A. Dejean. 1987. A novel steroid thyroid hormone receptor-related gene inappropriately expressed in human hepatocellular carcinoma. Nature 330:667-670. [DOI] [PubMed] [Google Scholar]
- 17.Dussault, I., M. Lin, K. Hollister, M. Fan, J. Termini, M. A. Sherman, and B. M. Forman. 2002. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol. Cell. Biol. 22:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangloff, M., M. Ruff, S. Eiler, S. Duclaud, J. M. Wurtz, and D. Moras. 2001. Crystal structure of a mutant hERα ligand-binding domain reveals key structural features for the mechanism of partial agonism. J. Biol. Chem. 276:15059-15065. [DOI] [PubMed] [Google Scholar]
- 19.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 20.Grignani, F., S. De Matteis, C. Nervi, L. Tomassoni, V. Gelmetti, M. Cioce, M. Fanelli, M. Ruthardt, F. F. Ferrara, I. Zamir, C. Seiser, M. A. Lazar, S. Minucci, and P. G. Pelicci. 1998. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391:815-818. [DOI] [PubMed] [Google Scholar]
- 21.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 22.Guidez, F., S. Ivins, J. Zhu, M. Söderström, S. Waxman, and A. Zelent. 1998. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARα underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood 91:2634-2642. [PubMed] [Google Scholar]
- 23.Hassig, C. A., and S. L. Schreiber. 1997. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1:300-308. [DOI] [PubMed] [Google Scholar]
- 24.Hauksdottir, H., and M. L. Privalsky. 2001. DNA recognition by the aberrant retinoic acid receptors implicated in human acute promyelocytic leukemia. Cell Growth Differ. 12:85-98. [PMC free article] [PubMed] [Google Scholar]
- 25.He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat. Genet. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann, B., J. M. Lehmann, X. K. Zhang, T. Hermann, M. Husmann, G. Graupner, and M. Pfahl. 1990. A retinoic acid receptor-specific element controls the retinoic acid receptor-beta promoter. Mol. Endocrinol. 4:1727-1736. [DOI] [PubMed] [Google Scholar]
- 27.Hong, S. H., G. David, C. W. Wong, A. Dejean, and M. L. Privalsky. 1997. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94:9028-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong, S. H., and M. L. Privalsky. 1999. Retinoid isomers differ in the ability to induce release of SMRT corepressor from retinoic acid receptor-alpha. J. Biol. Chem. 274:2885-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong, S. H., and M. L. Privalsky. 2000. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell. Biol. 20:6612-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horwitz, K. B., T. A. Jackson, D. L. Bain, J. K. Richer, G. S. Takimoto, and L. Tung. 1996. Nuclear receptor coactivators and corepressors. Mol. Endocrinol. 10:1167-1177. [DOI] [PubMed] [Google Scholar]
- 31.Hu, X., and M. A. Lazar. 1999. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402:93-96. [DOI] [PubMed] [Google Scholar]
- 32.Hu, X., Y. Li, and M. A. Lazar. 2001. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell. Biol. 21:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson, T. A., J. K. Richer, D. L. Bain, G. S. Takimoto, L. Tung, and K. B. Horwitz. 1997. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol. Endocrinol. 11:693-705. [DOI] [PubMed] [Google Scholar]
- 34.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 35.Koenig, R. J. 1998. Thyroid hormone receptor coactivators and corepressors. Thyroid 8:703-713. [DOI] [PubMed] [Google Scholar]
- 36.Kogan, S. C., S. H. Hong, D. B. Shultz, M. L. Privalsky, and J. M. Bishop. 2000. Leukemia initiated by PMLRARα: the PML domain plays a critical role while retinoic acid-mediated transactivation is dispensable. Blood 95:1541-1550. [PubMed] [Google Scholar]
- 37.Kralli, A., S. P. Bohen, and K. R. Yamamoto. 1995. LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc. Natl. Acad. Sci. USA 92:4701-4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, G., F. Elwood, J. McNally, J. Weiszmann, M. Lindstrom, K. Amaral, M. Nakamura, S. Miao, P. Cao, R. M. Learned, J. L. Chen, and Y. Li. 2002. T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J. Biol. Chem. 277:19649-19657. [DOI] [PubMed] [Google Scholar]
- 39.Lee, H., and P. M. Yen. 1999. Recent advances in understanding thyroid hormone receptor coregulators. J. Biomed. Sci. 6:71-78. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann, J. M., X.-K. Zhang, and M. Pfahl. 1992. RARγ2 expression is regulated through a retinoic acid response element embedded in Sp1 sites. Mol. Cell. Biol. 12:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leroy, P., H. Nakshatri, and P. Chambon. 1991. Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc. Natl. Acad. Sci. USA 88:10138-10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, B. C., S.-H. Hong, S. Krig, S. M. Yoh, and M. L. Privalsky. 1997. A conformational switch in nuclear hormone receptors is involved in coupling hormone binding to corepressor release. Mol. Cell. Biol. 17:6131-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 44.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 45.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schütz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marimuthu, A., W. Feng, T. Tagami, H. Nguyen, J. L. Jameson, R. J. Fletterick, J. D. Baxter, and B. L. West. 2002. TR surfaces and conformations required to bind nuclear receptor corepressor. Mol. Endocrinol. 16:271-286. [DOI] [PubMed] [Google Scholar]
- 47.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 48.McKenna, N. J., J. Xu, Z. Nawaz, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J. Steroid Biochem. Mol. Biol. 69:3-12. [DOI] [PubMed] [Google Scholar]
- 49.Muscat, G. E., L. J. Burke, and M. Downes. 1998. The corepressor N-CoR and its variants RIP13a and RIP13Delta1 directly interact with the basal transcription factors TFIIB, TAFII32 and TAFII70. Nucleic Acids Res. 26:2899-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagaya, T., M. Fujieda, and H. Seo. 1998. Requirement of corepressor binding of thyroid hormone receptor mutants for dominant negative inhibition. Biochem. Biophys. Res. Commun. 247:620-623. [DOI] [PubMed] [Google Scholar]
- 51.Nagy, L., H. Y. Kao, J. D. Love, C. Li, E. Banayo, J. T. Gooch, V. Krishna, K. Chatterjee, R. M. Evans, and J. W. R. Schwabe. 1999. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 13:3209-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 53.Ordentlich, R., M. Downes, and R. M. Evans. 2001. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 254:101-116. [DOI] [PubMed] [Google Scholar]
- 54.Paige, L. A., D. J. Christensen, H. Grøn, J. D. Norris, E. B. Gottlin, K. M. Padilla, C. Y. Chang, L. M. Ballas, P. T. Hamilton, D. P. McDonnell, and D. M. Fowlkes. 1999. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc. Natl. Acad. Sci. USA 96:3999-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 56.Perissi, V., L. M. Staszewski, E. M. McInerney, R. Kurokawa, A. Krones, D. W. Rose, M. H. Lambert, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1999. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 13:3198-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Privalsky, M. L. 2001. Regulation of SMRT and N-CoR corepressor function. Curr. Top. Microbiol. Immunol. 254:117-136. [DOI] [PubMed] [Google Scholar]
- 58.Renaud, J. P., N. Rochel, M. Ruff, V. Vivat, P. Chambon, H. Gronemeyer, and D. Moras. 1995. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acids. Nature 378:681-689. [DOI] [PubMed] [Google Scholar]
- 59.Ribeiro, R. C., P. J. Kushner, and J. D. Baxter. 1995. The nuclear hormone receptor gene superfamily. Annu. Rev. Med. 46:443-453. [DOI] [PubMed] [Google Scholar]
- 60.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 61.Safer, J. D., R. N. Cohen, A. N. Hollenberg, and F. E. Wondisford. 1998. Defective release of corepressor by hinge mutants of the thyroid hormone receptor found in patients with resistance to thyroid hormone. J. Biol. Chem. 273:30175-30182. [DOI] [PubMed] [Google Scholar]
- 62.Sande, S., and M. L. Privalsky. 1996. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol. Endocrinol. 10:813-825. [DOI] [PubMed] [Google Scholar]
- 63.Shao, W., L. Benedetti, W. W. Lamph, C. Nervi, and W. H. Miller, Jr. 1997. A retinoid-resistant acute promyelocytic leukemia subclone expresses a dominant negative PML-RAR alpha mutation. Blood 89:4282-4289. [PubMed] [Google Scholar]
- 64.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937. [DOI] [PubMed] [Google Scholar]
- 65.Shibata, H., T. E. Spencer, S. A. Oñate, G. Jenster, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog. Horm. Res. 52:141-165. [PubMed] [Google Scholar]
- 66.Smith, C. L., Z. Nawaz, and B. W. O'Malley. 1997. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol. Endocrinol. 11:657-666. [DOI] [PubMed] [Google Scholar]
- 67.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 68.Sucov, H. M., and R. M. Evans. 1995. Retinoic acid and retinoic acid receptors in development. Mol. Neurobiol. 10:169-184. [DOI] [PubMed] [Google Scholar]
- 69.Sucov, H. M., K. K. Murakami, and R. M. Evans. 1990. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc. Natl. Acad. Sci. USA 87:5392-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tagami, T., and J. L. Jameson. 1998. Nuclear corepressors enhance the dominant negative activity of mutant receptors that cause resistance to thyroid hormone. Endocrinology 139:640-650. [DOI] [PubMed] [Google Scholar]
- 71.Torchia, J., C. Glass, and M. G. Rosenfeld. 1998. Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 10:373-383. [DOI] [PubMed] [Google Scholar]
- 72.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 73.Wagner, B. L., J. D. Norris, T. A. Knotts, N. L. Weigel, and D. P. McDonnell. 1998. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol. Cell. Biol. 18:1369-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wagner, R. L., J. W. Apriletti, M. E. McGrath, B. L. West, J. D. Baxter, and R. J. Fletterick. 1995. A structural role for hormone in the thyroid hormone receptor. Nature 378:690-697. [DOI] [PubMed] [Google Scholar]
- 75.Wong, C. W., and M. L. Privalsky. 1998. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol. Cell. Biol. 18:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong, C. W., and M. L. Privalsky. 1998. Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors. Mol. Cell. Biol. 18:5724-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 78.Wu, C. 1997. Chromatin remodeling and the control of gene expression. J. Biol. Chem. 272:28171-28174. [DOI] [PubMed] [Google Scholar]
- 79.Xu, H. E., T. B. Stanley, V. G. Montana, M. H. Lambert, B. G. Shearer, J. E. Cobb, D. D. McKee, C. M. Galardi, K. D. Plunket, R. T. Nolte, D. J. Parks, J. T. Moore, S. A. Kliewer, T. M. Willson, and J. B. Stimmel. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature 415:813-817. [DOI] [PubMed] [Google Scholar]
- 80.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 81.Yang, Z., and M. L. Privalsky. 2001. Isoform-specific transcriptional regulation by thyroid hormone receptors: hormone-independent activation operates through a steroid receptor mode of co-activator interaction. Mol. Endocrinol. 15:1170-1185. [DOI] [PubMed] [Google Scholar]
- 82.Yoh, S. M., V. K. Chatterjee, and M. L. Privalsky. 1997. Thyroid hormone resistance syndrome manifests as an aberrant interaction between mutant T3 receptors and transcriptional corepressors. Mol. Endocrinol. 11:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoh, S. M., and M. L. Privalsky. 2002. Molecular analysis of human resistance to thyroid hormone syndrome. Methods Mol. Biol. 202:129-152. [DOI] [PubMed] [Google Scholar]
- 84.Yoh, S. M., and M. L. Privalsky. 2000. Resistance to thyroid hormone (RTH) syndrome reveals novel determinants regulating interaction of T3 receptor with corepressor. Mol. Cell. Endocrinol. 159:109-124. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, X., M. Jeyakumar, S. Petukhov, and M. K. Bagchi. 1998. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol. Endocrinol. 12:513-524. [DOI] [PubMed] [Google Scholar]