Abstract
The SWI/SNF family of chromatin-remodeling complexes has been discovered in many species and has been shown to regulate gene expression by assisting transcriptional machinery to gain access to their sites in chromatin. Several complexes of this family have been reported for humans. In this study, two additional complexes are described that belong to the same SWI/SNF family. These new complexes contain as many as eight subunits identical to those found in other SWI/SNF complexes, and they possess a similar ATP-dependent nucleosome disruption activity. But unlike known SWI/SNFs, the new complexes are low in abundance and contain an extra subunit conserved between human and yeast SWI/SNF complexes. This subunit, ENL, is a homolog of the yeast SWI/SNF subunit, ANC1/TFG3. Moreover, ENL is a fusion partner for the gene product of MLL that is a common target for chromosomal translocations in human acute leukemia. The resultant MLL-ENL fusion protein associates and cooperates with SWI/SNF complexes to activate transcription of the promoter of HoxA7, a downstream target essential for oncogenic activity of MLL-ENL. Our data suggest that human SWI/SNF complexes show considerable heterogeneity, and one or more may be involved in the etiology of leukemia by cooperating with MLL fusion proteins.
Chromatin plays a key role in the regulation of tissue-specific gene expression during development and differentiation in higher eucaryotes (23, 45). As the fertilized egg differentiates into various cells and tissues, the chromatin structures of tissue-specific genes are remodeled so that they become open in tissues where the genes are expressed, but they are closed in other tissues (42). How these chromatin structures are established and maintained has been a subject of intensive investigation.
Recently, molecules capable of remodeling chromatin structures have been identified. They play important roles in transcription, replication, chromatin assembly, and possibly repair (9, 17). These remodelers, often in multiprotein complexes, alter chromatin structures through two distinct mechanisms. One class covalently modifies histone tails, which directly alters the interaction between histones and DNA (33). The modified histone tails also serve as signals, termed histone codes, to recruit additional chromatin-remodeling molecules, leading to changes in higher-order chromatin structure. The other class alters chromatin structures through noncovalent interactions. It includes ATP-dependent chromatin-remodeling complexes that utilize the energy of ATP hydrolysis to disrupt nucleosome structures (38). The disruption can produce altered nucleosome states that are more accessible to transcription machinery (21, 30). The nucleosomes also become more mobile after disruption and can slide on the DNA (8, 43).
SWI/SNF complexes in mammals have been shown to participate in transcriptional regulation of many genes, some of which are critical for normal growth of organisms (16). SWI/SNF components have been found mutated in human tumors and cell lines derived from cancer (37, 44). In mice, heterozygous mutation of some SWI/SNF components results in an increased risk of cancer, whereas homozygous mutation causes embryonic lethality (3, 15). These studies suggest that SWI/SNF complexes play important roles during normal development and differentiation in mammals.
Several members of the human SWI/SNF family of ATP-dependent chromatin-remodeling complexes have previously been described, including BAF (SWI/SNF-A) and PBAF (SWI/SNF-B) (BAF stands for BRG1- or hbrm-associated factors, and PBAF is for polybromo-associated BAF) (18, 20, 32, 40). These complexes consist of 9 to 12 subunits, most of which have been identified. The complexes contain either BRG1 or hbrm (14, 22), which are ATPases capable of remodeling nucleosome structures by themselves (28). Here we describe two new human complexes that belong to the same SWI/SNF family. These complexes contain a unique subunit that has been previously identified as a fusion partner in mixed-lineage leukemia, suggesting an involvement of SWI/SNFs in the etiology of leukemia.
MATERIALS AND METHODS
Cloning of BAF250b.
Two expressed sequence tag (EST) clones, AA173396 and AI848081, were found to be homologous to BAF250a by BLAST searches of the National Center for Biotechnology Information database. They were used as probes to screen a cDNA library from human T cells. More than 10 cDNA clones were obtained and sequenced. They were found to be overlapping clones of the same gene. The assembled full-length cDNA is 6,366 bp, carrying a predicted open reading frame of 1,956 amino acids.
Immunopurification of the ENL-associated BAF-containing (EBAF) complexes.
A rabbit polyclonal antibody to BAF250b was raised against a fusion protein containing a region of this gene (amino acids 737 to 817) linked to the maltose-binding protein (New England Biolabs). The fusion protein was expressed in Escherichia coli and was purified according to the manufacturer's protocol. The antibody was affinity purified and used for immunopurification and immunoblotting. Anti-ENL and anti-MLL antibodies have been reported previously (4).
The immunopurification directly from nuclear extract followed the same procedure as that used for isolation of the BAF complex from Jurkat T cells that stably express hemagglutinin-tagged BAF57 (39). Fractionation of nuclear extract by ion-exchange columns and subsequent affinity purification followed the same procedure for purification of the BAF complex (40). Fractionation of nuclear extract by Superose 6 gel filtration chromatography has been described previously (41). For affinity purification, the affinity-purified BAF250b and ENL antibodies were cross-linked to protein A beads and were incubated at 4°C with either nuclear extract or the 0.3 M fraction from a DEAE column (see Fig. 5a). The beads were washed with 0.5 M buffer D. They were then either used for mononucleosome disruption assay directly or were eluted with 0.1 M glycine (pH 2.5) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. For mass spectrometry analysis, each complex was resolved by SDS-PAGE and was visualized by colloidal Coomassie blue staining. Each band was excised from the gel, digested with trypsin, and identified by peptide mapping with a matrix-assisted laser desorption ionization-time of flight instrument. Further confirmation by high-performance liquid chromatography-tandem mass spectrometry was obtained when necessary. The mass data are not shown but are available upon request.
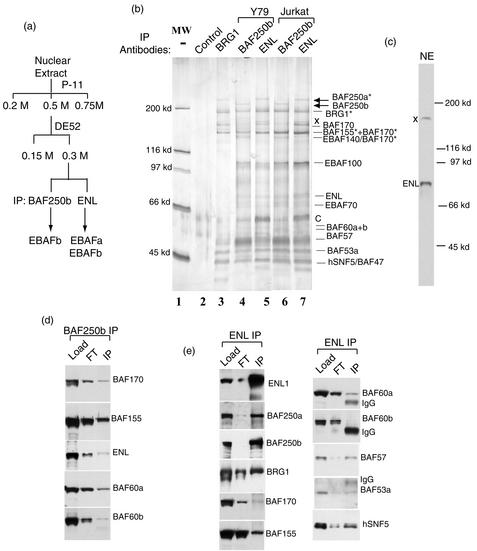
FIG. 5.
ENL also associates with a BAF250a-containing SWI/SNF complex. (a) A purification scheme for ENL-associated SWI/SNFs. (b) A silver-stained SDS-PAGE gel showing the complexes immunopurified by ENL and BAF250b antibodies from Y79 and Jurkat T cells. The arrows indicate BAF250a and BAF250b that have distinct mobility. The asterisks mark the polypeptides isolated by ENL antibody and identified by mass spectrometry. Some subunits, most notably BAF170 and ENL, are partially disassociated and degraded, which may explain the reason that peptides derived from BAF170 are found in the 155- and 140-kDa polypeptides. BAF complex isolated by BRG1 from HeLa cells is shown as a reference. A control immunopurification with preimmune serum is also shown (lane 2). A contaminant isolated by the control purification is marked C. A polypeptide (X) likely derived from ENL antibody cross-reactivity is indicated. (c) Immunoblotting analysis showing that the ENL antibody has a major cross-reactive polypeptide (marked X). (d) Immunoblot analysis showing the presence of different components in EBAF (IP) isolated by BAF250b antibody from the DEAE-0.3 M fraction. The DE52-0.3 M fraction (Load) and flowthrough fraction (FT) are shown. (e) Immunoblot analysis showing the presence of different components in complexes isolated by ENL antibody (IP).
The whole-cell extracts from HB(11;19) cells and the MLL control cell line, RCH-ACV, were prepared by extracting cells directly with Laemmli SDS gel loading buffer.
Analysis of BAF250b expression patterns.
Northern blot analysis was performed by using a fragment (bp 3059 to 3836) from BAF250b as a probe to hybridize with a membrane containing RNAs from multiple human tissues according to the manufacturer's protocol (Clontech). The probe for BAF250a has been previously described (25).
In situ hybridization was performed by using frozen sections of mouse embryos (C57 Black) at embryonic day 15.5 as described previously (36). One exception is that the hybridization and post-RNase treatment washes were carried out at 58 and 60°C, respectively. Mouse EST clone AA709949, which encodes a fragment of mouse BAF250b corresponding to the human gene between 5518 to 6022 bp, was used as antisense and sense probes.
Chromatin-remodeling assay.
The ATP-dependent mononucleosome disruption assay was performed as described previously (27, 39).
Cell culture and transfection.
SW13 cells were maintained as described previously (40). The leukemia cell line that carries MLL-ENL translocation, HB(11;19), was maintained in RPMI medium supplemented with 10% fetal calf serum. A control cell line that has no MLL chromosomal translocation, RCH-ACV, was grown under the same conditions. Transfections were performed by using Polyfect reagent (QIAGEN) mixed with DNA according to the manufacturer's protocol. Cells were harvested after 24 h and were analyzed for the luciferase activity as described previously (25).
RESULTS
Cloning of a homolog for human SWI/SNF subunit BAF250.
One component of BAF, BAF250 (25), was noted to have several homologous sequences in the human EST database. Several cDNA clones were isolated based on these EST sequences and were found to be overlapping fragments of the same gene. The gene is homologous to BAF250 throughout its length (over 60% identity) (Fig. 1a). It was named BAF250b (GenBank accession number AF253515; synonyms for this gene include p250R, hOsa2, and hELD/OSA1). The originally reported BAF250 was renamed BAF250a (synonyms for this gene include p270 and hOsa1). BAF250b contains all three evolutionarily conserved domains present in BAF250a as well as in the homologous subunits in SWI/SNFs of Drosophila melanogaster (Osa/eyelid) and yeast (SWI1). Therefore, BAF250b represents a novel member of this family of proteins (Fig. 1b).
FIG. 1.
BAF250b is highly homologous to BAF250a and belongs to the same family as yeast SWI1. (a) Sequence alignment of BAF250b and BAF250a. The three evolutionarily conserved regions are boxed and labeled ARID, C1, and C2. The predicted LXXLL motifs, which may be potential interaction domains for nuclear hormone receptors, are underlined. (b) Schematic representation of BAF250b and its orthologs from Drosophila and yeast. Each box represents a conserved region. The predicted LXXLL motifs are indicated by arrows. The letters A, N, P, and Q mark the regions rich in alanines, asparagines, prolines, and glutamines, respectively.
While this work was in progress, two labs reported partial cDNA sequences of BAF250b (10, 12). Their reported proteins are 1,486 and 1,740 amino acids long, respectively, whereas BAF250b described here is 1,956 amino acids long. As shown below, the apparent molecular mass of BAF250b is nearly 230 kDa (see Fig. 3b), close to the coding capacity reported here. Both previous studies suggested that BAF250b could interact with BRG1 and other BAFs, but neither distinguished the BAF250b complex from the more abundant BAF250a complex.
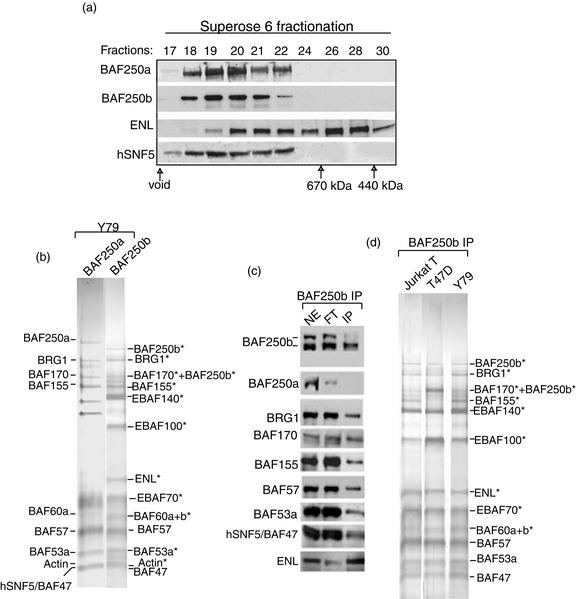
FIG. 3.
BAF250b is a subunit of a novel human SWI/SNF complex that contains ENL, a protein involved in chromosomal translocation in leukemia. (a) Immunoblot analysis of fractions from Superose 6 gel filtration showing that BAF250b is present in a 1- to 1.5-MDa complex. The antibodies used for the analysis are marked on the left. The fractions corresponding to proteins with known molecular size markers are shown at the bottom. (b) A silver-stained SDS gel showing BAF and EBAFb complexes immunopurified by BAF250a and BAF250b antibodies, respectively, from the retinoblastoma Y79 cell line. BAF and potential EBAFb components are marked. The EBAFb components identified by mass spectrometry are indicated by asterisks. The 170-kDa band was found containing not only BAF170 but also BAF250b, which is likely derived from proteolytic degradation. BAF53a is the new name for BAF53, which was used to distinguish it from its newly described homolog, BAF53b (26). (c) Immunoblotting analysis of nuclear extract (NE), flowthrough fraction (FT), and the EBAFb complex isolated by BAF250b antibody (IP). (d) Silver stain analysis of EBAFb complexes isolated from T47D (a breast cancer cell line lacking BAF250a), Jurkat T (human T-cell leukemia), and Y79 cells. The 155-kDa band in EBAFb of the T47D cell line is reproducibly substoichiometric compared to those isolated from other cell lines. This band was identified as BAF155 by mass spectrometry.
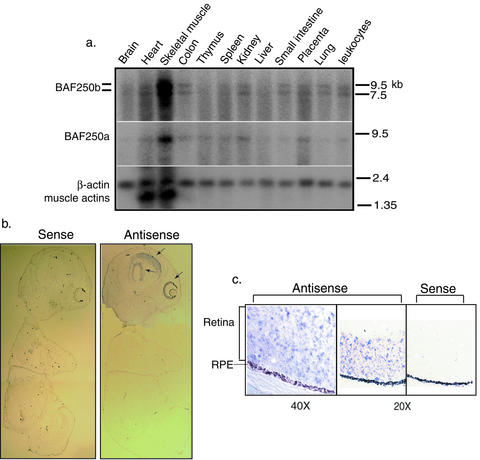
BAF250b mRNA was detectable in multiple human tissues by Northern blot analysis (Fig. 2a). Many partial cDNA sequences encoding BAF250b have also been identified in the dbEST database and are derived from a wide range of tissues, including retina, brain, lung, stomach, kidney, nerves, blood cells, and heart. These data suggest that BAF250b, like BAF250a (25), is widely expressed. In situ hybridization of day 16 mouse embryos further showed that BAF250b is highly expressed in selected regions of the brain and in retinal cells (Fig. 2b and c). This finding is consistent with those of a report that noted the abundance of this protein in brain extract and its low level in HeLa cells (10).
FIG. 2.
BAF250b is expressed in many different tissues, including retina cells. (a) Northern blot analyses of BAF250b expression by using RNAs from different human tissues. The expression patterns for BAF250a and β-actin are shown as controls. The molecular size markers (in kilobases) are indicated. (b and c) In situ hybridization analysis of BAF250b expression in mouse 15.5-day embryos (b) and eyes (c). Arrows indicate the regions in brain and eyes where BAF250b is highly expressed. The retina pigment epithelium (RPE) is marked.
A new human SWI/SNF complex containing BAF250b.
Based on the high level of BAF250b in retina cells (Fig. 2c), we initiated isolation of BAF250b from a retinoblastoma cell line, Y79. BAF250b was fractionated at 1 to 1.5 MDa on a Superose 6 sizing column (Fig. 3a). This property resembles that of BAF subunits, such as BAF250a and hSNF5, which peaked in the same fractions, implying that BAF250b may be part of a complex similar to BAF.
We attempted to purify the BAF250b-containing complex by a combination of chromatography and immunopurification, a strategy that has been used successfully to purify the BAF and PBAF (40). However, BAF250b was found to be subject to rapid proteolytic degradation even in the presence of protease inhibitors (Fig. 3b). In addition, several subunits of the BAF250b complex are partially disassociated and degraded by using such a protocol (see Fig. 5b). To circumvent these problems, we performed immunopurification directly from unfractionated nuclear extracts. But because a single-step procedure is used, some of the copurified polypeptides may be contaminants and require further analysis.
Fourteen major polypeptides were isolated by immunopurification by using an antibody to BAF250b. They displayed an SDS gel electrophoretic pattern different from that of BAF immunoisolated with a BAF250a antibody (Fig. 3b). Mass spectrometry analysis identified the 230-kDa polypeptide as BAF250b. In addition, peptides derived from BAF250b were also found in the 170-kDa band, which could represent a degradation product. BAF250a was not detected either by mass spectrometry analysis or immunoblotting (Fig. 3c), suggesting that the polypeptides isolated here specifically associate with BAF250b but not BAF250a. The other polypeptides were identified by a combination of mass spectrometry and immunoblotting. They included at least eight components previously identified in both BAF and PBAF: BRG1, BAF170, BAF155, BAF57, BAF60a, BAF60b, BAF53a, actin, and hSNF5/INI1 (Fig. 3a and b; also see Fig. 5d). These results indicate that the BAF250b-associated polypeptides represent a new human SWI/SNF complex. This complex is abbreviated to EBAFb for convenience purposes (for ENL-associated BAF-containing BAF250b). Notably, EBAFb has several potentially new components that are absent in BAF or PBAF (Fig. 3b). These new components, termed EBAFxx (where xx is molecular size), may confer unique properties on EBAFb compared to those of other SWI/SNFs.
Two EBAFb components are homologous but show some differences from BAF subunits. EBAFb contains only BRG1 as its ATPase, whereas BAF contains either BRG1 or hbrm. Also, EBAFb contains both BAF60a and BAF60b, whereas BAF contains only BAF60a. These results are consistent with the proposal that different homologs of BAF subunits assemble in a combinatory manner to produce a large number of SWI/SNF-related complexes to serve the needs of specialized tissues and cells (41).
Because of the sequence similarity between BAF250b and BAF250a, it was possible that the BAF250b antibody might cross-react with BAF250a, fortuitously isolating BAF components and leading to a false inference of association. To exclude this possibility the cell line T47D, known to be deficient for BAF250a, was utilized for BAF250b immunopurification (25). A complex similar to that of Y79 cells was isolated (Fig. 3d) and was found to include BAF250b, BRG1, and other EBAFb components by mass spectrometry and immunoblotting analyses (data not shown). The data thus show that BRG1 and other BAF components are not immunoisolated due to BAF250b antibody cross-reacting with BAF250a and suggest that these proteins form a specific complex with BAF250b. It was noticed that EBAFb from T47D cells reproducibly contain substoichiometric amounts of BAF155 compared to levels from other cells (data from Coomassie-stained gels are consistent with that from silver-stained gel; data not shown). BAF170 and BAF155 are homologous to each other and may form a heterodimeric structure within human SWI/SNF complexes (41). Possibly, the majority of EBAFb from T47D cells may have BAF170 homodimer rather than the BAF170/BAF155 heterodimer.
Identification of ENL as a component of the EBAFb complex.
The 80-kDa subunit of EBAFb was identified by mass spectrometry and immunoblot analysis as ENL, a chromosomal translocation partner protein involved in mixed-lineage leukemia (Fig. 3b and c) (35). EBAFb isolated from a T-cell leukemia cell line, Jurkat, showed a polypeptide composition indistinguishable from that of Y79 cells, including the presence of ENL (Fig. 3d). The data suggest that EBAFb is present in lymphocyte lineages.
ENL belongs to a conserved family of proteins present in yeast, Drosophila, and humans. One yeast member, TFG3/ANC1, has been identified as a component of yeast SWI/SNF (5). This protein interacts with the SNF5 component of the complex by protein interaction blots. The same study also noted the homology between yeast TFG3/ANC1 and two of its human homologs, ENL and AF9. Because both ENL and AF9 are fusion partners of MLL in mixed-lineage leukemia, the authors raised the possibility that human SWI/SNF complex is involved in leukemia. However, none of the human homologs of this family (ENL, AF9, and GAS41) has been purified in any transcriptional complexes. Our finding that ENL is a component of a human SWI/SNF is consistent with results with yeast and further suggests that this family of proteins could have an evolutionarily conserved function in SWI/SNFs.
Evidence that MLL-ENL associates with EBAFb.
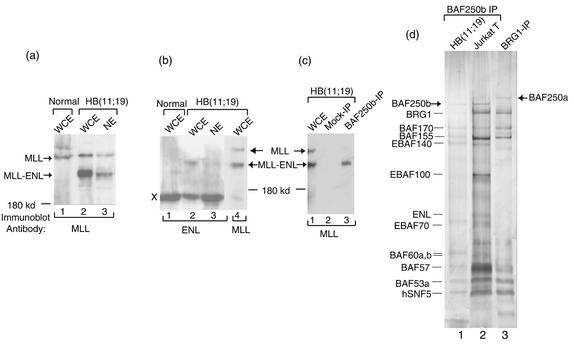
To explore the possible involvement of human SWI/SNF complex in leukemia, we determined whether MLL-ENL associates with the BAF250b-containing complex in a leukemia cell line, HB(11;19), which carries a chromosomal translocation encoding this protein. As expected, a polypeptide with the size (220 kDa) of MLL-ENL was detected by antibodies specific for either MLL or ENL in the whole-cell extract of HB(11;19) cells but not in extract from a control cell line that lacks the chromosomal translocation (Fig. 4a and b). Notably, a polypeptide of the same size was detected by using the anti-MLL antibody in EBAFb isolated by BAF250b antibody but not in polypeptides isolated by mock purification with preimmune serum (Fig. 4c). As an internal control, wild-type MLL protein was not detected in EBAFb (Fig. 4c), even though MLL is present in the same nuclear extract (Fig. 4a). The results suggest that the association between MLL-ENL and EBAFb is specific. However, silver staining analysis showed that the major polypeptides isolated by BAF250b antibody from this cell line, HB(11;19), resemble those obtained from Jurkat cells (Fig. 4d). The lack of an additional major polypeptide with the predicted size of MLL-ENL by this analysis implies that MLL-ENL may be present only in a proportion of EBAFb complexes. Taken together these data suggest that MLL-ENL, but not MLL, is present in a subset of EBAFb complexes in this leukemia cell line.
FIG. 4.
The MLL-ENL fusion protein, but not MLL itself, associates with EBAFb complex. (a and b) Immunoblotting analyses of whole-cell extract (WCE) and nuclear extract (NE) prepared from the HB(11;19) cell line as well as from a control cell line (Normal), RCH-ACV, that lacks the chromosomal translocation. The antibodies used are shown at the bottom. A polypeptide cross-reactive with ENL antibody was marked X (see the legend to Fig. 4c) and was used as a loading control. (c) Immunoblotting to show the presence of MLL-ENL in EBAFb isolated by BAF250b antibody from HB(11;19) cells. The mock immunopurification (IP) was done by using preimmune serum from normal rabbits. (d) A silver-stained SDS gel to show EBAFb isolated from the HB(11;19) leukemia cell line.
In nuclear extract prepared by using the standard high-salt extraction procedure, the level of MLL-ENL is very low (Fig. 4b; use the cross-reactive 180-kDa polypeptide as a reference). The finding is consistent with that of a previous report that the amino terminus of MLL targets itself or its fusion proteins to nuclear matrix and chromatin scaffold (6), which are not efficiently extracted with this procedure.
ENL also associates with a new BAF250a-containing SWI/SNF.
To further demonstrate that ENL is a component of EBAFb, we used an unbiased approach to independently isolate ENL-associated complexes and to determine whether they contain EBAFb. Superose 6 profiles of ENL in nuclear extract only partially overlapped those of BAF250b and other BAFs (Fig. 3a). This suggested that the majority of ENL does not associate with SWI/SNFs. Indeed, the major polypeptides immunoisolated by ENL antibody from nuclear extract are not BAF components, based on their different mobility on SDS gels (data not shown). Nevertheless, immunoblotting revealed the presence of EBAFb (data not shown), suggesting that EBAFb is a low-abundance complex among ENL-associated polypeptides. These results are consistent with yeast data that the homolog of ENL, TFG3/ANC1, is present in at least two other transcriptional complexes besides SWI/SNF (5).
To isolate ENL-associated SWI/SNFs away from other irrelevant ENL complexes, the nuclear extract was fractionated (Fig. 5a). After fractionation, most polypeptides immunoisolated by the ENL antibody display gel mobility similar to that of the components of EBAFb isolated by BAF250b antibody from the same fraction (Fig. 5b). Mass spectrometry and immunoblotting analyses confirmed that many of these polypeptides are BAF subunits and ENL (Fig. 5b and e). The fact that an ENL antibody can independently isolate all BAF subunits strongly supports the suggestion that ENL is a subunit of SWI/SNFs.
Importantly, the major polypeptide larger than 200 kDa isolated by ENL antibody has the gel mobility of BAF250a but not BAF250b (Fig. 5b). This polypeptide was identified as BAF250a by both mass spectrometry and immunoblotting (Fig. 5e). As shown above, BAF250a is absent in the EBAFb complex isolated by BAF250b antibody (Fig. 3b and c). Therefore, the major polypeptides isolated by ENL antibodies comprise a further variant SWI/SNF different from EBAFb. In addition, this complex should also be different from the other BAF250a-containing complex, BAF (Fig. 5b, lane 3), because it contains ENL, EBAF70, EBAF100, and other potential subunits that have not been found in BAF. This complex is abbreviated as EBAFa for convenience (for ENL-associated BAF containing BAF250a).
Although BAF250b was not found as a prominent polypeptide by immunopurification with the ENL antibody (Fig. 5b, lanes 5 and 7), immunoblotting confirmed that BAF250b is present in the ENL-associated polypeptides (Fig. 5e), consistent with the data above that ENL is also a component of the BAF250b-containing EBAFb complex (Fig. 3b and c). Therefore, ENL-associated polypeptides apparently include at least two different SWI/SNFs, EBAFa and EBAFb. Because BAF250a is more abundant than BAF250b in polypeptides isolated by ENL antibody (Fig. 5b), the results imply that the BAF250a-containing EBAFa is present at a higher level in cells than is the BAF250b-containing EBAFb.
EBAFa and EBAFb are less abundant complexes compared to BAF.
EBAFa and EBAFb are immunoisolated from the same fraction as that used for isolation of the BAF complex. The only difference is that the EBAF complexes were isolated by using antibodies for ENL and BAF250b, which are unique subunits of these complexes, whereas the BAF complex was isolated by antibodies against the common SWI/SNF subunits, including BRG1, hSNF5, and BAF57 (32, 39, 40), which are not only present in BAF but also are in the two EBAF complexes. Therefore, EBAF complexes should be present in the BAF preparation. However, all polypeptides in the BAF preparation that can be seen on the Coomassie blue-stained gel have been extensively analyzed by both Edman degradation microsequencing and mass spectrometry analysis, and neither ENL nor BAF250b has been detected. This is consistent with a low level of EBAF complexes compared to that of BAF.
Also consistent with this notion, immunopurification of SWI/SNFs from unfractionated nuclear extract by using antibodies against several common subunits of SWI/SNFs predominantly yielded BAF (32, 39, 48). In addition, purification of human SWI/SNFs by conventional chromatography obtained BAF and PBAF complexes (20). Again, the EBAF complexes described here must be in low abundance compared to BAF.
EBAF complexes contain a nucleosome disruption activity similar to that of other SWI/SNF complexes.
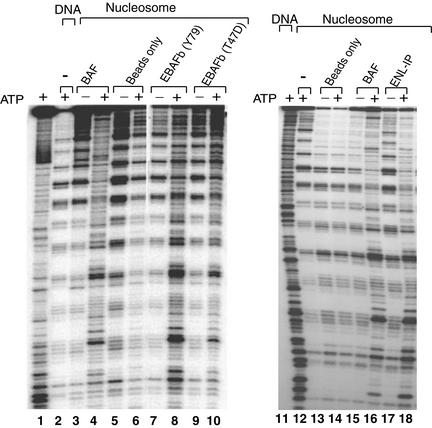
EBAFb immunoisolated by BAF250b antibody was found to exhibit an ATP-dependent mononucleosome disruption activity (Fig. 6). In this assay (27), a 176-bp DNA containing a nucleosomal positioning sequence was assembled into a rotationally phased mononucleosome. DNase I digestion of this nucleosome yields a distinctive pattern of 10-bp ladders. EBAFb strongly disrupted the 10-bp ladders in the presence of ATP, with a pattern similar to that generated by BAF. As a control, EBAFb isolated from the T47D cell line also displayed this activity. Because this cell line lacks BAF250a, the observed activity should be derived from EBAFb and not from the abundant BAF250a-containing BAF. Likewise, the mixture of EBAF complexes isolated by ENL antibody, containing much more EBAFa than EBAFb, was found to display a similar nucleosome disruption activity. These results suggest that these BAF250b- and ENL-containing complexes have chromatin-remodeling activity similar to that of other SWI/SNFs.
FIG. 6.
EBAF complexes have ATP-dependent nucleosome disruption activity. (a) Autoradiographs showing the mononucleosome disruption activity of EBAF complexes isolated by either BAF250b or ENL antibodies. The complexes used in these studies are shown on top. Antibody beads without the loaded complex were used as a negative control. The presence or absence of ATP is indicated.
MLL-ENL and SWI/SNF complexes coactivate HoxA7 promoter.
MLL is a common target for chromosomal translocations in human acute leukemia (2). More than 30 proteins have been identified as fusion partners of MLL. The resultant MLL fusion proteins always retain the DNA-binding domains of MLL. When individual components of SWI/SNF are tethered to a promoter through a DNA-binding domain, they can activate transcription in an SWI/SNF-dependent manner (19). We hypothesized that the MLL-ENL complex could activate transcription by a similar mechanism. To test this possibility, we used the HoxA7 gene promoter as a reporter in a transfection assay in the SW13 cell line. HoxA7 is essential for the oncogenic activities of MLL-ENL, since bone marrow cells deficient for HoxA7 are highly resistant to transformation by MLL-ENL (1). In addition, HoxA7 is a downstream target regulated by MLL and various MLL fusion proteins (1, 13, 31, 47). SW13 cells contain ENL at a level comparable to that of Jurkat T cells (data not shown). Importantly, SW13 cells have no hbrm and very low levels of BRG1, and transfection into this cell line has been frequently used in studies of human SWI/SNF-dependent genes (22, 40). However, the limitation of this assay is that BRG1 is a common subunit of many SWI/SNF complexes, so the assay cannot determine the specific SWI/SNF involved in activation of a given gene.
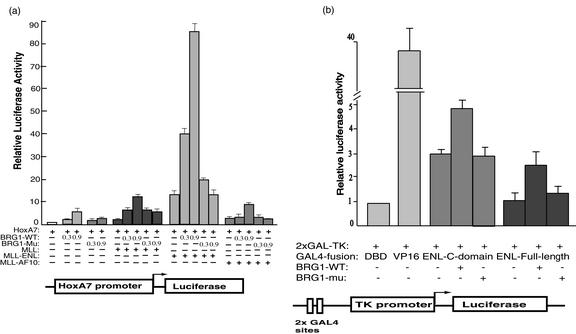
MLL-ENL was found to activate transcription of the HoxA7 promoter in the absence of added BRG1 (about 10-fold) (Fig. 7a). BRG1 also modestly stimulated this promoter (about fivefold), consistent with the findings for Drosophila that homeotic genes are positively regulated by fly homologs of SWI/SNF and MLL (the fly homolog of MLL is trithorax) (34). Importantly, cotransfection of MLL-ENL and BRG1 resulted in synergistic activation of the same promoter (40- to 90-fold), whereas cotransfection of MLL-ENL with a BRG1 mutant (K798R), which is defective in chromatin-remodeling activity, exhibited no synergistic activation. Moreover, transfection of either MLL or an unrelated MLL fusion protein, MLL-AF10, resulted in much less stimulation of the HoxA7 promoter compared to that with MLL-ENL. These results suggest that the MLL-ENL fusion protein and SWI/SNF complexes can cooperate to facilitate transcription of HoxA7.
FIG. 7.
MLL-ENL and BRG1 coactivate the promoter of the HoxA7 gene. (a) A graph showing transcriptional activation of the HoxA7 promoter by MLL, two MLL fusion proteins, and BRG1. All transfections contain a reporter plasmid of the HoxA7 promoter upstream of the luciferase gene (70 ng). The presence or absence of the plasmids expressing MLL, MLL-ENL fusion proteins, wild-type BRG1 (BRG1-WT), and a BRG1 K798R mutant (BRG1-mu) that inactivates its ATPase activity are indicated, with their amounts shown in micrograms. The amount of plasmid used for either MLL or MLL fusion protein is 100 ng. (b) A graph showing transcriptional activation of GAL4-ENL and BRG1 on a luciferase reporter containing two GAL4-binding sites upstream of the thymidine kinase (TK) promoter (bottom). All reactions contained 0.1 μg of the reporter plasmid, 0.1 μg of plasmid expressing GAL4 fusion protein, and 0.3 μg of either wild-type or mutant BRG1. Two GAL4-ENL fusion proteins, the full-length and the C-terminal conserved domain, were tested as shown. Plasmids expressing the GAL4-DNA-binding domain (DBD) and GAL4-VP16 were used as negative and positive controls, respectively.
The ENL portion of MLL-ENL activates transcription when it is tethered to a promoter through the GAL4 DNA-binding domain (29). By using the same GAL4 reporter system we found that transcriptional activation of GAL4-ENL was stimulated about twofold by BRG1 (Fig. 7b). This is a lesser effect than that of BRG1 stimulation of MLL-ENL on the HoxA7 promoter. The different effects of BRG1 on ENL-fusion proteins may be due to the different DNA-binding domains (MLL versus GAL4) and/or different local chromatin structures at the promoters (HoxA7 versus thymidine kinase).
DISCUSSION
Several complexes of the human SWI/SNF family, which include BAF and PBAF, have been previously characterized. Based on structural domains that are unique in each complex, BAF and yeast SWI/SNF have been classified into one subfamily, whereas PBAF and a different yeast complex, Rsc, are classified into the other subfamily (46). This study described two novel complexes, abbreviated as EBAFa and EBAFb, which contain all eight common subunits present in both BAF and PBAF, indicating that these complexes represent new members of the SWI/SNF family. Specifically, the two new complexes should belong to the BAF-SWI/SNF subfamily, because they contain either BAF250a or BAF250b, which are related to yeast SWI1 and which distinguishes them from the PBAF-Rsc subfamily. Notably, the new complexes reported here not only contain all the evolutionarily conserved subunits of BAF but also have an extra conserved subunit, ENL, which is absent in BAF but has a homolog in yeast SWI/SNF. The new complexes also resemble yeast SWI/SNF in their low abundance (BAF is abundant and resembles yeast Rsc in this regard). This suggests that the new complexes identified here may be functionally more similar to yeast SWI/SNF than is BAF.
Why are there so many human SWI/SNF complexes? A recent study showed that BAF and PBAF cannot replace each other during transcription mediated by specific activators, despite the fact that these complexes are highly similar in structure and chromatin-remodeling activities (20). Thus, SWI/SNFs must have certain degrees of specificity when they cooperate with specific activators. One complex may cooperate with certain types of activators but not others. Because the numbers and types of activators are large in human cells, multiple SWI/SNFs may be needed for proper coordination with activators in regulating gene expression. If so, there may exist other low-level SWI/SNF complexes waiting to be discovered.
It should be pointed out that in vivo functional studies of the human SWI/SNFs so far (including this one) have focused on the common subunits shared by many SWI/SNFs. They cannot distinguish the specific SWI/SNF involved in a particular process. Given the heterogeneity of human SWI/SNFs, highly discriminating assays will be required to identify the precise SWI/SNF regulating a specific gene.
Concerning involvement of SWI/SNFs in leukemia, all three members of the human ENL family have been implicated to participate in leukemogenesis. Both ENL and AF9 are direct fusion partners of the MLL gene (11, 24, 35). The third member of the ENL family, GAS41, has been suggested to interact with a different MLL fusion partner, AF10 (7). It remains to be determined whether AF9 and GAS41 are components of other less abundant SWI/SNF complexes. Nevertheless, our data that MLL-ENL associates with EBAFb and synergistically activates transcription with BRG1 support a model that MLL-ENL recruits SWI/SNFs to genes normally controlled by MLL, such as HoxA7, and upregulate their expression. The aberrant expression of these target genes may account for mixed-lineage leukemia.
Acknowledgments
We greatly appreciate the support by O. Sundin. We thank D. Zack and D. Otteson for materials and D. Schlessinger for critical reading of the manuscript. We thank the National Cell Culture Center for providing cells.
W.W. has received grants from the Ellison Medical Foundation and Rett Syndrome Research Foundation.
REFERENCES
- 1.Ayton, P. M., and M. L. Cleary. 2001. Transformation of myeloid progenitors by MLL fusion proteins requires HoxA7 and HoxA9 and their DNA binding partner Pbx2. Blood 98:800. [Google Scholar]
- 2.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 3.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 4.Butler, L. H., R. Slany, X. Cui, M. L. Cleary, and D. Y. Mason. 1997. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood 89:3361-3370. [PubMed] [Google Scholar]
- 5.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caslini, C., A. S. Alarcon, J. L. Hess, R. Tanaka, K. G. Murti, and A. Biondi. 2000. The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia 14:1898-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debernardi, S., A. Bassini, L. K. Jones, T. Chaplin, B. Linder, D. R. de Bruijn, E. Meese, and B. D. Young. 2002. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood 99:275-281. [DOI] [PubMed] [Google Scholar]
- 8.Hamiche, A., R. Sandaltzopoulos, D. A. Gdula, and C. Wu. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833-842. [DOI] [PubMed] [Google Scholar]
- 9.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 10.Hurlstone, A. F., I. A. Olave, N. Barker, M. Van Noort, and H. Clevers. 2002. Cloning and characterization of hELD/OSA1, a novel BRG1 interacting protein. Biochem. J. 364:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iida, S., M. Seto, K. Yamamoto, H. Komatsu, A. Tojo, S. Asano, N. Kamada, Y. Ariyoshi, T. Takahashi, and R. Ueda. 1993. MLLT3 gene on 9p22 involved in t(9;11) leukemia encodes a serine/proline rich protein homologous to MLLT1 on 19p13. Oncogene 8:3085-3092. [PubMed] [Google Scholar]
- 12.Kato, H., A. Tjernberg, W. Zhang, A. N. Krutchinsky, W. An, T. Takeuchi, Y. Ohtsuki, S. Sugano, B. T. Chait, and R. G. Roeder. 2001. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J. Biol. Chem. 277:5498-5505. [DOI] [PubMed] [Google Scholar]
- 13.Kawagoe, H., R. Kawagoe, and K. Sano. 2001. Targeted down-regulation of MLL-AF9 with antisense oligodeoxyribonucleotide reduces the expression of the HOXA7 and -A10 genes and induces apoptosis in a human leukemia cell line, THP-1. Leukemia 15:1743-1749. [DOI] [PubMed] [Google Scholar]
- 14.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 15.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klochendler-Yeivin, A., C. Muchardt, and M. Yaniv. 2002. SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 12:73-79. [DOI] [PubMed] [Google Scholar]
- 17.Krebs, J. E., and C. L. Peterson. 2000. Understanding “active” chromatin: a historical perspective of chromatin remodeling. Crit. Rev. Eukaryot. Gene Expr. 10:1-12. [PubMed] [Google Scholar]
- 18.Kwon, H., A. N. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 19.Laurent, B. C., M. A. Treitel, and M. Carlson. 1991. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl. Acad. Sci. USA 88:2687-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924-928. [DOI] [PubMed] [Google Scholar]
- 21.Lorch, Y., B. R. Cairns, M. Zhang, and R. D. Kornberg. 1998. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell 94:29-34. [DOI] [PubMed] [Google Scholar]
- 22.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller, C., and A. Leutz. 2001. Chromatin remodeling in development and differentiation. Curr. Opin. Genet. Dev. 11:167-174. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, T., H. Alder, Y. Gu, R. Prasad, O. Canaani, N. Kamada, R. P. Gale, B. Lange, W. M. Crist, P. C. Nowell, et al. 1993. Genes on chromosomes 4, 9, and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Proc. Natl. Acad. Sci. USA 90:4631-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olave, I., W. Wang, Y. Xue, A. Kuo, and G. R. Crabtree. 2002. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16:2509-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owen-Hughes, T., R. T. Utley, J. Cote, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273:513-516. [DOI] [PubMed] [Google Scholar]
- 28.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 3:247-253. [DOI] [PubMed] [Google Scholar]
- 29.Rubnitz, J. E., J. Morrissey, P. A. Savage, and M. L. Cleary. 1994. ENL, the gene fused with HRX in t(11;19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells. Blood 84:1747-1752. [PubMed] [Google Scholar]
- 30.Schnitzler, G., S. Sif, and R. E. Kingston. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 94:17-27. [DOI] [PubMed] [Google Scholar]
- 31.Schreiner, S. A., M. P. Garcia-Cuellar, G. H. Fey, and R. K. Slany. 1999. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia 13:1525-1533. [DOI] [PubMed] [Google Scholar]
- 32.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 34.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 35.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691-700. [DOI] [PubMed] [Google Scholar]
- 36.Toy, J., J. M. Yang, G. S. Leppert, and O. H. Sundin. 1998. The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl. Acad. Sci. USA 95:10643-10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 38.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, W., T. Chi, Y. Xue, S. Zhou, A. Kuo, and G. R. Crabtree. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 42.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848-856. [DOI] [PubMed] [Google Scholar]
- 43.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 44.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 45.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 46.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505-508. [DOI] [PubMed] [Google Scholar]
- 48.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]