Abstract
The c-Jun NH2-terminal kinase (JNK) is activated by the cytokine tumor necrosis factor (TNF). This pathway is implicated in the regulation of AP-1-dependent gene expression by TNF. To examine the role of the JNK signaling pathway, we compared the effects of TNF on wild-type and Jnk1−/− Jnk2−/− murine embryo fibroblasts. We show that JNK is required for the normal regulation of AP-1 by TNF. The JNK-deficient cells exhibited decreased expression of c-Jun, JunD, c-Fos, Fra1, and Fra2; decreased phosphorylation of c-Jun and JunD; and decreased AP-1 DNA binding activity. The JNK-deficient cells also exhibited defects in the regulation of the AP-1-related transcription factor ATF2. These changes were associated with marked defects in TNF-regulated gene expression. The JNK signal transduction pathway is therefore essential for AP-1 transcription factor regulation in cells exposed to TNF.
Tumor necrosis factor alpha (TNF-α) is a potent cytokine that influences immune response, proliferation, differentiation, and apoptosis (6). These effects of TNF-α are mediated by several signal transduction pathways, including the activation of caspases, the NF-κB pathway, and mitogen-activated protein kinases (MAPK). Dysregulated TNF-α function is implicated in the pathology of many diseases, including rheumatoid arthritis, Crohn's disease, and several neuropathologies. Understanding the mechanism of signal transduction by TNF-α is therefore important, because this knowledge will allow the design of novel therapeutic strategies for these diseases.
The binding of TNF to TNF receptor-1 (TNF-R1) causes trimerization of the receptor and recruitment of the adapter protein TRADD to the cytoplasmic domain of the receptor (6). TRADD acts as a scaffold protein that recruits FADD, RIP1, and TRAF2. In contrast, TNF-R2 directly binds TRAF1, which subsequently recruits TRAF2. These adapter proteins act to initiate signal transduction pathways within a complex that is bound to the receptor. Thus, apoptosis and anti-inflammatory responses are mediated by the activation of caspase 8 by TRADD (43). In contrast, antiapoptosis and inflammatory responses are mediated by activation of MAPK pathways by TRAF2 (44) and the NF-κB pathway by TRAF2 plus RIP1 (11, 19).
The inflammatory response of cells to TNF is mediated, in part, by the regulation of gene expression by the AP-1 and NF-κB groups of transcription factors (2). The NF-κB signal transduction pathway is directly engaged by the TNF receptors. In contrast, the AP-1 group of transcription factors is activated by an indirect mechanism that involves MAPK. The activation of AP-1 is mediated by at least two regulatory events. First, some AP-1 proteins (e.g., c-Jun) are encoded by immediate-early genes that are transcriptionally induced following treatment of cells with TNF. Second, some AP-1 proteins (e.g., c-Jun) are posttranslationally modified to increase transcription activity. The relative roles of different MAPK in these processes is unclear. For example, the transcriptional induction of c-Jun expression may be mediated by several MAPK-responsive elements in the promoter, including MEF2, a consensus AP-1 site that binds c-Jun, and a nonconsensus AP-1 site (Jun2 TRE) that binds c-Jun/ATF2 heterodimers (30). The p38 MAPK can phosphorylate and activate MEF2 (15), c-Jun NH2-terminal kinase (JNK) can phosphorylate and activate c-Jun (10, 20), and both p38 MAPK and JNK can phosphorylate and activate ATF2 (14, 22, 27, 28, 37). Together, these data suggest that the regulation of AP-1 activity by TNF may involve the concerted actions of MAPK.
The purpose of this study was to examine the role of JNK in cells treated with TNF. It is known that TNF activates JNK by a pathway that is initiated by TRAF2 (44). The MAPK kinase kinase in this pathway has not been defined, although a role for ASK1 has been proposed at late times of TNF signaling (33). It is established that the MAPK kinase MKK7 is essential for JNK activation caused by TNF (34). However, the role of activated JNK in TNF-treated cells is unclear. Gene disruption studies demonstrate that JNK is not required for TNF-stimulated apoptosis (35). Here we demonstrate that JNK is critical for TNF regulation of the AP-1 group of transcription factors. Loss of AP-1 activation in JNK-deficient cells causes marked defects in the response of cells following treatment with TNF.
MATERIALS AND METHODS
Materials.
Recombinant murine TNF-α, the p38 inhibitor PD 169316, myelin basic protein, and [γ-32P]ATP were obtained from R&D Systems, Calbiochem, Sigma, and Pharmacia-Amersham, respectively. Recombinant glutathione S-transferase (GST)-c-Jun (residues 1 to 79) (10) and GST-ATF2 (residues 1 to 109) (14) were expressed in bacteria and purified by affinity chromatography.
Tissue culture and preparation of cell extracts.
Wild-type (WT), Jnk1−/− Jnk2−/− (Jnk−/−), and Mkk4−/− Mkk7−/− (Mkk−/−) fibroblasts were established from embryonic day 13.5 mouse embryos (34, 35). The cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Life Technologies) at 37°C in a humidified atmosphere with 5% CO2. JNK was expressed in the cells in transient-transfection assays using plasmid expression vectors and the Lipofectamine reagent (Invitrogen). Retroviral transduction experiments were performed using mouse stem cell retroviral vector (MSCV)-internal ribosome entry site-green fluorescent protein vectors, and the transduced cells were isolated by flow cytometry. The JNK1 and JNK2 retroviral vectors were constructed by subcloning the cDNAs as blunt-ended fragments into the EcoRI site of the vector MCSV-internal ribosome entry site-green fluorescent protein (46).
Cell lysates were prepared from cells starved of serum (2 h) prior to treatment with TNF-α (10 ng/ml). The cells were washed twice with phosphate-buffered saline (PBS), and extracted (10 min at 4°C) with lysis buffer (25 mM HEPES [pH 7.5], 0.3 M NaCl, 0.2 mM EDTA, 0.1% Triton X-100, 0.5 mM dithiothreitol, 20 mM β-glycerophosphate, 0.1 mM vanadate, leupeptin [2 μg/ml], aprotinin [2 μg/ml], 1 mM phenylmethylsulfonyl fluoride). The extracts were clarified using an Eppendorf microcentrifuge (14,000 rpm for 10 min at 4°C). The concentration of total soluble protein in the supernatant was quantitated by the Bradford method (Bio-Rad). These extracts were employed for immunoblot analysis and protein kinase assays.
Luciferase reporter gene assays.
Transfection assays were performed using Lipofectamine reagent (Invitrogen) according to manufacturer's recommendations. The cells were transfected with 0.2 μg of the GAL4 fusion protein expression vector pGAL4, pGAL4-ATF2, or pGAL4-ATF2-Thr69Ala/Thr-71Ala (14), 0.5 μg of the luciferase reporter plasmid pG5E1bLuc (14), and 0.05 μg of the control vector pRL-null (Promega). The effect of cotransfection with 0.2 μg of pCMV-MEKK1 (40), pCDNA3-Flag-MKK6(Ser207Glu/Thr211Glu) (28), or pCDNA3-HA-MLK3 (40) was examined. Control studies were performed using the empty expression vectors. Cell extracts and luciferase quantitation were performed using the Dual-Luciferase Reporter assay system (Promega Corp., Madison, Wis.).
Measurement of protein kinase activity.
The activities of JNK, p38, and extracellular signal-regulated kinase (ERK) were determined by immunoprecipitation-kinase assays using antibodies to JNK (FL; Santa Cruz Biotechnology, Inc.), p38 MAPK (27), and ERK1/2 (C16/C14; Santa Cruz Biotechnology, Inc.). Cell lysates were incubated for 3 to 4 h at 4°C with antibodies and then isolated by incubation with protein G- or protein A (Pharmacia-LKB Biotechnology)-Sepharose beads for 2 h at 4°C. Complexes were washed three times with a solution containing 1× PBS, 1% NP-40, and 2 mM vanadate; once with 100 mM Tris (pH 7.5)-0.5 M LiCl, and once with kinase buffer (12.5 mM morpholinepropanesulfonic acid [pH 7.5], 12.5 mM β-glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM NaF, 0.5 mM vanadate). The activity was measured by adding 30 μl of kinase buffer containing 50 μM [γ-32P]ATP (10 Ci/mmol) and 1 μg of GST-c-Jun (JNK), 1 μg of GST-ATF2 (p38 MAPK), or 1 μg of MBP (ERK). The reactions were terminated by addition of Laemmli sample buffer. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12 to 15% polyacrylamide gel) and identified by autoradiography. The incorporation of [32P]phosphate was quantitated by PhosphorImager analysis.
Electrophoretic mobility shift assays.
Cytosol and nuclear extracts were prepared (1) and DNA binding activity was examined using oligonucleotide probes for AP-1 and Jun2 TRE. The AP-1 probe was a double-stranded oligonucleotide (5′-CGCTTGATGACTCAGCCGGAA-3′) and was labeled with T4 polynucleotide kinase and [γ-32P]ATP. The Jun2 TRE probe was made by annealing 5′-AGCTAGCATTACCTCATCCC-3′ and 5′-GATCGGGATGAGGTAATGCT-3′ and was labeled with Klenow and [α-32P]ATP. Binding assays were performed (for 30 min at 4°C) and examined on a 6% nondenaturing polyacrylamide gel. Competition analysis was performed using a 100-fold excess of unlabeled probe.
Immunoblot analysis.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide gel) and electrophoretically transferred to an Immobilon-P membrane (Millipore, Inc.). The membranes were incubated with 5% nonfat dry milk (at 4°C for 5 h) and then probed with antibodies to ATF2 (N-96 and F2BR-1; Santa Cruz), phospho(Thr-71)-ATF2 (F1; Santa Cruz), c-Jun (H-79; Santa Cruz), JunD (329; Santa Cruz), phospho (Ser-73)-Jun (Upstate Biotechnology), JNK 1/2 (PharMingen), ERK 1/2 (C14/16; Santa Cruz) and p38 MAPK (N-20; Santa Cruz). Immune complexes were detected by enhanced chemiluminescence (NEN).
Immunofluorescence microscopy.
Cells grown on glass coverslips were fixed at −20°C in methanol (5 min) and acetone (2 min). The cells were then incubated (30 min) in blocking buffer (3% bovine serum albumin in PBS) and subsequently incubated (60 min) in blocking buffer with primary antibodies, including mouse monoclonal anti-phospho(Thr-71) ATF2 (F1 [1/100]; SantaCruz), rabbit anti-ATF2 (N-96 [1/100]; SantaCruz), rabbit anti-phospho(Thr183 and Tyr 185) JNK (anti-active JNK [1/100]; Promega) and anti-JNK1/2 (1/100; Pharmingen). Immune complexes were detected with Texas red-conjugated anti-rabbit immunoglobulin (Ig) and fluorescein isothiocyanate (FITC)-conjugated anti-mouse Ig antibodies (1/300; Jackson Immunoresearch, Inc.) in blocking buffer. The coverslips were mounted in Vectashield with DAPI (4′,6′-diamino-2-phenylindole) (Vector Laboratories) and examined by immunofluorescence microscopy using a conventional Axioplan 2 microscope with a MicroImager charge-coupled device camera (Carl Zeiss).
Flow cytometry.
The cells were detached in PBS with 0.53 mM EDTA and washed once in PBS with 1% FBS, and 106 cells were treated (5 min on ice) with Fc Block (anti-mouse CD16/CD32; BD Pharmingen). The cells were then incubated (1 h on ice) with an antibody to TNF-R1 (559915; BD Pharmingen). TNF-R1 immunocomplexes were detected with biotin-conjugated mouse anti-hamster IgG antibody (BD Pharmingen) followed by phycoerythrin-conjugated streptavidin (BD Pharmingen). The cells were incubated (60 min on ice) and washed in PBS with 1% FBS between incubations. The cells were fixed in 100 μl of 4% paraformaldehyde (Ted Pella, Inc.) in PBS (for 20 min at 25°C), washed, and resuspended in PBS with 1% FBS prior to analysis by flow cytometry.
RNase protection assays.
Total RNA (5 μg) was examined using the Multi-probe RNase protection assay (Pharmingen) with the template sets mCK3, mCK4, mCR4, and mFos/Jun following the manufacturer's recommendations. The products were separated on a 5% sequencing gel, detected by autoradiography, and quantitated by PhosphorImager analysis (Molecular Dynamics).
RESULTS
WT and Jnk−/− murine embryo fibroblasts were examined in culture. Control experiments demonstrated equal expression of TNF receptor mRNA (TNF-R1 and TNF-R2) in RNase protection assays (data not shown). In addition, equal amounts of TNF-R1 were detected by flow cytometry on the surface of WT and Jnk−/− fibroblasts (data not shown). Furthermore, the rate and extent of TNF-stimulated degradation of IκB and nuclear accumulation of NF-κB was similar in WT and Jnk−/− cells (data not shown). These data indicate that the TNF signaling pathway is functional in both WT and Jnk−/− fibroblasts. This is consistent with the results of previous studies which indicate that TNF (in the presence of protein or mRNA synthesis inhibitors) causes apoptosis of both WT and Jnk−/− cells (35). Together, these data indicate that JNK deficiency does not cause generalized defects in TNF receptor activation in fibroblasts. In contrast, a marked and selective defect in MAPK activation was observed in JNK-deficient cells.
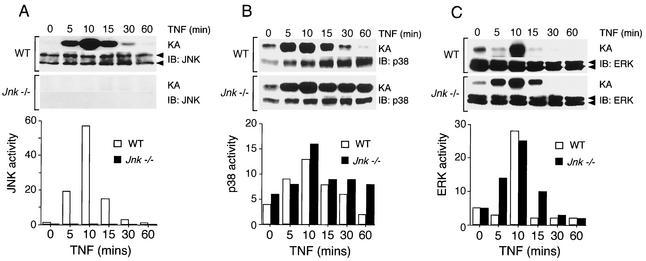
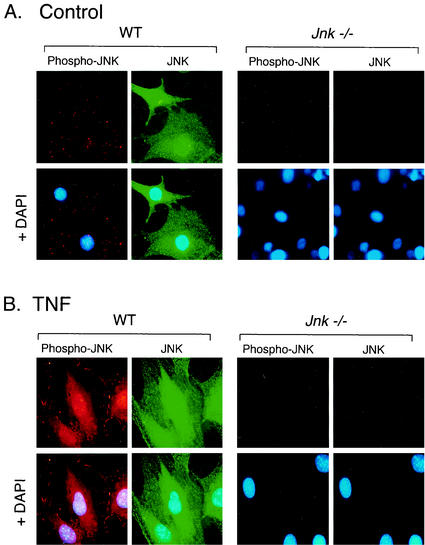
Treatment of WT fibroblasts with TNF caused increased activity of the ERK, JNK, and p38 MAPK (Fig. 1). Comparative studies of WT and Jnk−/− fibroblasts indicated that the regulation of ERK activity and the regulation of p38 MAPK activity were similar. In contrast, both JNK protein and activity were absent in the Jnk−/− fibroblasts (Fig. 1). To confirm the loss of JNK in the Jnk−/− fibroblasts, we examined the expression of JNK by immunofluorescence analysis (Fig. 2). In WT fibroblasts, JNK was detected in both the cytoplasm and the nucleus. Treatment with TNF caused a marked increase in the amount of activated JNK that was detected using an antibody that binds phospho-Thr-185/Tyr-185 JNK (Fig. 2). Neither JNK nor activated JNK was observed in Jnk−/− cells (Fig. 2). Together, these data indicate that the Jnk−/− cells appear to have a selective defect in the JNK signaling pathway.
FIG. 1.
Effect of TNF on the activation of MAPK in WT and Jnk−/− fibroblasts. The activities of JNK (A), p38 MAPK (B), and ERK (C) in cells treated with TNF-α (10 ng/ml) for different times were measured in an immunocomplex protein kinase assay (KA). The amount of MAPK was examined by immunoblot (IB) analysis. The substrate phosphorylation in the kinase assays was quantitated using a PhosphorImager and is presented graphically in relative units. The data shown were derived from a single experiment and are representative of three independent experiments.
FIG. 2.
Comparison of JNK expression and activation in WT and Jnk−/− fibroblasts. JNK and activated JNK (Phospho-JNK) were examined by immunofluorescence microscopy. The cells were treated without (A) and with (B) TNF-α (10 ng/ml for 15 min), fixed, and stained with antibodies to JNK (FITC [green]) and phospho-JNK (Texas red [red]). DNA was stained with DAPI (blue). The cells were imaged by conventional fluorescence microscopy.
JNK is required for TNF-stimulated expression of AP-1 transcription factors.
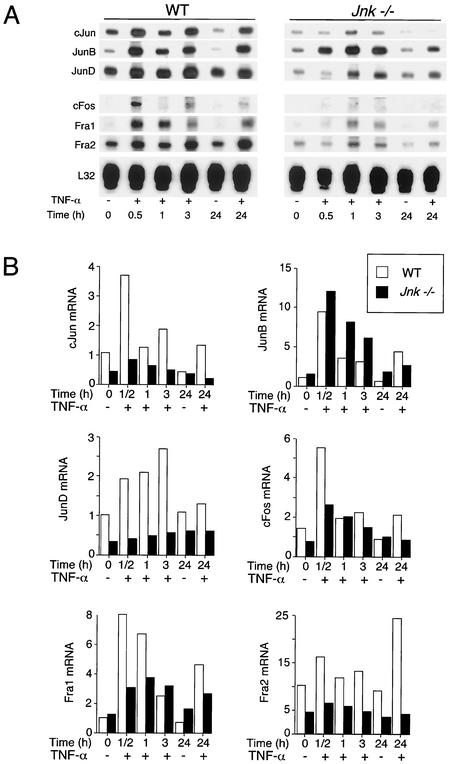
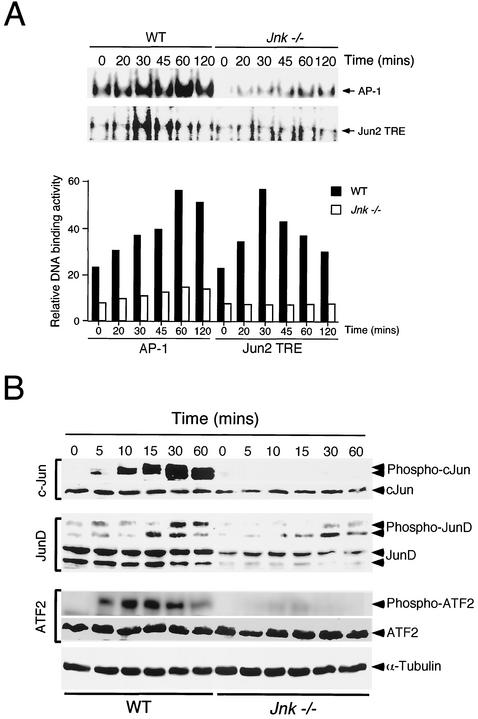
TNF regulates AP-1 transcription factor activity, in part, by increasing the expression of members of the Jun and Fos families. Studies of WT cells indicated that TNF caused increased expression of c-Jun, JunB, JunD, c-Fos, Fra1, and Fra2 (Fig. 3). Marked changes in the expression of these transcription factors were observed in Jnk−/− fibroblasts, with the exception of JunB, which was expressed at similar levels in WT and Jnk−/− fibroblasts. The TNF-stimulated increase in expression of c-Jun, JunD, c-Fos, Fra1, and Fra2 was strongly suppressed in the Jnk−/− cells (Fig. 3). In addition, the basal expression of c-Jun, JunD, c-Fos, and Fra2 in cells treated without TNF was greatly reduced in Jnk−/− cells compared to WT cells. These data indicate that JNK-deficient fibroblasts have severe defects in the expression of the AP-1 group of transcription factors. To confirm this conclusion, we examined AP-1 DNA binding activity in electrophoretic mobility shift assays. Studies using a consensus AP-1 probe demonstrated that TNF caused a time-dependent increase in AP-1 DNA binding activity, which was strongly suppressed in Jnk−/− fibroblasts (Fig. 4A). Similar observations were made in experiments using a nonconsensus AP-1 probe (Jun2 TRE) that preferentially binds heterodimeric complexes of Jun proteins with ATF2 (Fig. 4A). These data demonstrate that Jnk−/− cells exhibit a severe defect in TNF-stimulated AP-1 transcription factor expression.
FIG. 3.
AP-1 transcription factor expression in WT and Jnk−/− fibroblasts. WT and Jnk−/− cells were treated without (−) and with (+) TNF-α (10 ng/ml). The cells were harvested after different times, and total RNA was isolated. The c-Jun, JunB, JunD, c-Fos, Fra1, Fra2, and ribosomal protein L32 mRNAs were examined in an RNase protection assay. The mRNAs were detected by autoradiography (A) and quantitated by PhosphorImager analysis (B). The normalized AP-1 mRNA/L32 mRNA ratio is presented. The data shown are representative of data obtained in three independent experiments.
FIG. 4.
AP-1 transcription factors in WT and Jnk−/− fibroblasts. (A) WT and Jnk−/− cells were treated without and with TNF-α (10 ng/ml). Nuclear extracts were prepared at different times and used to examine DNA binding activity with an electrophoretic mobility shift assay using probes containing a consensus AP-1 site (TRE) and a nonconsensus site (Jun2 TRE). The DNA binding activity was detected by autoradiography (upper panel) and was quantitated by PhosphorImager analysis (lower panel). (B) WT and Jnk−/− cells were treated without and with TNF-α (10 ng/ml). The cells were harvested after different times. Cell lysates were examined by immunoblot analysis by probing with antibodies to ATF2, c-Jun, and JunD. The phosphorylation of these transcription factors on sites that are phosphorylated by JNK was examined using phospho-specific antibodies. Blots were probed with an antibody to α-tubulin to monitor protein loading on each lane of the gel.
JNK is required for TNF-stimulated phosphorylation of AP-1 transcription factors.
It is established that TNF regulates AP-1 activity, in part, by increasing the expression of members of the Jun and Fos families. However, AP-1 activity can also be regulated by posttranslational modification of these transcription factors, including phosphorylation by MAPK. Previous studies indicate that the NH2-terminal phosphorylation of c-Jun, JunD, and ATF2 contributes to AP-1 activation (8). We therefore examined the phosphorylation of endogenous AP-1 proteins in WT and Jnk−/− fibroblasts.
TNF caused a marked increase in the NH2-terminal phosphorylation of c-Jun in WT fibroblasts, but no phosphorylation of c-Jun was detected in Jnk−/− cells (Fig. 4B). These data are consistent with previous studies that indicate a role for JNK in the NH2-terminal phosphorylation of c-Jun (8). In contrast to the essential requirement of JNK for the TNF-stimulated phosphorylation of c-Jun, the NH2-terminal phosphorylation of JunD was only partially suppressed in Jnk−/− fibroblasts (Fig. 4B). The presence of TNF-stimulated JunD phosphorylation in Jnk−/− cells is particularly striking because these cells expressed lower amounts of JunD protein than WT fibroblasts. Together, these data establish that JunD phosphorylation is partially controlled by a JNK-independent mechanism (e.g., ERK or p38 MAPK) in TNF-treated cells. However, JNK is essential for TNF-stimulated NH2-terminal phosphorylation of c-Jun.
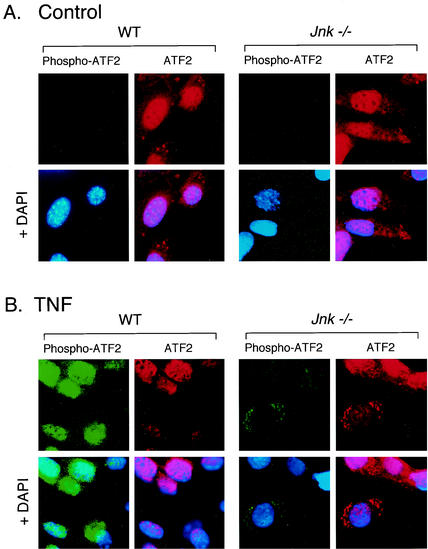
The NH2-terminal phosphorylation of the ATF2 transcription factor has been reported to be regulated by JNK (14, 22), and also by other MAPK, including ERK (23) and p38 MAPK (27). The role of JNK in the NH2-terminal phosphorylation of ATF2 is therefore unclear. TNF caused a marked increase in ATF2 phosphorylation in WT cells, but only a very low level of ATF2 phosphorylation was detected in Jnk−/− cells. These data indicate that while several TNF-stimulated MAPK are capable of phosphorylating ATF2 in vitro, the major TNF-stimulated activity in vivo is JNK. This conclusion was confirmed by immunofluorescence analysis (Fig. 5). Studies of WT cells demonstrated that ATF2 was detected in the cytoplasm but was predominantly localized to the nucleus. A similar distribution of ATF2 was observed in Jnk−/− cells. Treatment of WT cells with TNF caused the accumulation of ATF2 in the nucleus and the disappearance of this transcription factor from the cytoplasm. This change in subcellular localization was associated with a marked increase in the NH2-terminal phosphorylation of ATF2 (Fig. 5). In contrast, studies of TNF-treated Jnk−/− cells demonstrated markedly different ATF2 phosphorylation and subcellular localization compared with TNF-treated WT cells. First, cytoplasmic ATF2 was detected in TNF-treated Jnk−/− cells, although the majority of the ATF2 was localized to the nucleus. Second, very little NH2-terminally phosphorylated ATF2 was detected in TNF-treated Jnk−/− cells. Interestingly, this phosphorylated ATF2 localized to the perinuclear region in the cytoplasm rather than in the nucleus (Fig. 5). Studies using the small-molecule inhibitor PD 169316 indicated that this residual phosphorylation of ATF2 in Jnk−/− cells was accounted for by p38 MAPK (data not shown). These data indicate that JNK is essential for TNF-stimulated NH2-terminal phosphorylation of ATF2 in the nucleus.
FIG. 5.
Effect of JNK deficiency on phosphorylation and subcellular location of ATF2. ATF2 and phospho-ATF2 were examined by immunofluorescence microscopy. Fibroblasts were treated without (A) and with (B) TNF-α (10 ng/ml for 15 min), fixed, and stained with antibodies to ATF2 (Texas red [red]) and phospho-ATF2 (FITC [green]). DNA was stained with DAPI (blue). The cells were imaged by conventional fluorescence microscopy.
Expression of JNK in Jnk−/− cells corrects the defect in TNF-stimulated NH2-terminal phosphorylation of ATF2 and c-Jun.
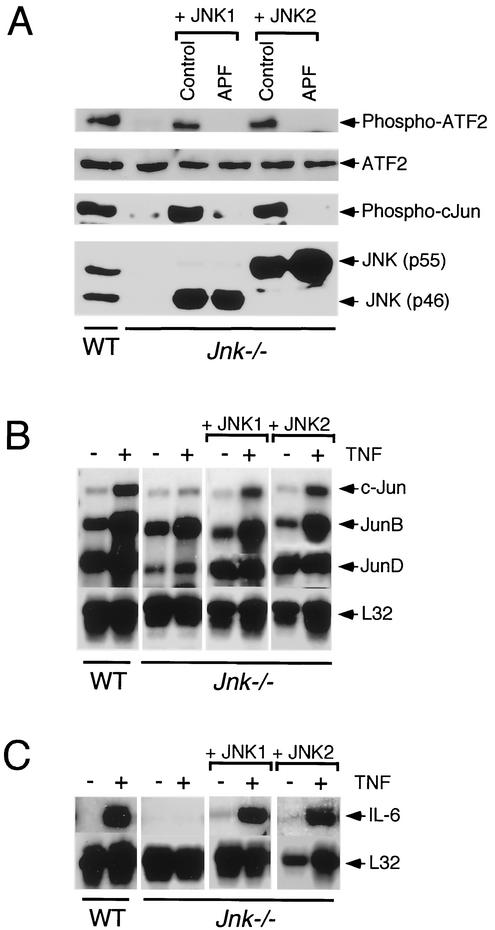
To test whether the defect in TNF-stimulated NH2-terminal phosphorylation of ATF2 and c-Jun in Jnk−/− cells was caused by JNK deficiency, we performed complementation analysis by expression of JNK1 and JNK2 in the Jnk−/− cells. Control experiments were performed using kinase-negative JNK1 and JNK2 (replacement of the activating sites of Thr and Tyr phosphorylation with Ala and Phe, respectively). These studies were performed using cells derived from pools of clones that were obtained following transduction using retroviral vectors. Immunoblot analysis of WT cells demonstrated the expression of p46 and p55 forms of JNK1 and JNK2 (Fig. 6A). No JNK was detected in Jnk−/− cells. Complemented Jnk−/− cells expressing WT or kinase-negative p46 JNK1 or p55 JNK2 were obtained. Immunoblot analysis demonstrated that the defect in endogenous ATF2 and c-Jun NH2-terminal phosphorylation was corrected by expression of WT JNK1 or JNK2, but not by expression of kinase-negative JNK1 or JNK2 (Fig. 6A). These data demonstrate that the defective NH2-terminal phosphorylation of ATF2 and c-Jun in Jnk−/− cells was caused by the absence of JNK in these cells.
FIG. 6.
Phosphorylation of ATF2 and c-Jun is restored in Jnk−/− fibroblasts following ectopic expression of JNK1 or JNK2. (A) Complementation analysis of Jnk−/− cells by ectopic expression of JNK1 or JNK2. Jnk−/− cells were transduced with retroviruses that expressed WT JNK1 or JNK2. Studies were also performed using retroviruses expressing kinase-inactive JNK1 or JNK2 (APF, replacement of the tripeptide dual phosphorylation motif Thr-Pro-Tyr with Ala-Pro-Phe). The WT cells, Jnk−/− cells, and the four complemented Jnk−/− cell populations were treated with TNF (15 min) and harvested to prepare lysates that were probed with antibodies to JNK, ATF2, phospho-ATF2, and phospho-c-Jun. (B and C) WT cells, Jnk−/− cells, and Jnk−/− cells complemented with JNK1 or JNK2 were treated without (−) and with (+) TNF-α (10 ng/ml for 60 min). Total RNA (5 μg) was isolated and examined using an RNase protection assay to measure the expression of c-Jun, JunB, and JunD (B) and IL-6 mRNA (C). Control assays were performed to measure the expression of the ribosomal protein L32.
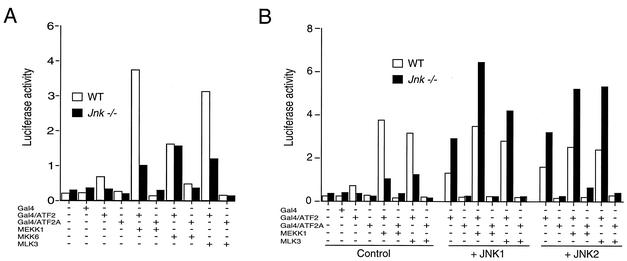
The observation that TNF stimulates JNK, ERK, and p38 MAPK (Fig. 1) and that Jnk−/− cells have severe defects in ATF2 NH2-terminal phosphorylation (Fig. 4 to 6) is intriguing. This finding indicates that while other MAPK can phosphorylate ATF2, the physiologically relevant kinase in TNF-treated cells is JNK. This conclusion led us to reexamine the role of p38 MAPK and JNK in the regulation of ATF2. Studies of ATF2 transcription activity were performed with a GAL4 reporter assay (14) using WT and Jnk−/− fibroblasts (Fig. 7). Cotransfection assays using activators of the JNK signaling pathway (MEK kinase 1 or mixed-lineage kinase 3) strongly increased ATF2-dependent reporter gene expression in WT cells, and this transcription activity was suppressed in Jnk−/− cells (Fig. 7A). Complementation assays demonstrated that the defect in ATF2 transcription activity detected in Jnk−/− cells could be corrected by ectopic expression of JNK1 or JNK2 (Fig. 7B). Together, these data are consistent with a role for the JNK signaling pathway in the activation of ATF2. Studies performed using a strong activator of the p38 MAPK pathway (MKK6) also demonstrated increased ATF2-dependent transcription activity, but this activity was not suppressed in the Jnk−/− cells (Fig. 7A). These data confirm previous conclusions that the transcription activity of ATF2 can be regulated by a JNK-independent mechanism, including the p38 MAPK signaling pathway. The selective requirement of JNK for TNF action therefore appears to reflect physiologically relevant signal transduction specificity.
FIG. 7.
ATF2 transcription activity in WT and Jnk−/− fibroblasts. ATF2 activity was examined in a luciferase reporter assay using the activation domain of ATF2 fused to the GAL4 DNA binding domain (14). The effect of replacement of the sites of activating phosphorylation (Thr-69 and Thr-71) with Ala was investigated (GAL4/ATF2A). The relative firefly luciferase activity was measured and was normalized to the amount of activity detected for a cotransfected control Renilla luciferase reporter plasmid. (A) The effect of coexpression of MEK kinase 1 (MEKK1), MKK6 (Ser208Glu/Thr-211Glu), and mixed-lineage kinase 3 (MLK3) was examined. (B) Complementation analysis of Jnk−/− cells by ectopic expression of JNK1 or JNK2. The cells were cotransfected with an expression vector for JNK1 or JNK2. Control studies were performed by cotransfecting the empty expression plasmid.
JNK is required for TNF-stimulated cytokine gene expression.
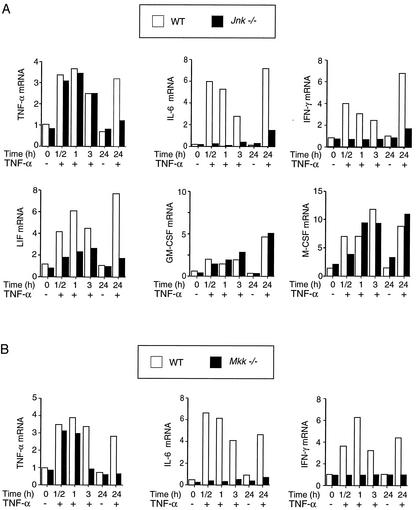
A hallmark of the cellular response to TNF is an increase in cytokine expression. To examine the role of JNK in this process, we measured the amount of cytokine mRNA present within WT and Jnk−/− cells using an RNase protection assay (Fig. 8A). TNF was found to increase the expression of TNF, interleukin-6 (IL-6), gamma interferon (IFN-γ), leukemia inhibitory factor (LIF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and macrophage colony-stimulating factor (M-CSF) in WT cells. JNK deficiency caused no marked changes in GM-CSF and M-CSF expression but did cause some decrease in TNF-stimulated LIF expression. In contrast, the TNF-stimulated expression of IL-6 and IFN-γ was severely reduced in JNK-deficient cells. These data suggest that JNK is selectively required for TNF-stimulated expression of the inflammatory cytokines IL-6 and IFN-γ. To confirm this conclusion, we examined TNF-stimulated IL-6 and IFN-γ expression in Mkk−/− cells which lack a functional JNK signal transduction pathway (34). Like JNK-deficient cells, the MKK-deficient cells exhibited marked defects in TNF-stimulated IL-6 and IFN-γ expression (Fig. 8B). Together, these data indicate that the JNK signaling pathway is essential for TNF-stimulated expression of IL-6 and IFN-γ.
FIG. 8.
Role of JNK in TNF-stimulated cytokine expression. (A) WT and Jnk−/− fibroblasts were starved for 3 h and then treated with TNF-α (10 ng/ml). Total RNA was purified, and 5 μg was used for RNase protection assays to measure the amount of TNF-α, IL-6, IFN-γ, LIF, GM-CSF, M-CSF, and ribosomal protein L32 mRNA. The ratio of cytokine mRNA/L32 mRNA was calculated and is presented as the relative amount of cytokine expression. The data shown are representative of three independent experiments. (B) RNase protection assays were used to measure the amount of TNF-α, IL-6, and IFN-γ mRNA expressed by WT and Mkk−/− cells.
A role for the JNK signaling pathway in the TNF-stimulated expression of TNF-α was also identified, but it appears that JNK was only required for sustained signaling (Fig. 8). Neither JNK deficiency (Fig. 8A) nor MKK deficiency (Fig. 8B) prevented the immediate increase in TNF-α expression caused by treatment of cells with TNF. However, sustained increase in TNF-α expression (24 h) caused by TNF was observed in WT cells, but not in the JNK-deficient or MKK-deficient cells (Fig. 8). This selective requirement of the JNK signaling pathway for sustained TNF-α expression (Fig. 8) differs from observations obtained in studies of IL-6 and IFN-γ expression where JNK signaling was required for both the sustained response and the immediate response of cells to TNF (Fig. 8).
To test whether the observed defect in gene expression in Jnk−/− cells was caused by JNK deficiency, we performed complementation analysis by expression of JNK1 and JNK2 in Jnk−/− cells. In control studies, the expression of members of the Jun family was examined (Fig. 6B). JNK deficiency caused no change in JunB expression but caused decreased expression of c-Jun and JunD. These defects in c-Jun and JunD expression were restored by ectopic expression of JNK1 or JNK2 in the Jnk−/− cells (Fig. 6B). Similarly, the loss of TNF-stimulated IL-6 expression in Jnk−/− cells was restored by ectopic expression of JNK1 or JNK2 (Fig. 6C). Together, these data indicate that JNK is an important regulator of AP-1-dependent gene expression in cells exposed to TNF.
DISCUSSION
TNF-α is a critical mediator of the stress response that influences immune cell function, proliferation, differentiation, and apoptosis (6). TNF-α can cause apoptosis by activating caspases. However, TNF-α can also promote inflammation by suppressing apoptosis by a mechanism that involves both the NF-κB and AP-1 groups of transcription factors (2). Since dysregulation of TNF-α expression or signaling is implicated in the pathology of many diseases, including rheumatoid arthritis, Crohn's disease, and several neuropathologies (e.g., stroke, multiple sclerosis, and Alzheimer's disease), an understanding of the mechanism of proinflammatory signaling by TNF-α is essential for the design of novel therapeutic strategies.
Substantial evidence implicates a role for the AP-1 group of transcription factors in many of the nonapoptotic cellular responses to TNF-α (2). The mechanism of AP-1 activation involves increased expression (5) and phosphorylation (20, 32, 39) of AP-1 proteins. The mechanism by which TNF regulates these functions of AP-1 proteins is unclear. However, an important role for the JNK signaling pathway in AP-1 activation by TNF has been proposed (8). The studies we present were designed to test this hypothesis. Our approach was to compare the effects of TNF on WT fibroblasts and compound knockout fibroblasts that lack expression of both Jnk1 and Jnk2. The JNK-deficient fibroblasts expressed similar amounts of TNF-R1 and demonstrated no defects in the TNF-stimulated activation of NF-κB. In addition, TNF caused an efficient apoptotic response in both the WT and the JNK-deficient cells (in the presence of inhibitors of protein or mRNA synthesis). These data demonstrate that JNK-deficient fibroblasts respond to treatment with TNF. However, these cells exhibited marked defects in TNF-stimulated JNK activation and AP-1 transcription factor expression, phosphorylation, and function. These data confirm the hypothesis that JNK is essential for the activation of AP-1 in cells treated with TNF-α.
Role of JNK in AP-1 transcription factor expression.
WT and JNK-deficient fibroblasts exhibited similar basal and TNF-stimulated expression of JunB (Fig. 3). The absence of a requirement of JNK for JunB expression is consistent with the results of previous studies that indicate a critical role for ERK-regulated Ets transcription factors in the expression of JunB (7). Similarly, the expression of the AP-1 related transcription factor ATF2 was not altered in JNK-deficient cells. However, the JNK-deficient cells exhibited marked defects in the TNF-stimulated expression of c-Jun, JunD, c-Fos, Fra1, and Fra2 (Fig. 3). These defects in AP-1 expression were confirmed by measurement of AP-1 DNA binding activity in TNF-treated cells (Fig. 4A). These data indicate that JNK is required for the normal regulation of AP-1 transcription factor expression by TNF.
It is interesting that JNK is not required for TNF-stimulated expression of JunB, an AP-1 protein that has previously been implicated as an antagonist of c-Jun transcription activity (9). The expression of the c-Jun antagonist JunB and the selective loss of expression of other AP-1 proteins indicate that TNF-treated JNK-deficient cells have profound defects in AP-1 function.
Role of JNK in the NH2-terminal phosphorylation of AP-1 transcription factors.
Treatment of WT fibroblasts with TNF-α caused a marked increase in the NH2-terminal phosphorylation of c-Jun, JunD, and ATF2 (Fig. 4B). Interestingly, the effect of JNK deficiency on this phosphorylation was different for each transcription factor. Phosphorylated c-Jun was not detected in JNK-deficient cells, indicating that JNK is the physiologically relevant NH2-terminal c-Jun kinase in TNF-treated cells. This finding contrasts with the conclusions drawn in previous studies that indicate a role for multiple MAPK in the NH2-terminal phosphorylation of c-Jun, including members of both the JNK and ERK groups of MAPK (21, 26). The selective role of JNK in the NH2-terminal phosphorylation of c-Jun in TNF-treated cells provides evidence for the specificity of protein phosphorylation by MAPK in vivo.
JunD phosphorylation was only partially affected in the absence of JNK. Thus, JNK is not a major contributor to the NH2-terminal phosphorylation of JunD in TNF-treated cells. This phosphorylation is therefore largely mediated by other protein kinases. Candidate protein kinases that may phosphorylate JunD include the ERK group of MAPK. The absence of an important role for JNK in the NH2-terminal phosphorylation of JunD is consistent with the results of previous studies that demonstrate a requirement for a docking site in c-Jun for NH2-terminal phosphorylation by JNK (10, 17). This docking site is absent in JunD and in vitro studies demonstrate that JunD is a poor JNK substrate, although the sites of c-Jun phosphorylation are conserved in JunD (13, 18).
Studies of ATF2 demonstrated marked defects in the NH2-terminal phosphorylation of this transcription factor in TNF-treated JNK-deficient cells. Thus, JNK is essential for TNF-stimulated phosphorylation of ATF2. This conclusion was surprising because we have previously reported that ATF2 is phosphorylated and activated by both JNK and p38 MAPK (27, 28). Furthermore, both JNK and p38 MAPK are activated by TNF in these fibroblasts (Fig. 1). Indeed, control studies demonstrated that activated p38 MAPK can stimulate ATF2 in both WT and JNK-deficient cells (Fig. 7). Together, these data suggest that while p38 MAPK can phosphorylate ATF2, JNK is the physiologically relevant TNF-stimulated ATF2 NH2-terminal kinase. The mechanism that accounts for the selective requirement of JNK for ATF2 phosphorylation in TNF-treated cells is unclear. However, there are several mechanisms that could account for this observation. Among these possibilities, the simplest hypothesis appears to be that p38 MAPK only leads to ATF2 phosphorylation when p38 MAPK is markedly activated and that lower levels of p38 MAPK activation are insufficient for ATF2 phosphorylation. Such a mechanism would account for the observation that high levels of sustained p38 MAPK activation (e.g., in transfection assays using MKK6) lead to ATF2 phosphorylation but treatment of cells with TNF (which causes only a transient low level of p38 MAPK activation) does not lead to marked ATF2 phosphorylation. This mechanism of high-activity-dependent phosphorylation of a transcription factor by p38 MAPK has been previously reported for phosphorylation of the Ets protein Elk-1 by p38 MAPK (42). This hypothesis predicts that TNF should cause a low level of p38 MAPK-dependent ATF2 phosphorylation in JNK-deficient cells. Consistent with this prediction, the low level of residual ATF2 phosphorylation caused by TNF in JNK-deficient cells was blocked by a small molecule inhibitor of p38 MAPK. A rigorous test of the physiological role of p38 MAPK in ATF2 phosphorylation remains to be obtained. Nevertheless, the data we present firmly establish JNK as a physiologically relevant ATF2 kinase in TNF-treated cells.
JNK is essential for TNF-induced expression of inflammatory cytokines.
An important aspect of the response to TNF-α is the production of cytokines that are secreted from the responding cell. These cytokines act by signaling to other cell types and serve to amplify the initial response. Previous studies have strongly implicated roles for the NF-κB and AP-1 groups of transcription factors (2) and MAPK pathways (45). However, the specific role of JNK in the cytokine response of cells to TNF-α was unclear. Comparison of WT and JNK-deficient cells demonstrated that JNK was essential for TNF-stimulated expression of the inflammatory cytokines IL-6 and IFN-γ.
The IL-6 promoter contains several cis-acting elements, including NF-κB, C/EBPβ, AP-1, and CRE (38). Activation of NF-κB is important, but is not sufficient, for IL-6 gene expression (24, 31). A role for p38 MAPK in IL-6 expression has also been reported (4, 41). The results of this study indicate that JNK is essential for TNF-stimulated IL-6 expression. This role of JNK may include activation of factors that bind to the AP-1 and CRE sites in the IL-6 promoter. It is possible that ATF2 is a major contributor to this response since ATF2 binding to the CRE site is critical for IL-6 expression (12) and IL-6 expression is severely reduced in ATF2-deficient cells (29). An important role for ATF2 is also consistent with the observation that JNK-deficient cells exhibit marked defects in TNF-stimulated IFN-γ expression, since the IFN-γ promoter contains two cis-acting elements (CRE and AP-1) that bind ATF2 (25).
JNK was also found to influence TNF-stimulated TNF-α expression (Fig. 8). JNK did not affect the immediate increase in TNF-α expression caused by TNF, but JNK was required for the sustained TNF-stimulated expression of TNF-α. The mechanism that accounts for this selective requirement of JNK is unclear. The TNF-α promoter is complex and has been shown to contain binding sites for NFAT and an enhanceosome with CBP/p300 in a complex with Ets/Elk-1, Egr-1, Sp1, and ATF2/c-Jun (36). It is likely that the requirement of JNK for sustained TNF-stimulated TNF-α expression is mediated, in part, by the phosphorylation of ATF2 and AP-1 transcription factors that bind to the promoter.
Conclusions.
This study establishes that JNK is an essential component of the response of cells to TNF. JNK is required for the TNF-regulated expression and phosphorylation of AP-1 transcription factors and for the phosphorylation of the AP-1-related transcription factor ATF2. Loss of JNK causes marked defects in the effect of TNF to induce proinflammatory cytokine expression. Consequently, JNK inhibition represents a potential therapy for the treatment of TNF-mediated inflammatory diseases (3, 16).
Acknowledgments
We thank T. Barrett and S. Stone for technical assistance and K. Gemme for administrative assistance.
This study was supported, in part, by grant CA65861 from the National Cancer Institute. R. J. Davis and R. A. Flavell are investigators of the Howard Hughes Medical Institute.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baud, V., and M. Karin. 2001. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11:372-377. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyaert, R., A. Cuenda, W. Vanden Berghe, S. Plaisance, J. C. Lee, G. Haegeman, P. Cohen, and W. Fiers. 1996. The p38/RK mitogen-activated protein kinase pathway regulates interleukin-6 synthesis response to tumor necrosis factor. EMBO J. 15:1914-1923. [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner, D. A., M. O'Hara, P. Angel, M. Chojkier, and M. Karin. 1989. Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature 337:661-663. [DOI] [PubMed] [Google Scholar]
- 6.Chen, G., and D. V. Goeddel. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634-1635. [DOI] [PubMed] [Google Scholar]
- 7.Coffer, P., M. de Jonge, A. Mettouchi, B. Binetruy, J. Ghysdael, and W. Kruijer. 1994. junB promoter regulation: Ras mediated transactivation by c-Ets-1 and c-Ets-2. Oncogene 9:911-921. [PubMed] [Google Scholar]
- 8.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 9.Deng, T., and M. Karin. 1993. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 7:479-490. [DOI] [PubMed] [Google Scholar]
- 10.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 11.Devin, A., A. Cook, Y. Lin, Y. Rodriguez, M. Kelliher, and Z. Liu. 2000. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity 12:419-429. [DOI] [PubMed] [Google Scholar]
- 12.Franchimont, N., D. Durant, S. Rydziel, and E. Canalis. 1999. Platelet-derived growth factor induces interleukin-6 transcription in osteoblasts through the activator protein-1 complex and activating transcription factor-2. J. Biol. Chem. 274:6783-6789. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, S., T. Barrett, A. J. Whitmarsh, J. Cavanagh, H. K. Sluss, B. Derijard, and R. J. Davis. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760-2770. [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, S., D. Campbell, B. Derijard, and R. J. Davis. 1995. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267:389-393. [DOI] [PubMed] [Google Scholar]
- 15.Han, J., Y. Jiang, Z. Li, V. V. Kravchenko, and R. J. Ulevitch. 1997. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386:296-299. [DOI] [PubMed] [Google Scholar]
- 16.Han, Z., D. L. Boyle, L. Chang, B. Bennett, M. Karin, L. Yang, A. M. Manning, and G. S. Firestein. 2001. c-Jun N-terminal kinase is required for metalloproteinase expression and joint destruction in inflammatory arthritis. J. Clin. Investig. 108:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hibi, M., A. Lin, T. Smeal, A. Minden, and M. Karin. 1993. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7:2135-2148. [DOI] [PubMed] [Google Scholar]
- 18.Kallunki, T., T. Deng, M. Hibi, and M. Karin. 1996. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87:929-939. [DOI] [PubMed] [Google Scholar]
- 19.Kelliher, M. A., S. Grimm, Y. Ishida, F. Kuo, B. Z. Stanger, and P. Leder. 1998. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity 8:297-303. [DOI] [PubMed] [Google Scholar]
- 20.Kyriakis, J. M., P. Banerjee, E. Nikolakaki, T. Dai, E. A. Rubie, M. F. Ahmad, J. Avruch, and J. R. Woodgett. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature 369:156-160. [DOI] [PubMed] [Google Scholar]
- 21.Leppa, S., R. Saffrich, W. Ansorge, and D. Bohmann. 1998. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 17:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livingstone, C., G. Patel, and N. Jones. 1995. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 14:1785-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouwens, D. M., N. D. De Ruiter, G. C. Van Der Zon, A. P. Carter, J. Schouten, C. Van Der Burgt, K. Kooistra, J. L. Bos, J. A. Maassen, and H. Van Dam. 2002. Growth factors can activate ATF2 via a two-step mechanism: phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 21:3782-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patestos, N. P., G. Haegeman, V. Vandevoorde, and W. Fiers. 1993. Activation of the nuclear factor kappa B is not sufficient for regulation of tumor necrosis factor-induced interleukin-6 gene expression. Biochimie 75:1007-1018. [DOI] [PubMed] [Google Scholar]
- 25.Penix, L. A., M. T. Sweetser, W. M. Weaver, J. P. Hoeffler, T. K. Kerppola, and C. B. Wilson. 1996. The proximal regulatory element of the interferon-gamma promoter mediates selective expression in T cells. J. Biol. Chem. 271:31964-31972. [DOI] [PubMed] [Google Scholar]
- 26.Pulverer, B. J., J. M. Kyriakis, J. Avruch, E. Nikolakaki, and J. R. Woodgett. 1991. Phosphorylation of c-jun mediated by MAP kinases. Nature 353:670-674. [DOI] [PubMed] [Google Scholar]
- 27.Raingeaud, J., S. Gupta, J. S. Rogers, M. Dickens, J. Han, R. J. Ulevitch, and R. J. Davis. 1995. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 270:7420-7426. [DOI] [PubMed] [Google Scholar]
- 28.Raingeaud, J., A. J. Whitmarsh, T. Barrett, B. Derijard, and R. J. Davis. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimold, A. M., J. Kim, R. Finberg, and L. H. Glimcher. 2001. Decreased immediate inflammatory gene induction in activating transcription factor-2 mutant mice. Int. Immunol. 13:241-248. [DOI] [PubMed] [Google Scholar]
- 30.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu, H., K. Mitomo, T. Watanabe, S. Okamoto, and K. Yamamoto. 1990. Involvement of a NF-κB-like transcription factor in the activation of the interleukin-6 gene by inflammatory lymphokines. Mol. Cell. Biol. 10:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sluss, H. K., T. Barrett, B. Derijard, and R. J. Davis. 1994. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol. Cell. Biol. 14:8376-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobiume, K., A. Matsuzawa, T. Takahashi, H. Nishitoh, K. Morita, K. Takeda, O. Minowa, K. Miyazono, T. Noda, and H. Ichijo. 2001. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tournier, C., C. Dong, T. K. Turner, S. N. Jones, R. A. Flavell, and R. J. Davis. 2001. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 15:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 36.Tsai, E. Y., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanden Berghe, W., L. Vermeulen, G. De Wilde, K. De Bosscher, E. Boone, and G. Haegeman. 2000. Signal transduction by tumor necrosis factor and gene regulation of the inflammatory cytokine interleukin-6. Biochem. Pharmacol. 60:1185-1195. [DOI] [PubMed] [Google Scholar]
- 39.Westwick, J. K., C. Weitzel, A. Minden, M. Karin, and D. A. Brenner. 1994. Tumor necrosis factor alpha stimulates AP-1 activity through prolonged activation of the c-Jun kinase. J. Biol. Chem. 269:26396-26401. [PubMed] [Google Scholar]
- 40.Whitmarsh, A. J., J. Cavanagh, C. Tournier, J. Yasuda, and R. J. Davis. 1998. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science 281:1671-1674. [DOI] [PubMed] [Google Scholar]
- 41.Wysk, M., D. D. Yang, H. T. Lu, R. A. Flavell, and R. J. Davis. 1999. Requirement of mitogen-activated protein kinase kinase 3 (MKK3) for tumor necrosis factor-induced cytokine expression. Proc. Natl. Acad. Sci. USA 96:3763-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, S. H., A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 17:1740-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh, W. C., J. L. Pompa, M. E. McCurrach, H. B. Shu, A. J. Elia, A. Shahinian, M. Ng, A. Wakeham, W. Khoo, K. Mitchell, W. S. El-Deiry, S. W. Lowe, D. V. Goeddel, and T. W. Mak. 1998. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 279:1954-1958. [DOI] [PubMed] [Google Scholar]
- 44.Yeh, W. C., A. Shahinian, D. Speiser, J. Kraunus, F. Billia, A. Wakeham, J. L. de la Pompa, D. Ferrick, B. Hum, N. Iscove, P. Ohashi, M. Rothe, D. V. Goeddel, and T. W. Mak. 1997. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity 7:715-725. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, W., J. S. Downey, J. Gu, F. Di Padova, H. Gram, and J. Han. 2000. Regulation of TNF expression by multiple mitogen-activated protein kinase pathways. J. Immunol. 164:6349-6358. [DOI] [PubMed] [Google Scholar]
- 46.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]