Abstract
Many eukaryotic mRNAs exhibit regulated decay in response to cellular signals. AU-rich elements (AREs) identified in the 3′ untranslated region (3′-UTR) of several such mRNAs play a critical role in controlling the half-lives of these transcripts. The yeast ARE-containing mRNA, MFA2, has been studied extensively and is degraded by a deadenylation-dependent mechanism. However, the trans-acting factors that promote the rapid decay of MFA2 have not been identified. Our results suggest that the chaperone protein Hsp70, encoded by the SSA family of genes, is involved in modulating MFA2 mRNA decay. MFA2 is specifically stabilized in a strain bearing a temperature-sensitive mutation in the SSA1 gene. Furthermore, an AU-rich region within the 3′-UTR of the message is both necessary and sufficient to confer this regulation. Stabilization occurs as a result of slower deadenylation in the ssa1ts strain, suggesting that Hsp70 is required for activation of the turnover pathway.
Gene expression is a highly controlled process involving regulation at both transcriptional and posttranscriptional levels. In recent years mRNA turnover has emerged as an important target for the regulation of gene expression (33, 54). A large number of mRNAs encoding cytokines, growth factors, and proto-oncogenes display regulated decay in response to external signals (44, 50). Hence, understanding the pathways regulating transcript stability is of critical importance. Selective mRNA degradation is mediated by a number of different cis-acting sequences, including the most-investigated class, called AU-rich elements (AREs), present in the 3′ untranslated region (3′-UTR) of a variety of mammalian and yeast mRNAs (44, 49, 54). Recent experiments have shown that the pathways and factors involved in ARE-mediated mRNA decay are conserved between yeast and higher eukaryotes, making yeast an ideal system for the study of this phenomenon (49).
The ARE is a stability determinant whose sequence is loosely defined and ranges in size from 50 to 150 nucleotides. These elements are typically found in the 3′-UTR and contain one or more copies of the pentameric sequence AUUUA flanked by a high content of U's and A's (8, 47). An important feature of many AREs is that they modulate the stability of transcripts in response to cellular stimuli. They can cause instability under some conditions by enhancing the rate of removal of the poly(A) tail and the subsequent degradation of the body of the transcript (7, 38, 48). In contrast, under stabilizing conditions AREs can inhibit the decay process (see references 49 and 54 and references therein). In yeast, at least two classes of ARE-containing mRNAs have been identified, represented by the MFA2 and TIF51A/HYP2 transcripts (49). In both cases decay proceeds through poly(A) tail shortening followed by decapping and 5′→3′ exonucleolytic decay (36, 49). The stability of the TIF51A transcript is modulated in response to changes in carbon source (11, 49). The mRNA is stable in cells grown in glucose and unstable in cells grown under nonglucose conditions. This regulation is mediated by the 3′-UTR of the TIF51A transcript, which harbors putative AREs (49). Unlike TIF51A and most mammalian AREs, the MFA2 3′-UTR promotes instability under all conditions tested thus far (36, 37, 49). The sequences that mediate the instability of MFA2 have been extensively studied (37). The 3′-UTR of the transcript can be divided into two domains (Fig. 1B). Domain I harbors two AUUUA motifs and promotes rapid decay of the mRNA on its own. Domain II is pyrimidine rich and cannot mediate rapid decay in the absence of domain I. However, when critical residues in domain I are mutated, domain II can compensate for these mutations and promote turnover of the mRNA. Significantly, the AUUUA pentamer motifs are not required for the instability of the MFA2 transcript, as simultaneous mutation of both motifs in domain I has no effect on the rate of decay of this mRNA (37). This suggests that the surrounding context or secondary structure is also important for recognition of the ARE of MFA2 by regulatory factors. Several ARE-binding factors have been identified both in mammalian and yeast systems, but it remains unclear how binding of these proteins leads to modulation of mRNA decay rates (22, 27, 49).
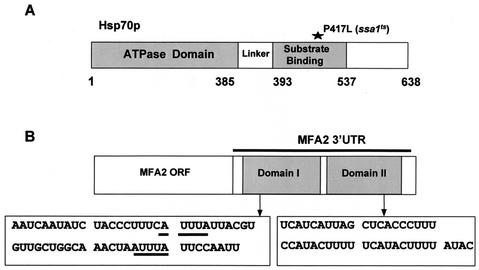
FIG. 1.
(A) Structure of the Hsp70 protein. Hsp70s consist of an N-terminal ATPase domain and linker and variable C-terminal domains of unknown function. The position of the point mutation (P417L), which makes the SSA1 gene temperature sensitive, is indicated. (B) Structure of the MFA2 mRNA. The MFA2 mRNA consists of the 117-nt coding region and a 169-nt 3′-UTR. The 3′-UTR is divided into a 58-nt domain, domain I (nt 184 to 241), and a 44-nt domain, domain II (nt 245 to 288). The putative AREs are underlined.
A number of studies have implicated the Hsp70 (heat shock protein 70) family of chaperone proteins in regulating decay of ARE-containing transcripts in mammalian systems (19, 26, 52, 57). The Hsp70 family comprises highly conserved, essential ATP-binding proteins involved in diverse cellular functions during both stress and nonstress conditions (24, 31, 39). In the absence of cellular stress these functions include, among others, protection against apoptosis (3, 35), protein degradation (16), complex formation (12, 34), translation (20), vesicle uncoating (15), protein folding, and protein translocation (2, 18, 43). During heat stress the synthesis of a subset of Hsp70 proteins is up-regulated to protect heat-denatured proteins from aggregation (24, 31, 39). Structurally, the Hsp70s are defined with an N-terminal ATP-binding domain followed by a substrate-binding domain and a variable C terminus of unknown function (Fig. 1A) (6). The substrate-binding domain binds exposed hydrophobic residues of unfolded proteins and through repetitive cycles of ATP hydrolysis refolds them until the correct functional configuration is reached. The cochaperone activity of another heat shock protein, Hsp40, drives the chaperone cycles by modulating Hsp70 ATP hydrolysis (9, 23, 39). In Saccharomyces cerevisiae there are at least 14 homologues of Hsp70, which are divided into five subfamilies based on similarities in location, structure, and function (21, 24, 39, 51).The SSA (stress seventy A) subfamily contains four redundant members (SSA1 to SSA4), which are abundant cytoplasmic proteins and exhibit the highest identity (76%) to the mammalian Hsp70s. They are essential for cell viability and can functionally substitute for each other but are differentially regulated (2, 24, 39, 51). The SSA1 gene product is overexpressed during heat shock, and a temperature-sensitive mutation confers both mitochondrial and endoplasmic reticulum translocation defects (2).
Intriguingly, Hsp70 has been suggested to function in ARE-mediated mRNA decay by two different mechanisms. AUF1, an established ARE-binding protein, has been found complexed with Hsp70/Hsc70, poly(A) binding protein and the translation initiation factor eIF4G. This complex is affected by heat shock, which results in stabilization of an ARE-containing reporter transcript (26). In this case, Hsp70 may act in its classical role as a modulator of protein complex formation on the ARE. Additionally, recent studies have demonstrated that Hsp70 can bind directly to AU-rich 3′-UTR sequences of various lymphokine and proto-oncogenic mRNAs in vitro (19, 52, 57). This binding occurs through the N-terminal ATP-binding domain of the protein and can be regulated by physiological concentrations of ATP (19, 57). These data invoke the interesting hypothesis that Hsp70 can regulate gene expression by targeting RNA molecules directly to control protein expression. However, direct evidence for specific regulation of a cellular RNA substrate by Hsp70 has so far not been demonstrated.
In this study the role of the yeast Hsp70 protein Ssa1p in regulating the stability of the yeast ARE-containing transcripts MFA2 and TIF51A has been investigated. The MFA2 mRNA is normally unstable, decaying with a half-life of 3.5 min (37). However, we find that it is significantly stabilized in a strain bearing a temperature-sensitive mutation in the SSA1 gene. Intriguingly, this effect is mediated through an AU-rich domain within the 3′-UTR of the transcript. In contrast, the ssa1ts mutation had no effect on TIF51A mRNA stability, suggesting that the effect of the thermo-sensitive mutation is MFA2 specific.
Further analysis demonstrated that stabilization of MFA2 mRNA occurs by a reduction in deadenylation rates, indicating that the ssa1ts mutation interferes with the first step of the deadenylation-dependent mRNA decay pathway. A model is presented, proposing that Hsp70 is required to remodel the protein complexes associated with the mRNA in order to allow access of the deadenylase to the poly(A) tail.
MATERIALS AND METHODS
Yeast strains and growth conditions.
S. cerevisiae strains used in this study were Y516 (SSA1: MATahis 3-11,3-15 leu2-3,2-112 ura3-52 trp1-Δ1 lys2 SSA1 ssa2-1 ssa3-1 ssa4-2), Y449 (ssa1ts: MATahis 3-11,3-15 leu2-3,2-112 ura3-52 trp1-Δ1 lys2 ssa1-45 BKD ssa2-1 ssa3-1 ssa4-2 [2]), Y517 (ydj1Δ) (32), Y518 (YDJ1) (32), Y418 (SIS1) (56), and Y419 (sis1ts) (56). Y516 and Y449 were grown in synthetic complete medium supplemented with either 2% dextrose or 2% glycerol, while Y418, Y419, Y517, and Y518 were grown in complete minimal medium lacking Leu containing 2% dextrose following standard protocols (1).
Plasmid constructs and DNA manipulation.
General DNA manipulations were carried out using standard protocols (1, 42). The following plasmids were used in this study. p4437 contained the GCN4 leader region with upstream ORF 2 (uORF2), uORF3, and uORF4 inactivated by mutation of the AUGs (45) fused to the 169-nucleotide (-nt) MFA2 3′-UTR (this study). p4036 containing the mini-PGK gene (PGK1-1 Δ 1 cloned in pRIP1) has been described previously (55). MFA2 constructs obtained from Muhlrad and Parker (37) consisted of the MFA2 coding region and the 169-nt 3′-UTR containing the following modifications: deletion of domain I producing pRP324 (MFA2-Δ1) and deletion of domain II producing pRP323 (MFA2-Δ2). The TRP1 markers of these plasmids were replaced by URA3 to give rise to p5059 and p5060, respectively. Plasmid p5062 containing the MFA2 gene with a poly(G) tract in the beginning of the 3′-UTR and under the control of the CTR1 (copper transporter 1) promoter was constructed as follows: a fragment containing the MFA2 ORF with the 3′-UTR sequence and poly(G) tract was obtained by PCR using p4034 (10) as the template and primers 418, 5′ TATTCTAGATACCAACCTTAATGC 3′, with an XbaI site and 290, 5′ ATAAAGCTTCGAATGTAATGGGTG 3′, with an HindIII site. The PCR products were digested with XbaI and HindIII, and the fragments were ligated to plasmid p5053 (ATCC 87737) linearized with SpeI and HindIII. For overexpression of Ssa1p plasmid, p5065 containing SSA1 under the ADH1 promoter was used (40).
mRNA decay measurements.
mRNA decay rates of strains Y516 (SSA1) and Y449 (ssa1ts) were measured by Northern blot analysis as previously described (41, 49) with the following modifications. Briefly, 100 ml of cells was grown at 24°C to mid-logarithmic phase, shifted to 37°C for 20 min, treated with a 150 μM concentration of the copper chelator bathocuprioinedisulphonic acid (BCS), and incubated for a further 10 min. Transcription was shut off by addition of thiolutin (a gift from Pfizer, Groton, Conn.) to a final concentration of 10 μg/ml, as well as 150 nM CuSO4. Aliquots of cells were removed at various times points, total cellular RNA was extracted from both strains, and decay rates were analyzed by Northern blotting. Addition of BCS and CuSO4 was found to enhance the response of the cells to thiolutin. For strains Y418, Y419, Y517, and Y518, mid-log cells were shifted to 30°C for 20 min before transcription was shut off using thiolutin (15 μg/ml). The cells were then harvested and analyzed as before. DNA probes were prepared by labeling appropriate fragments of MFA2, PGK1, GCN4, TIF51A, and HTB1 gene with [α-32P]dCTP as described by Hagan et al. (17). Alternatively for MFA2, the transcript was detected by RNA probes that were transcribed antisense to MFA2 as described in Ma et al. (29). The results of hybridization were normalized to the loading control U3 RNA and quantitated by PhosphorImager using ImageQuant software (Molecular Dynamics PSI-PC, Sunnyvale, Calif.). For each experiment transcriptional shutoff was confirmed by probing for the unstable transcript HTB1. Each analysis was repeated at least three times, and the half-lives shown represent an average.
Pulse-chase analysis.
Transcriptional pulse-chase analysis was performed using a protocol based on that described by Decker and Parker (10). Plasmid p5062, in which MFA2 is under the control of the CTR1 promoter, was used for the pulse-chase assay (25, 49). Briefly, Y516 (SSA1) and Y449 (ssa1ts) strains transformed with this plasmid were grown to mid-logarithmic phase at 24°C in complete minimal medium lacking Ura supplemented with 2% dextrose. The cultures were incubated with 350 nM CuSO4 for 20 min, and an aliquot of preinduced sample was taken. Cells were then transferred to 37°C for 3 min and simultaneously treated with a 150 μM concentration of the copper chelator BCS in order to turn on the promoter. After 10 min both 150 nM CuSO4 and 10-μg/ml thiolutin were added to achieve tighter and faster control over transcriptional repression. Aliquots of cells were collected at different time points, and total cellular RNA was harvested. Twenty micrograms of total RNA was separated on a 6% denaturing polyacrylamide gel and analyzed by Northern blotting. An aliquot of RNA (20 μg) was annealed to oligo(dT) primer and cleaved by RNase H to yield the deadenylated transcripts. Transcripts were detected by using oligonucleotide probes synthesized as described by Decker et al. (10). The results were quantitated and analyzed by PhosphorImager.
RESULTS
MFA2 mRNA is stabilized in a strain bearing a temperature-sensitive SSA1 allele.
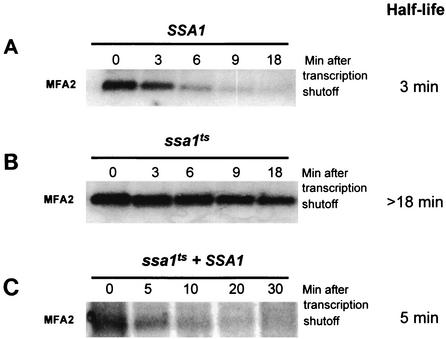
The role of Hsp70 in regulating ARE-mediated mRNA decay in yeast was investigated using strains that have all members of the SSA family of Hsp70 proteins deleted except for the wild-type SSA1 gene (SSA1 ssa2-1, ssa3-1 ssa4-2), or an isogenic ssa1ts allele harboring a point mutation (P417L) in the peptide binding domain (ssa1-45 ssa2-1, ssa3-1 ssa4-2) (Fig. 1A). The strain harboring the wild-type SSA1 allele is functionally wild type; Ssa1p compensates for the absence of the other members of the SSA family (2, 51). The effect of the ssa1ts allele on the decay of the unstable MFA2 transcript was characterized. The MFA2 transcript harbors AUUUA motifs in its 3′-UTR, and the sequences required for its decay have been characterized in detail (Fig. 1B) (37). Prior to shutting off transcription each strain was shifted to 37°C for 20 min to induce the mutant phenotype. The results demonstrated that in the wild-type strain the endogenous MFA2 transcript was predictably unstable, decaying with a half-life of 3 min (Fig. 2A). This result shows that heat shock per se does not affect the stability of the transcript. In the strain harboring the ssa1ts allele, however, the MFA2 mRNA was dramatically stabilized, with a half-life greater than 18 min (Fig. 2B). This result indicates that turnover of MFA2 transcript requires Ssa1p function.
FIG. 2.
Stabilization of MFA2 mRNA occurs as a result of a loss-of-function mutation in SSA1. The half-life of MFA2 was assessed by Northern blotting in a wild-type strain (A), an ssa1ts strain (B), and an ssa1ts strain overexpressing SSA1 (C).
The ssa1ts mutation is recessive and does not destabilize the protein.
It is possible that the effect of the ssa1ts mutant on MFA2 mRNA is due to a novel function of the protein induced by the mutation. If this were the case we would expect the mutant protein to act in a dominant-negative manner. We expressed a wild-type copy of SSA1 in the ssa1ts strain, rescuing the temperature-sensitive phenotype. The half-life of MFA2, when measured in this strain, was 5 min, indicating that the thermo-sensitive mutation in Ssa1p was not dominant negative in its function for regulating MFA2 turnover (Fig. 2C). Furthermore, the wild-type and mutant proteins are equally abundant at the restrictive temperature, suggesting that the temperature-sensitive mutation results in a loss of function of Ssa1p (data not shown).
Transcripts lacking AREs are not affected by the ssa1ts mutation.
We next determined whether other endogenous transcripts are affected by the loss of SSA1 function. The unstable HTB1 mRNA, the stable PGK1 mRNA, and the moderately stable GCN4 transcript were analyzed. None of these mRNAs contain AREs in their 3′-UTRs. The half-lives of these transcripts were unaffected in cells harboring the ssa1ts allele (Fig. 3). This result suggests that the effect of the ssa1ts mutation on mRNA stability is specific for MFA2 mRNA.
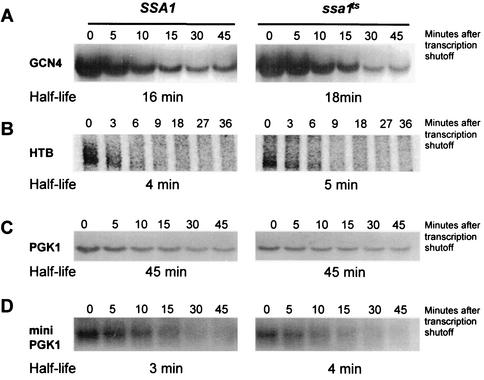
FIG. 3.
Non-ARE-containing transcripts are not affected by the ssa1ts mutation. GCN4 (A), HTB (B), and PGK1 (C) transcripts were tested for altered decay in the ssa1ts strain by Northern blot analysis. None of these transcripts were differentially stabilized, indicating non-ARE-containing transcripts are not affected. (D) A nonsense-containing transcript, mini-PGK1, is also immune to ssa1ts, with no demonstrable difference in half-life, indicating that NMD substrates are not targeted for regulation by ssa1ts.
Hsp70 does not regulate transcripts decaying through the nonsense-mediated mRNA decay pathway.
Nonsense-mediated decay (NMD) is a distinct, deadenylation-independent decay pathway acting on transcripts harboring premature stop codons (17, 30). These RNAs represent a class of transcripts distinct from those undergoing decay by the deadenylation-dependent decapping mechanism. In order to analyze if the ssa1ts allele affects NMD substrates, the half-life of a nonsense-containing mini-PGK1 allele (55) was measured. This mRNA harbors a premature stop codon and codes for a truncated Pgk1p protein. We found that the ssa1ts mutant failed to stabilize this transcript, indicating that the NMD decay route was not affected (Fig. 3D).
Hsp70 does not regulate TIF51A mRNA stability.
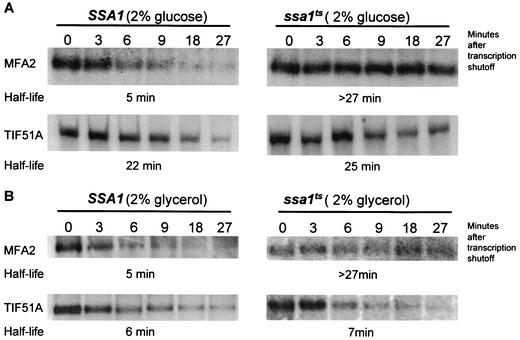
In order to determine whether stabilization by the ssa1ts allele is a general feature of ARE-containing mRNAs, we measured the half-life of another ARE-containing transcript, TIF51A, in the mutant strain. The TIF51A message, like MFA2, has an AU-rich 3′-UTR with AUUUA motifs. However, unlike MFA2, the TIF51A transcript is regulated by changes in carbon source (49). This transcript is unstable under nonglucose conditions, decaying with a half-life of 7 min, and is stabilized significantly in the presence of glucose (49). The half-lives of both the TIF51A and MFA2 mRNAs were monitored in SSA1 and ssa1ts strains grown in the presence of glucose and glycerol by Northern analysis as described above. The MFA2 transcript was unstable in both carbon sources in the wild-type strain as expected (49) and stabilized both in glucose and glycerol conditions in the ssa1ts mutant (Fig. 4). Interestingly, the decay of the TIF51A mRNA was not affected by the ssa1ts mutation, and the transcript remained stable in glucose and unstable in glycerol conditions. These results suggest that the effect of the ssa1ts mutation is specific for the MFA2 mRNA and is independent of the carbon source in which the cells were grown. This is consistent with the fact that the MFA2 transcript represents an independent class of ARE-containing mRNA, distinct from TIF51A (49).
FIG. 4.
Regulation of stability by SSA1 is specific for the MFA2 mRNA and does not affect TIF51A. (A) The ARE-containing TIF51A mRNA is not further stabilized by the ssa1ts mutation under glucose conditions. (B) The ssa1ts mutation regulates MFA2 but not TIF51A stability under nonglucose conditions. Northern blot profiles show that TIF51A is unstable in the ssa1ts mutant, decaying at a rate similar to that observed for the wild type.
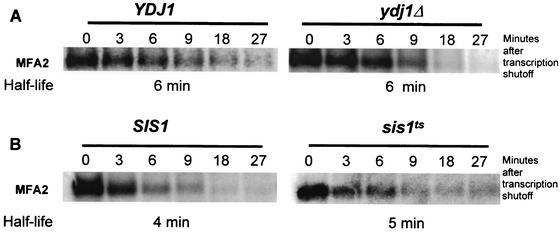
Hsp70 partner proteins Sis1p and Ydj1p do not affect MFA2 mRNA stability.
Chaperone proteins are known to interact with cochaperones (Hsp40s) that drive the folding cycle by stimulating ATP hydrolysis (9, 23, 39). In addition, Hsp40 cochaperones can also enhance Hsp70-RNA interaction in vitro (57). We have monitored the effect of mutations in Hsp40 partner proteins on MFA2 mRNA stability. In S. cerevisiae there are at least 16 different Hsp40 proteins, and Ssa1p has been shown to functionally interact with 2: Ydj1p and Sis1p (2, 20, 28). Both Ydj1p and Sis1p can stimulate Ssa1p ATPase activity (28). We have investigated the effects of mutations in these SSA1 partner proteins on the half-life of the MFA2 transcript. The results demonstrated that neither a deletion of YDJ1 nor a temperature-sensitive mutation in SIS1 is able to significantly stabilize MFA2 (Fig. 5). In both strains the MFA2 mRNA was unstable, decaying with a very similar half-life of 4 to 6 min. Taken together, the results indicate that these cochaperones are not required for SSA1 function in regulating MFA2 stability. It is possible that a different Hsp40 partner protein might be involved with Ssa1p for regulating this function, or alternatively Ssa1p can function independently of cochaperones to regulate MFA2 mRNA decay.
FIG. 5.
MFA2 mRNA decay is not affected by the Ssa1p partner proteins Ydj1p and Sis1p. Northern blots showing the decay profile of the MFA2 transcript in wild-type and mutant YDJ1 (A) and SIS1 (B) strains. MFA2 stability is not affected by either of these mutants.
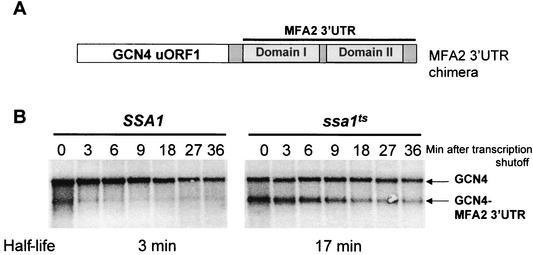
The 3′-UTR sequence of MFA2 harboring the AREs is sufficient to mediate Hsp70-dependent regulation.
We next determined whether the alteration of the half-life of the MFA2 transcript in the ssa1ts strain is mediated by its 3′-UTR. To accomplish this, a chimeric plasmid was used in which the MFA2 3′-UTR was fused downstream of the GCN4 uORF1 (Fig. 6A). The GCN4-MFA2 chimera was transformed into strains harboring either the SSA1 or the ssa1ts allele, and the half-lives of both endogenous and hybrid transcripts were monitored as described before. As shown previously for a PGK1-MFA2 hybrid mRNA (46), the GCN4-MFA2 chimera was significantly destabilized in the wild-type strain, decaying with a half-life of 3 min (Fig. 6B). However, in the ssa1ts mutant strain the chimera was stabilized approximately sixfold, decaying with a half-life of 17 min. Thus, the fusion transcript recapitulates the decay rates of the endogenous MFA2 transcript. This result defines the role of the AU-rich 3′-UTR as the primary sequence element in MFA2 that responds to regulation by SSA1. Furthermore, it also suggests that the MFA2 3′-UTR has the ability to confer regulation by SSA1 onto a heterologous transcript.
FIG. 6.
The AU-rich 3′-UTR sequence of MFA2 is sufficient to mediate Hsp70-dependent regulation. (A) Map of the GCN4-MFA2 3′-UTR chimera. This chimeric construct contains the functional GCN4 uORF1 fused to the MFA2 3′-UTR. (B) MFA2 3′-UTR can stabilize a heterologous mRNA in the ssa1ts mutant. Northern blot analysis of the chimeric construct in SSA1 and ssa1ts strains shows that GCN4, which is normally not affected by ssa1ts, can now respond to loss of Ssa1p activity, and behaves in the same manner as MFA2.
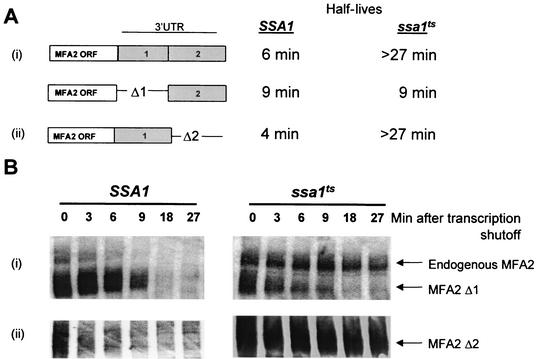
SSA1 regulates MFA2 mRNA stability through the AU-rich domain I of the MFA2 3′-UTR.
The 3′-UTR of MFA2 can be divided into two regions, each of which exerts a different effect on the stability of MFA2 transcript (37). Domain I has been previously demonstrated to be sufficient for promoting decay and contains AUUUA motifs. Domain II, however, is not required for instability but can act as a destabilizing element if domain I function is eliminated by point mutation. Simultaneous mutation of critical residues in both domains results in an increase in half-life of the transcript. To determine whether the effect of SSA1 on MFA2 stability is mediated through domain I or II, the stability of mRNAs containing domain I and domain II deletions were monitored in SSA1 and ssa1ts strains. Plasmids encoding MFA2 that lack either domain I (MFA2-Δ1) or domain II (MFA2-Δ2) were transformed into SSA1 and ssa1ts strains, and their half-lives were assessed as described above (Fig. 7). The MFA2 mRNA lacking domain I (MFA2-Δ1) decayed at a similar rate (half-life = 9 min) in both the wild-type and ssa1ts mutant strain (Fig. 7A). Deletion of domain II (MFA2-Δ2) did not affect the stability of the MFA2 transcript in the wild-type strain as previously observed (37). In the ssa1ts strain however, the domain II deletion mutant was stabilized to the same extent as a transcript bearing the full 3′-UTR (Fig. 7). These results indicate that the AU-rich domain I of the MFA2 3′-UTR, which has been previously established as being sufficient for stimulating decay of the RNA (37), is critical for regulation by SSA1. Domain II cannot substitute for domain I in this regulation since the transcript harboring domain II only, remains unstable in an ssa1ts strain.
FIG. 7.
SSA1 regulates MFA2 mRNA stability through the AU-rich domain I of the MFA2 3′-UTR. (A) Shown are half-life measurements of wild-type and mutant (Δ1 or Δ2) MFA2 transcripts analyzed in SSA1 and ssa1ts strains after shifting to nonpermissive temperature. (B) Northern blots depicting the decay profiles of the transcripts. A domain II deletion (MFA2-Δ2) is stabilized similarly to the endogenous transcript in an ssa1ts mutant. A domain I deletion (MFA2-Δ1), however, makes the transcript unstable in an ssa1ts mutant, indicating that the domain I region is critical for the regulation of MFA2 stability by SSA1.
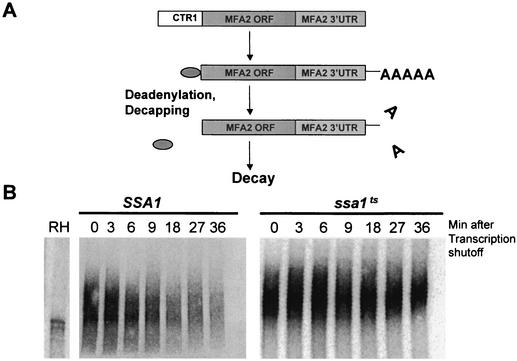
The ssa1ts mutation stabilizes MFA2 by affecting deadenylation.
We next determined whether poly(A) shortening rates were affected in an ssa1ts strain. To accomplish this we analyzed decay intermediates using a pulse-chase approach. The MFA2 gene was cloned downstream of the copper transporter promoter CTR1 (Fig. 8A). This promoter is repressed in the presence of copper and activated under copper starvation conditions. Total RNA was extracted at different times following transcription shutoff and was analyzed. Rates of deadenylation were estimated by comparing the time required for the poly(A) tail to shorten completely compared to control samples treated with oligo(dT) and RNase H. In the wild-type strain the MFA2 transcript deadenylates rapidly. In the ssa1ts strain, however, there was little shortening of the poly(A) tail, suggesting a major block to the deadenylation process due to inactivation of SSA1 (Fig. 8B). These results indicate that the stabilization of MFA2 mRNA observed in the ssa1ts strain is a consequence of reducing the deadenylation rates.
FIG. 8.
Deadenylation of MFA2 mRNA is inhibited in the ssa1ts strain. (A) Pathway of mRNA decay. In yeast most mRNAs degrade by the deadenylation-dependent decay pathway where deadenylation is followed by decapping and finally by predominantly 5′→3′ exonucleolytic decay. The entire MFA2 gene was fused downstream of the inducible CTR1 promoter in order to dissect which step of the deadenylation-dependent decay pathway was affected by the ssa1ts mutant. (B) ssa1ts affects deadenylation rates. The polyacrylamide Northern blot of MFA2 mRNA shows that deadenylation is dramatically reduced in the ssa1ts mutant. RH denotes samples annealed with oligo(dT) and treated with RNase H prior to loading, in order to indicate the size of the deadenylated transcript.
DISCUSSION
The 70-kDa heat shock proteins (Hsp70s) are ubiquitous chaperones with well-characterized roles in protein folding, trafficking, translocation, and protection against cellular stress (24). Recently, several studies have implicated Hsp70 proteins as regulators of ARE-mediated mRNA decay. In vitro experiments have described a novel interaction of Hsp70s with cellular targets other than proteins. Several studies have demonstrated the ability of recombinant mammalian Hsp70 to specifically bind AU-rich mRNA sequences (19, 52, 57). Additionally, it has been shown that heat shock, which stimulates the production of Hsp70, can significantly stabilize ARE-containing reporter transcripts in HeLa cells (26). These reports implicate Hsp70 proteins in the regulation of ARE-mediated mRNA decay. However, to date no results have defined the exact role of the Hsp70 proteins in this process. In this study, we have used the yeast S. cerevisiae as a model system to study the role of Hsp70 in ARE-mediated mRNA decay. Both the Hsp70 proteins and mRNA turnover pathways have been extensively studied in this organism. We have utilized a temperature-sensitive mutant of the SSA1 gene to explore the role of the Hsp70 protein, Ssa1p, in regulating the stability of two naturally occurring ARE-containing transcripts, MFA2 and TIF51A. Our results demonstrate that Ssa1p is specifically required for the rapid decay of the MFA2 mRNA (Fig. 2A and B). In addition, the stabilization of MFA2 by the ssa1ts mutant can be rescued by expression of wild-type SSA1 (Fig. 2C). This suggests that the thermo-sensitive SSA1 mutant does not function in a dominant-negative manner. To our knowledge this is the first demonstration of specific, in vivo regulation of an ARE-containing transcript by this class of chaperone proteins and also identifies the first trans-acting factor involved in regulating MFA2 mRNA stability.
The decay of the MFA2 mRNA has been studied extensively and is known to be modulated by sequences in the 3′-UTR harboring AREs. (36, 37, 49). In particular, domain I of the 3′-UTR, which contains two AUUUA motifs, is sufficient to promote rapid deadenylation-dependent decay of the transcript. Domain II promotes decay if the function of domain I is eliminated by mutation. The AUUUA motifs are not necessary as long as sequences in the surrounding region remains intact, suggesting that for regulated decay both the AREs and their context are important (37). Interestingly, the 3′-UTR of MFA2 can confer regulation by SSA1 on a heterologous RNA (Fig. 6). This result suggests that this region, which contains important stability determinants, is also essential for the specific effect of ssa1ts. Further analysis of deletions within the MFA2 3′-UTR has revealed that the AU-rich domain I contains the essential sequence elements required for SSA1 function (Fig. 7), while domain II appears to be dispensable.
It is known that the pathway of ARE-mediated decay in yeast initiates with rapid removal of the poly(A) tail, followed by decapping and finally 5′→3′ exonucleolytic degradation of the mRNA body (49). Therefore, we have examined which step of the decay process is affected by the ssa1ts mutation. A transcriptional pulse-chase analysis revealed that in the ssa1ts mutant there is a major reduction in deadenylation rates. (Fig. 8). This observation indicates that poly(A) tail shortening, which is the rate-limiting step of the decay process, is inhibited when SSA1 function is disabled (Fig. 8).
Our recent results in yeast suggest that decay of one class of ARE-containing mRNAs can be regulated by carbon source (49). MFA2 appears to represent an alternate class as it is unstable both in glucose and under nonglucose conditions (49). Here, we show that MFA2 is dramatically stabilized both in glucose and under nonglucose conditions by mutation of SSA1 (Fig. 4). Significantly, the stability of TIF51A mRNA, which is a carbon source-regulated ARE-containing mRNA, is not affected by ssa1ts in either glucose or glycerol medium (Fig. 4). This indicates that the Ssa1p mutant protein functions independently of carbon source to specifically regulate MFA2. The different response of the TIF51A mRNA to ssa1ts is not unexpected, as the 3′-UTRs of MFA2 and TIF51A differ greatly with respect to the context of AUUUA motifs. Our findings therefore emphasize that in yeast, as in higher eukaryotes, different AREs require different trans-acting factors to respond to cellular signals.
In principle, SSA1 could be acting by binding directly to the mRNA and promoting decay, or alternatively by modulating the conformation of the MFA2 mRNP to allow degradation. We have not observed any specific binding of either the wild-type or mutant Ssa1p to RNA either in extracts or using recombinant Ssa1p. This result therefore suggests that the S. cerevisiae Ssa1p either does not bind RNA or does so dynamically, beyond detection limits. As binding affinity of Hsp70 to AU-rich sequences is strongest in proteins of mammalian origin (57), it is possible that RNA-binding capacity is a feature of Hsp70s from higher eukaryotes only. One explanation is that Ssa1p regulates decay not by binding to the AU-rich sequences in the mRNA but by modulating the configuration of the mRNP complex.
Experiments utilizing in vitro mRNA decay systems from both mammalian and yeast cells have demonstrated that the deadenylation process can be activated by removal of the poly(A) binding protein (Pab1p) from the poly(A) tail by competition with exogenous poly(A) (13, 14, 53). This observation has led to the hypothesis that the onset of deadenylation in cells is triggered by dissociation of Pab1p from the poly(A) tail (4, 5). Therefore, the fact that deadenylation of MFA2 mRNA is inhibited in the ssa1ts mutant might indicate that removal of Pab1p from the poly(A) tail is affected. Intriguingly, Hsp70 has been demonstrated to interact with Pab1p in both yeast and mammalian systems (20, 26), suggesting a model where Hsp70 might facilitate dissociation of an mRNP complex formed between poly(A) binding protein, translation initiation factors, and ARE-binding proteins. In this model, in the absence of functional Hsp70 the mRNP complex would remain tightly associated and thereby prevent the rapid degradation of the mRNA.
Future experiments will focus on dissecting the mechanism by which mutation of SSA1 leads to stabilization of MFA2 mRNA and identification of other factors involved, including perhaps an Hsp40 partner for this process. It will be interesting to determine whether stabilization of other putative ARE-containing yeast mRNAs is observed in this mutant. The results of this study also demonstrate the potential of the yeast system to assess the effects of heat shock on regulation of mRNA decay.
Acknowledgments
We are grateful to Terri Goss Kinzy, Gerald Wilson, Gary Brewer, and Jeffrey Wilusz for critical reading of the manuscript and helpful discussion. We also thank Denise Muhlrad, Kenji Kohno, and Roy Parker for plasmids and Pfizer Inc. for the kind gift of thiolutin. We also thank Kim Arndt, Douglas Cyr, and Elizabeth Craig for yeast strains and Nader Mahmood for assistance.
C.J.W. is supported by a Scientist Development Award (0130470T) from the American Heart Association. S.W.P. is the recipient of a grant (GM 58276) from the National Institutes of Health.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1992. Current protocols in molecular biology, vol. 2, p. 1-13. John Wiley and Sons, New York, N.Y.
- 2.Becker, J., W. Walter, W. Yan, and E. A. Craig. 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16:4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beere, H. M., and D. R. Green. 2001. Stress management—heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol. 11:6-10. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, P., S. W. Peltz, and J. Ross. 1989. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 9:659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, P., and J. Ross. 1989. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem. Sci. 14:373-377. [DOI] [PubMed] [Google Scholar]
- 6.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Cyr, D. M., T. Langer, and M. G. Douglas. 1994. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19:176-181. [DOI] [PubMed] [Google Scholar]
- 10.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 11.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 12.Dittmar, K. D., M. Banach, M. D. Galigniana, and W. B. Pratt. 1998. The role of DnaJ-like proteins in glucocorticoid receptor·hsp90 heterocomplex assembly by the reconstituted hsp90·p60·hsp70 foldosome complex. J. Biol. Chem. 273:7358-7366. [DOI] [PubMed] [Google Scholar]
- 13.Ford, L. P., J. Watson, J. D. Keene, and J. Wilusz. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford, L. P., and J. Wilusz. 1999. An in vitro system using HeLa cytoplasmic extracts that reproduces regulated mRNA stability. Methods 17:21-27. [DOI] [PubMed] [Google Scholar]
- 15.Gao, B. C., J. Biosca, E. A. Craig, L. E. Greene, and E. Eisenberg. 1991. Uncoating of coated vesicles by yeast hsp70 proteins. J. Biol. Chem. 266:19565-19571. [PubMed] [Google Scholar]
- 16.Gusarova, V., A. J. Caplan, J. L. Brodsky, and E. A. Fisher. 2001. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J. Biol. Chem. 276:24891-24900. [DOI] [PubMed] [Google Scholar]
- 17.Hagan, K. W., M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz. 1995. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 15:809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-579. [DOI] [PubMed] [Google Scholar]
- 19.Henics, T., E. Nagy, H. J. Oh, P. Csermely, A. von Gabain, and J. R. Subjeck. 1999. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. J. Biol. Chem. 274:17318-17324. [DOI] [PubMed] [Google Scholar]
- 20.Horton, L. E., P. James, E. A. Craig, and J. O. Hensold. 2001. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J. Biol. Chem. 276:14426-14433. [DOI] [PubMed] [Google Scholar]
- 21.James, P., C. Pfund, and E. A. Craig. 1997. Functional specificity among Hsp70 molecular chaperones. Science 275:387-389. [DOI] [PubMed] [Google Scholar]
- 22.Jarzembowski, J. A., and J. S. Malter. 1997. Cytoplasmic fate of eukaryotic mRNA: identification and characterization of AU-binding proteins. Prog. Mol. Subcell. Biol. 18:141-172. [DOI] [PubMed] [Google Scholar]
- 23.Kelley, W. L. 1999. Molecular chaperones: how J domains turn on Hsp70s. Curr. Biol. 9:R305-R308. [DOI] [PubMed] [Google Scholar]
- 24.Kiang, J. G., and G. C. Tsokos. 1998. Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology. Pharmacol. Ther. 80:183-201. [DOI] [PubMed] [Google Scholar]
- 25.Labbe, S., and D. J. Thiele. 1999. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306:145-153. [DOI] [PubMed] [Google Scholar]
- 26.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 27.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273:27824-27830. [DOI] [PubMed] [Google Scholar]
- 29.Ma, Y. J., G. A. Dissen, F. Rage, and S. R. Ojeda. 1996. RNase protection assay. Methods 10:273-278. [DOI] [PubMed] [Google Scholar]
- 30.Maquat, L. E., and G. G. Carmichael. 2001. Quality control of mRNA function. Cell 104:173-176. [DOI] [PubMed] [Google Scholar]
- 31.Mayer, M. P., and B. Bukau. 1998. Hsp70 chaperone systems: diversity of cellular functions and mechanism of action. Biol. Chem. 379:261-268. [PubMed] [Google Scholar]
- 32.Meacham, G. C., B. L. Browne, W. Zhang, R. Kellermayer, D. M. Bedwell, and D. M. Cyr. 1999. Mutations in the yeast Hsp40 chaperone protein Ydj1 cause defects in Axl1 biogenesis and pro-a-factor processing. J. Biol. Chem. 274:34396-34402. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, P., and D. Tollervey. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10:193-198. [DOI] [PubMed] [Google Scholar]
- 34.Morishima, Y., P. J. Murphy, D. P. Li, E. R. Sanchez, and W. B. Pratt. 2000. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J. Biol. Chem. 275:18054-18060. [DOI] [PubMed] [Google Scholar]
- 35.Mosser, D. D., A. W. Caron, L. Bourget, A. B. Meriin, M. Y. Sherman, R. I. Morimoto, and B. Massie. 2000. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol. Cell. Biol. 20:7146-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 37.Muhlrad, D., and R. Parker. 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6:2100-2111. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee, D., M. Gao, P. J. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtsuka, K., and M. Hata. 2000. Molecular chaperone function of mammalian Hsp70 and Hsp40—a review. Int. J. Hyperthermia 16:231-245. [DOI] [PubMed] [Google Scholar]
- 40.Oka, M., M. Nakai, T. Endo, C. R. Lim, Y. Kimata, and K. Kohno. 1998. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 273:29727-29737. [DOI] [PubMed] [Google Scholar]
- 41.Parker, R., D. Herrick, S. W. Peltz, and A. Jacobson. 1991. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 194:415-423. [DOI] [PubMed] [Google Scholar]
- 42.Philippsen, P., A. Stotz, and C. Scherf. 1991. DNA of Saccharomyces cerevisiae. Methods Enzymol. 194:169-182. [DOI] [PubMed] [Google Scholar]
- 43.Rassow, J., and N. Pfanner. 1995. Molecular chaperones and intracellular protein translocation. Rev. Physiol. Biochem. Pharmacol. 126:199-264. [DOI] [PubMed] [Google Scholar]
- 44.Ross, J. 1996. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 12:171-175. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Echevarria, M. J., C. I. Gonzalez, and S. W. Peltz. 1998. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 17:575-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Echevarria, M. J., R. Munshi, J. Tomback, T. G. Kinzy, and S. W. Peltz. 2001. Characterization of a general stabilizer element that blocks deadenylation-dependent mRNA decay. J. Biol. Chem. 276:30995-31003. [DOI] [PubMed] [Google Scholar]
- 47.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 48.Shyu, A. B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 49.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7:1191-1200. [DOI] [PubMed] [Google Scholar]
- 50.Veyrune, J. L., J. Hesketh, and J. M. Blanchard. 1997. 3′ untranslated regions of c-myc and c-fos mRNAs: multifunctional elements regulating mRNA translation, degradation and subcellular localization. Prog. Mol. Subcell. Biol. 18:35-63. [DOI] [PubMed] [Google Scholar]
- 51.Werner-Washburne, M., D. E. Stone, and E. A. Craig. 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, G. M., K. Sutphen, S. Bolikal, K. Y. Chuang Ky, and G. Brewer. 2001. Thermodynamics and kinetics of Hsp70 association with A + U-rich mRNA-destabilizing sequences. J. Biol. Chem. 276:44450-44456. [DOI] [PubMed] [Google Scholar]
- 53.Wilusz, C. J., M. Gao, C. L. Jones, J. Wilusz, and S. W. Peltz. 2001. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 7:1416-1424. [PMC free article] [PubMed] [Google Scholar]
- 54.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, S., M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz. 1995. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 15:2231-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, T., and K. T. Arndt. 1993. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell 73:1175-1186. [DOI] [PubMed] [Google Scholar]
- 57.Zimmer, C., A. von Gabain, and T. Henics. 2001. Analysis of sequence-specific binding of RNA to Hsp70 and its various homologs indicates the involvement of N- and C-terminal interactions. RNA 7:1628-1637. [PMC free article] [PubMed] [Google Scholar]