Abstract
Different isoforms of a protein complex termed the apoptosis- and splicing-associated protein (ASAP) were isolated from HeLa cell extract. ASAP complexes are composed of the polypeptides SAP18 and RNPS1 and different isoforms of the Acinus protein. While Acinus had previously been implicated in apoptosis and was recently identified as a component of the spliceosome, RNPS1 has been described as a general activator of RNA processing. Addition of ASAP isoforms to in vitro splicing reactions inhibits RNA processing mediated by ASF/SF2, by SC35, or by RNPS1. Additionally, microinjection of ASAP complexes into mammalian cells resulted in acceleration of cell death. Importantly, after induction of apoptosis the ASAP complex disassembles. Taken together, our results suggest an important role for the ASAP complexes in linking RNA processing and apoptosis.
Gene expression in eukaryotic cells is a multistep process that begins in the nucleus with the generation of an mRNA precursor by gene transcription. In subsequent steps the mRNA transcript is subject to processing and export into the cytoplasm, where it undergoes quality control, translation, and finally decay. All of these metabolic steps are performed by large multisubunit machineries, appear to be mechanistically linked with each other, and are subject to regulation (12, 21, 27).
During mRNA processing, the removal of introns is facilitated by the spliceosome, which is composed of various small nuclear ribonucleoprotein particles (snRNPs) and several non-snRNP protein components (15, 29, 36), including members of the serine- and arginine-rich (SR) protein family (10, 23). In mammalian cells, SR proteins and other splicing factors exist in nuclear regions called splicing factor compartments, nuclear speckles, or interchromatin granules and have proposed functions in several steps of the gene expression process (26, 31). For example, a spliceosome-associated RNA-binding protein with a serine-rich domain, RNPS1, is thought to be involved in the regulation of splicing, mRNA export, and nonsense-mediated mRNA decay (16, 17, 19, 20, 25). Regulation of RNA processing allows alternative splicing of transcripts, a process that enables the cell to generate multiple distinct transcript species from a common precursor. Alternative splicing adds an additional layer of control to the gene expression process, since it leads to the production of discrete protein isoforms that can have distinct functions in cellular events (18, 22).
One important cellular event is the process of programmed cell death or apoptosis (32). Interestingly, numerous apoptotic genes, including death receptors and intracellular components of the death machinery, are subject to alternative splicing (3, 13). Apoptosis is distinguished from other forms of cell death, such as necrosis, on the basis of morphological criteria, which include membrane blebbing, fragmentation of nuclear DNA, and chromatin condensation (11). Most of the apoptotic morphological changes are caused by a set of cysteinyl aspartate-specific proteinases (caspases), which are specifically activated in apoptotic cells and cleave selected target proteins (6, 14). Among these target proteins is a nuclear protein named Acinus (for apoptotic chromatin condensation inducer in the nucleus) (30). Acinus is expressed in different isoforms, termed Acinus-L, Acinus-S, and Acinus-S′, with molecular masses of ca. 220, 98, and 94 kDa, respectively (Fig. 1a). Acinus is cleaved during apoptosis by caspase 3 and by an unidentified protease to produce a 23-kDa isoform (p23) that has been implicated in mediating apoptotic chromatin condensation prior to DNA fragmentation (30).
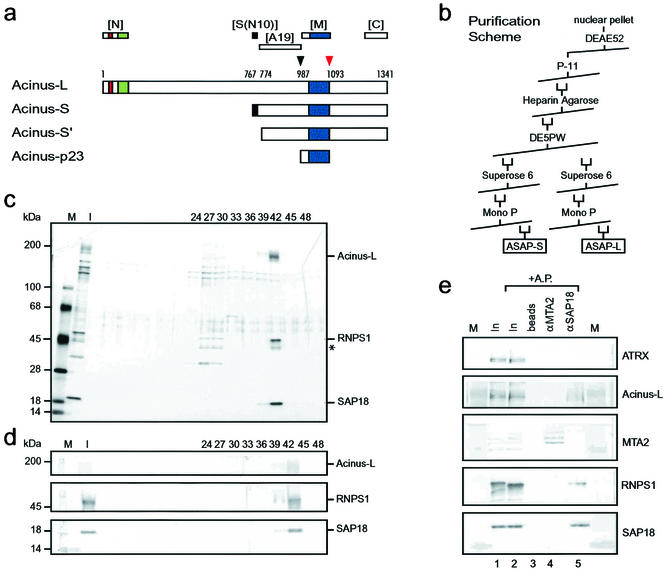
FIG. 1.
Purification of a complex consisting of SAP18, RNPS1, and Acinus-L. (a) Schematic representation of Acinus isoforms. Described protein domains as well as protease cleavage sites and their corresponding amino acids (1, 30) are indicated as follows: red box, P-loop for nucleotide binding; green box, SAP domain; dark blue box, region similar to RNA recognition motif of Drosophila Sxl; black box, region specific for Acinus-S; black arrowhead, cleavage site for unknown protease; red arrowhead, cleavage site for caspase 3. Regions recognized by polyclonal antibodies against Acinus ([N], [S(N10)], [A19], [M], and [C]) (see also Materials and Methods) are shown above the representation of Acinus-L. (b) Purification scheme used to isolate the ASAP complexes from HeLa cell nuclear pellets. (c) Silver stain of an SDS-4 to 15% (wt/vol) polyacrylamide gradient gel containing aliquots of fractions (input [I] and the indicated column fractions) derived from the MonoP column. High-molecular-mass standards [M] are indicated at the left, and components of the ASAP complex are indicated at the right side of the panel. A putative alternatively phosphorylated or partially degraded form of RNPS1 is marked with an asterisk. (d) Western blot analysis of the samples used in panel c. The antibodies used in the Western blot are indicated at the right, and high-molecular-mass standards [M] are indicated at the left side of the panel. (e) Verification of the complex composition by coimmunoprecipitation. Antibodies against SAP18 (lane 5) but not MTA-2 (lane 4) coimmunoprecipitate RNPS1 and Acinus-L from a protein fraction derived from nuclear pellet [In]. Samples in lanes 2 to 5 were treated with alkaline phosphatase [AP] after immunoprecipitation in an attempt to increase the signal for Acinus-L by focusing putative differently phosphorylated species. Ten percent of the input is loaded in lanes 1 and 2. Immunoprecipitated proteins were separated on an SDS-4 to 15% (wt/vol) polyacrylamide gradient gel and analyzed by Western blotting with the antibodies indicated at the right. Lanes labeled [M] contain high-molecular-mass protein standards.
All three nonapoptotic isoforms of Acinus contain a region whose function is unknown but which is similar to the RNA-binding domain of the Drosophila splicing regulator sex-lethal (Sxl), suggesting another role for Acinus in RNA metabolism (30). In fact, recent publications have shown that Acinus is a component of functional spliceosomes (29, 36).
Here we describe the purification and characterization of a novel protein comprised of the proteins SAP18, RNPS1, and distinct isoforms of Acinus, which we term the apoptosis- and splicing-associated protein (ASAP) complex. Addition of the complex to in vitro splicing assays inhibited RNA processing, and microinjection of ASAP into mammalian cells accelerated the progress of cell death after induction of apoptosis.
MATERIALS AND METHODS
Purification of the ASAP complex.
Solubilized HeLa cell nuclear pellets (9 g) in buffer D (50 mM Tris [pH 7.9], 0.1 mM EDTA, 10% glycerol, 1 mM dithiothreitol [DTT], and protease inhibitors) were adjusted to a 0.1 M concentration of (NH4)2SO4 and loaded onto a 900-ml DEAE-52 column equilibrated in buffer D-0.1 M (NH4)2SO4. The ASAP-containing flow-through of this column (4 g) was loaded onto a 400-ml P-11 column equilibrated in buffer D-0.1 M (NH4)2SO4. The column was washed with 4 column volumes of buffer D-0.1 M (NH4)2SO4 and subsequently eluted with a 10-column-volume linear gradient of (NH4)2SO4 from a concentration of 0.1 to 0.7 M in buffer D. ASAP-containing fractions eluting around 0.125 M (NH4)2SO4 (200 mg) were pooled, adjusted to a 0.1 M concentration of (NH4)2SO4, and loaded onto a 25-ml heparin agarose column equilibrated in buffer D-0.1 M (NH4)2SO4. The column was washed with 10 column volumes of buffer D-0.1 M (NH4)2SO4 and then eluted with a linear gradient of (NH4)2SO4 from 0.1 to 1 M in buffer D over 10 column volumes. ASAP-containing fractions eluting around 0.225 M (NH4)2SO4 (45 mg) were pooled, adjusted to 0.05 M (NH4)2SO4, and loaded onto a high-performance liquid chromatography DEAE-5PW column equilibrated in buffer D-0.05 M (NH4)2SO4. After washing with ca. 5 column volumes of buffer D-0.05 M (NH4)2SO4, the column was eluted with a 15-column-volume linear gradient of (NH4)2SO4 from 0.05 to 0.7 M in buffer D. ASAP-L and ASAP-S were further purified employing the following protocol. Fractions containing either ASAP-L (1.2 mg) or ASAP-S (1.8 mg) were pooled and concentrated with Centricon concentrators (Millipore). The concentrated samples were each subjected to gel filtration on a Pharmacia Superose 6 HR 10/30 column that was previously equilibrated in buffer D-0.1 M (NH4)2SO4. Fractions containing ASAP complexes were pooled and each loaded onto a Pharmacia Mono P 5/5 column equilibrated in buffer D-0.1 M (NH4)2SO4. The column was eluted with a linear gradient from 0.1 to 1 M (NH4)2SO4 in buffer D. Highly pure ASAP complexes eluted around 0.475 M (NH4)2SO4. Column fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blot analysis and silver staining.
Protein identification.
Gel-resolved proteins were digested with trypsin, and the mixtures were fractionated on a Poros 50 R2 RP micro-tip (7). Resulting peptide pools were then analyzed by matrix-assisted laser desorption ionization-reflectron time-of-flight mass spectrometry (MS) using a Reflex III instrument (Brüker Franzen; Bremen, Germany); and by electrospray ionization MS/MS on an API 300 triple quadrupole instrument (PE-SCIEX; Thornhill, Canada), modified with an ultra-fine ionization source (8). Selected mass values from the matrix-assisted laser desorption ionization-time-of-flight experiments were taken to search the protein nonredundant database (National Center for Biotechnology Information, Bethesda, Md.) using the PeptideSearch (24) algorithm. MS/MS spectra were inspected for y′′ ion series for comparison with the computer-generated fragment ion series of the predicted tryptic peptides.
Immunoprecipitations.
Goat polyclonal antibodies (1.2 μg; Santa Cruz) were bound to 10 μl of protein G-agarose beads (Roche), equilibrated in buffer C (20 mM Tris [pH 7.9], 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride) containing 0.1 M KCl, and incubated with 100 μl of input material for ca. 12 h at 4°C. Immunoprecipitates were collected by centrifugation in an Eppendorf 5417R centrifuge (1 min, 4,000 rpm, 4°C) and washed four times with 1 ml of buffer C containing 0.5 M KCl and 0.05% (vol/vol) NP-40. Protein complexes were eluted from the beads with SDS sample buffer and analyzed by SDS-PAGE followed by Western blot analysis.
For the immunoprecipitation experiments described in Fig. 6b, extracts were prepared as follows: cell pellets were resuspended in buffer containing 10 mM HEPES (pH 7.9), 5 mM MgCl2, 0.25 M sucrose, 5 mM NaF, 10 mM β-mercaptoethanol, and protease inhibitors. NP-40 was added to a final concentration of 0.1%, and cells were lysed by freezing and thawing followed by incubation on ice for 10 min. Lysates were spun in an Eppendorf 5417R centrifuge (5 min, 14,000 rpm, 4°C), and the pellet was resuspended in a solution containing 10 mM HEPES (pH 7.9), 25% glycerol, 1.5 mM MgCl2, 0.1 mM EDTA, 5 mM NaF, 10 mM β-mercaptoethanol, and protease inhibitors. After incubation on ice for 30 min, the samples were frozen and thawed and spun in an Eppendorf 5417R centrifuge (15 min, 14,000 rpm, 4°C). Supernatants were used as input material for immunoprecipitations, which were performed as described above.
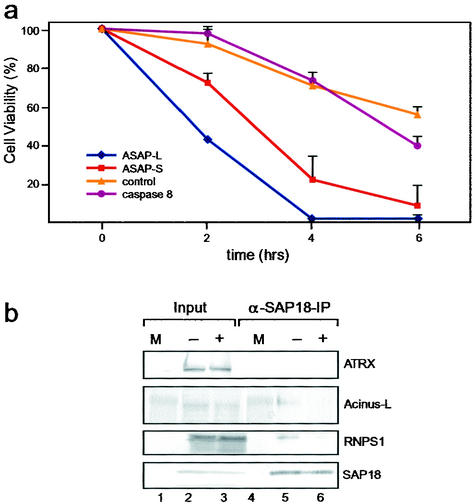
FIG. 6.
Function of ASAP during apoptosis. (a) Microinjection of ASAP complexes into HeLa cells. HeLa cells were microinjected with a caspase 8 expression construct (purple line), highly purified ASAP complexes (ASAP-L, blue line; ASAP-S, red line), or with buffer only (control, orange line). After injection the cells were allowed to recover for >2 h at 37°C in 7% CO2. Cell death was subsequently induced with 1 μM staurosporine. Apoptosis was determined on the basis of morphological criteria at the indicated time points. Equimolar amounts (determined by Western blot analyses; data not shown) of ASAP-L and ASAP-S were injected. Experiments were performed in triplicate, and standard deviations are shown. (b) Western blot analysis of the immunoprecipitates obtained with antibodies against SAP18. Proteins were immunoprecipitated from extracts prepared from HeLa cells, which were either untreated (−; lanes 2 and 5) or treated with 5 μM staurosporine for 8 h (+; lanes 3 and 6). Precipitated proteins are loaded in lanes 5 and 6, and 10% of the input is loaded in lanes 2 and 3. Immunoprecipitates were analyzed as in Fig. 1e, with the antibodies indicated at the right. Lanes labeled M contain high-molecular-mass protein standards (lanes 1 and 4).
Expression of recombinant proteins and production of polyclonal antibodies against Acinus.
Recombinant RNPS1 was expressed in insect cells as described previously (25). Recombinant SAP18 was produced in Escherichia coli as histidine-tagged fusion protein. For production of Flag-tagged Acinus-L in the baculovirus-system, SF9 cells were infected with a baculovirus encoding Flag-Acinus-L. After 3 days, SF9 cells were harvested, washed one time with phosphate-buffered saline (PBS), and resuspended in lysis buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 4 mM MgCl2, 0.4 mM EDTA, 10 mM β-glycerophosphate, 20% glycerol, 1 mM DTT, and protease inhibitors). After lysis by freezing and thawing (three times), insoluble material was pelleted by centrifugation in a Sorvall SA-600 rotor (15 min, 4°C, 8,000 rpm). The supernatant was combined with M2-agarose (Sigma) equilibrated in lysis buffer and incubated with rotation for 4 h at 4°C. The resin was washed three times with lysis buffer, and bound proteins were eluted with lysis buffer containing 200 μg of Flag peptide (Sigma)/ml. For expression of SAP18 in the baculovirus system, extracts from SF9 cells infected with a baculovirus encoding His-SAP18 were prepared as described for Flag-Acinus-L (see above). After pelleting of insoluble material by centrifugation in a Sorvall SA-600 rotor (15 min, 4°C, 8,000 rpm), the supernatant was dialyzed against buffer C (20 mM Tris [pH 7.9], 0.2 mM EDTA, 10% glycerol, 1 mM DTT, protease inhibitors), adjusted to 250 mM KCl, and bound to a P-11 column equilibrated in buffer C adjusted to a 250 mM concentration of KCl. After washing with five column volumes of buffer C adjusted to 250 mM KCl, the column was eluted with buffer C adjusted to 1 M KCl. Fractions containing His-SAP18 were pooled, dialyzed against buffer C (10 mM imidazole, no EDTA, no DTT), adjusted to 500 mM KCl, and bound to a Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) column equilibrated in buffer C (10 mM imidazole, no EDTA, no DTT) adjusted to 500 mM KCl. After washing with five column volumes of buffer C (20 mM imidazole, no EDTA, no DTT) adjusted to 500 mM KCl, bound proteins were eluted with buffer C (250 mM imidazole, no EDTA, no DTT) adjusted to 500 mM KCl.
For production of recombinant Acinus polypeptides as histidine-tagged fusion proteins in E. coli, DNA sequences encoding amino acids 1 to 108, 987 to 1093, and 1233 to 1341 of Acinus-L were amplified by PCR and subcloned into the pET-30a(+) expression plasmid (Novagen). Native and denaturing purification of the recombinant proteins was accomplished with Ni-NTA agarose following the instructions of the manufacturer (Qiagen). For production of polyclonal antibodies against Acinus, the recombinant polypeptides were injected into rabbits. Antibodies were affinity purified from serum by use of the recombinant polypeptides cross-linked to Affi-Gel 10 or Affi-Gel 15 according to the manufacturer's protocol (Bio-Rad).
Tissue culture and immunofluorescence.
HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% calf serum-200 mM l-glutamine and transfected according to the calcium phosphate protocol. For immunofluorescence, cells were fixed and permeabilized with 100% methanol, stained by sequential incubation with primary and secondary antibodies, and visualized using a Zeiss Axiovert 200 M system.
In vitro splicing assays.
Preparation of human β-globin substrate and in vitro splicing assays were carried out essentially as described previously (28). Briefly, 40 ng (see Fig. 4) or 50 ng (see Fig. 5) of His-tagged ASF/SF2 purified from recombinant baculovirus-infected cells was added to 8 μl of S100 extract. The final concentrations of components during the splicing reactions were as follows: 12 mM HEPES-KOH (pH 7.9), 10 mM creatine phosphate, 60 mM KCl, 0.2 mM EDTA, 2.5 U of RNasin, 2.6 mM MgCl2, 6% glycerol, 0.5 mM DTT, 2.6% polyvinyl alcohol (average molecular weight, 24,000; Sigma). In Fig. 5, splicing assays with suboptimal amounts of ASF/SF2 were performed with 5 ng of ASF/SF2. In RNPS1-dependent splicing activation assays (Fig. 4), 3 ng of ASF/SF2 was added to 8 μl of S100 extract in the presence of 360 ng of RNPS1 purified from recombinant baculovirus-infected cells. Splicing reactions were incubated at 30°C for 90 min. They were subsequently deproteinized and RNA precipitated with ethanol. RNA was fractionated by denaturing PAGE, and splicing products were visualized by autoradiography.
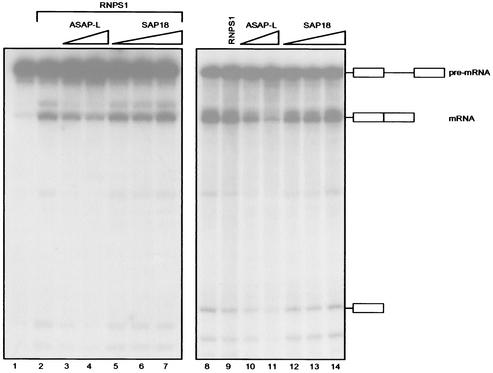
FIG. 4.
ASAP-L complex inhibits RNPS1-activated or ASF/SF2-mediated splicing in S100 extracts. Increasing amounts of purified ASAP-L complex (80 and 320 ng, 1.25 and 5 μl; lanes 3, 4, 10, and 11) or recombinant SAP18 (3, 9, and 27 ng; lanes 5 to 7 and 12 to 14) were added to S100 extracts complemented with limiting (3 ng; lanes 1 to 7) or optimal (40 ng; lanes 8 to 14) amounts of ASF/SF2 in the presence of RNPS1 (360 ng; lanes 2 to 7). Lane 1, S100 plus 3 ng of ASF/SF2 only; lane 2, S100 plus 3 ng of ASF/SF2 and 360 ng of RNPS1; lane 8, S100 plus 40 ng of ASF/SF2 only; lane 9, S100 plus 40 ng of ASF/SF2 and 360 ng of RNPS1. As determined by Western blot analyses (data not shown), 1.5 μl of purified ASAP-L contained amounts of recombinant RNPS1 and SAP18, which were stoichiometric to 14.4 ng of RNPS1 and 6.5 ng of SAP18, respectively. RNA was fractionated on a denaturing polyacrylamide gel, and splicing products, indicated schematically on the right, were visualized by autoradiography.
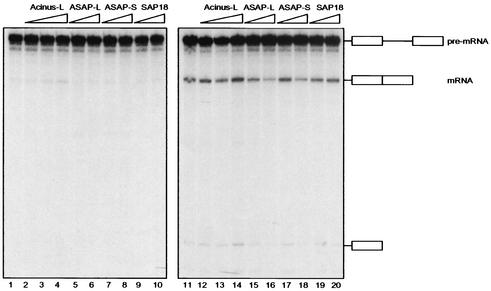
FIG. 5.
Analysis of ASAP-S, Acinus-L, and SAP18 in the in vitro splicing assay. Increasing amounts of purified ASAP-L (1.25 and 5 μl; lanes 5, 6, 15, and 16), ASAP-S (0.875 μl and 3.5 μl; lanes 7, 8, 17, and 18), and baculovirus-expressed recombinant Acinus-L (30, 90, and 260 ng; lanes 2, 3, 4, 12, 13, and 14) or SAP18 (4 and 12 ng; lanes 9, 10, 19, and 20) were added to S100 extracts complemented with limiting (5 ng; lanes 1 to 10) or optimal (50 ng; lanes 11 to 20) amounts of ASF/SF2. Lane 1, S100 plus 5 ng of ASF/SF2 only; lane 11, S100 plus 50 ng of ASF/SF2 only. As determined by Western blot analyses (data not shown), 5 μl of purified ASAP-L contained amounts of Acinus-L and SAP18, which were stoichiometric to 260 ng of recombinant Acinus-L and 12 ng of recombinant SAP18, respectively. Purified ASAP-S (3.5 μl) contained stoichiometric amounts of ASAP to 5 μl of ASAP-L. RNA was fractionated on a denaturing polyacrylamide gel, and splicing products, indicated schematically on the right, were visualized by autoradiography.
Microinjection experiments.
Cells were plated on polylysine-coated Matex dishes containing an embedded glass coverslip 24 h prior to injection and grown as described above. For injection the purified complex was mixed with either Alexa Fluor 568-dextran or tetramethyl rhodamine isocyanate-labeled dextran (Molecular Probes, Eugene, Ore.) so that the final concentrations for the complex and fluorescent dextran were 55 and 25 ng/μl, respectively. For each experiment, 100 to 200 cells were injected using a Zeiss Axiovert 135 TV inverted microscope with a heated stage and an Eppendorf transjector (5246) and micromanipulator at 20 to 40 hPa for 0.5 s. Following injection the cells were allowed to recover for 1 to 2 h, and fluorescent cells were counted. Apoptosis was induced using 1 μM staurosporine. Healthy nonapoptotic microinjected cells were identified by fluorescence and enumerated at 2-h intervals. In some experiments healthy cells were also identified by staining them with 10 μM FITC-ZVAD-FMK, a fluorescent caspase inhibitor (Promega, Madison, Wis.), as described by the manufacturer. The fluorescent cells were also monitored for morphological changes, which were documented by photography. Control cells overexpressing caspase 8 were generated by microinjection of a murine stem cell virus-caspase 8 internal ribosome entry site-green fluorescent protein expression construct 24 h prior to injecting other dishes with ASAP and control fractions. This resulted in two- to threefold increase in the level of caspase 8 protein over that for uninjected control cells but was not sufficient to induce apoptosis without additional stimuli. For examination of nuclear DNA condensation, the cells were fixed with 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100 for 5 min, stained with PBS containing 10 ng of 4′,6′-diamidino-2-phenylindole/ml, and photographed.
Plasmids, baculoviruses, and antibodies.
Plasmids containing the cDNA of Acinus-L (30) were a gift of Y. Tsujimoto and S. Sahara; the Flag-RNPS1 expression construct has been described before (19). A baculovirus encoding human RNPS1 (25) was a generous gift of A. Mayeda. For generation of a baculovirus transfer-vector encoding His-SAP18, a His-tagged human SAP18 cDNA was inserted into the plasmid pVL1393 (Pharmingen). For generation of a baculovirus transfer-vector encoding Flag-Acinus-L, a Flag-tagged human Acinus-L cDNA was inserted into the plasmid pVL1392 (Pharmingen). Baculoviruses for expression of His-SAP18 and Flag-Acinus-L were generated by Orbigen. If not indicated differently, antibodies against Acinus, Sin3, MTA2, and SAP18 and secondary antibodies for immunofluorescence analyses were obtained from Santa Cruz. The fluorescein isothiocyanate-coupled antibody against the Flag tag was purchased from Upstate Biotechnology. An antibody against ATRX was a gift of D. R. Higgs. The antibody against RNPS1 has been described before (19).
RESULTS
Identification of a novel protein complex that consists of SAP18, RNPS1, and Acinus-L.
Previous work in our laboratory resulted in the isolation of a Sin3 histone deacetylase (HDAC) complex (33, 34). The isolated Sin3-HDAC complex was found to be composed of a four-subunit core complex, which includes HDAC1, HDAC2, RbAp46, and RbAp48, and the associated polypeptides Sin3 and SAP30. During these studies a novel protein of 18 kDa was identified and suggested to be associated with the Sin3-HDAC complex. Accordingly this protein was termed SAP18, (for Sin3-associated protein of 18 kDa) (33).
To investigate the possible existence of additional protein complexes containing SAP18, we followed SAP18 during biochemical fractionation of an extract derived from HeLa nuclei by Western blotting. This procedure led to the discovery of a new putative protein complex containing SAP18 and several additional subunits. Identification of subunits of the complex by mass spectrometry of tryptic peptides confirmed the presence of SAP18 and identified the ca. 50-kDa protein RNPS1 and the ca. 220-kDa isoform of Acinus (Acinus-L) as additional subunits. No components of the Sin3-HDAC complex could be detected.
Following Western blot activity against SAP18, RNPS1, and Acinus-L, we repurified this putative novel complex according to the protocol shown in Fig. 1b. Analysis of fractions from the final MonoP column by SDS-PAGE and silver staining demonstrates purification of a three-subunit protein complex to near-homogeneity (Fig. 1c). The presence of SAP18, RNPS1, and Acinus-L was confirmed by Western blot analyses (Fig. 1d). In immunoprecipitation (IP) experiments using a partially purified fraction derived from a HeLa nuclear pellet as input (Fig. 1e), antibodies against SAP18 specifically immunoprecipitated SAP18, RNPS1, and Acinus-L but not the unrelated proteins MTA2, a component of the NuRD complex (35), or ATRX, a member of the SNF2 family of helicases and ATPases (9) (lane 5). Control experiments performed with empty beads (lane 3) or with antibodies against MTA2 (lane 4) precipitated either no proteins or specifically MTA2, respectively. IP experiments using crude nuclear pellet as input gave identical results (data not shown). Coimmunoprecipitation of SAP18, RNPS1, and Acinus-L using a crude nuclear extract as input is shown below (see Fig. 6b, lane 5). These analyses confirm the presence of the novel complex in cells and verify the identities of the components of this complex.
SAP18 and RNPS1 exist in distinct complexes containing different Acinus isoforms.
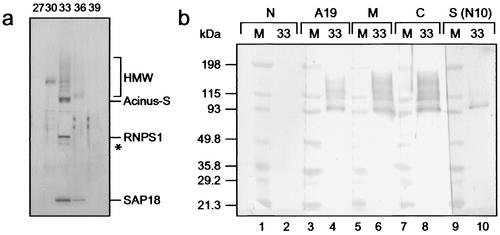
During separation on the 5PW column (the fourth purification step), we obtained two protein peaks containing SAP18 and RNPS1. One of these protein peaks comprised the Acinus-L-containing fraction. Purification of the second protein peak following the protocol presented in Fig. 1b led to the identification of a protein complex consisting of three major bands of ca. 20, 50, and 98 kDa as well as higher-molecular-mass bands between 98 and 220 kDa (Fig. 2a). While the 20- and 50-kDa subunits were confirmed as SAP18 and RNPS1 by Western blot analyses (data not shown), the 98-kDa band reacted specifically with an antibody against a unique peptide sequence at the N terminus of the Acinus-S isoform of Acinus (Fig. 2b, lane 10), which is not contained in any of the other described isoforms of Acinus (30), indicating that SAP18 and RNPS1 are in a second complex with Acinus-S.
FIG. 2.
A second ASAP complex containing Acinus-S. (a) Silver stain of an SDS-4 to 15% (wt/vol) polyacrylamide gradient gel containing aliquots of the indicated fractions derived from the final MonoP column. Components of the ASAP-S complex are indicated at the right side of the panel. A putative alternatively phosphorylated or partially degraded form of RNPS1 is marked with an asterisk. HMW, higher-molecular-weight bands. (b) Western blot analysis of fraction 33 of the MonoP column (lanes 2, 4, 6, 8, and 10). The Acinus antibodies used in the Western blot are indicated at the top (compare with Fig. 1a). High-molecular-mass protein standards [M] (lanes 1, 3, 5, 7, and 9) are indicated at the left.
To confirm the identity of the 98-kDa subunit and to analyze the higher-molecular-mass bands, we employed a set of antibodies recognizing different regions of Acinus (Fig. 1a) (see also Materials and Methods). For that purpose we generated an antibody against the first 108 amino acids of Acinus-L (Fig. 1a, [N]) which specifically recognizes Acinus-L and none of the other Acinus isoforms (data not shown). This antibody did not produce any signal in the peak fraction of the final MonoP column (Fig. 2b, lane 2), indicating the absence of Acinus-L. In contrast, antibodies recognizing regions contained in all nonapoptotic isoforms of Acinus (Fig. 1a, [A19], [M], and [C]) reacted with the 98-kDa subunit as well as with the higher-molecular-mass bands. These data suggest that the higher-molecular-mass bands represent additional Acinus isoforms and/or products of Acinus-L derived by proteolysis that are missing the N-terminal region of Acinus-L, which is recognized by the antibody specific for Acinus-L ([N]).
Currently we cannot distinguish if fraction 33 is composed of distinct complexes containing different Acinus isoforms and/or if a single complex contains more than one Acinus molecule. The separation of crude and partly purified nuclear pellets by gel filtration chromatography demonstrated that major fractions of the cellular SAP18 and RNPS1 are present in multiprotein complexes of different sizes and copurify with distinct isoforms of Acinus (data not shown). While a rough estimation of Western blot data obtained during purifications of ASAP indicates that more than 80% of SAP18 and RNPS1 in the extract are associated with each other, it is likely that fractions of the proteins are also components of other protein complexes. Accordingly, previous data have shown that RNPS1 is a component of several multiprotein complexes with distinct functions (16, 17, 19, 20, 25). While we have not analyzed the distribution of other Acinus isoforms during the purification steps, Western blot analyses indicate that more than 90% of Acinus-L in the extract copurified with SAP18 and RNPS1 (data not shown). Taken together, our data establish that SAP18 is a component of distinct isoforms of a novel protein complex consisting of SAP18, RNPS1, and Acinus. Based on the roles of the subunits in apoptosis and RNA processing, we have termed the complex the ASAP complex. To distinguish between the different isoforms of ASAP we refer to them as ASAP-L and ASAP-S, dependent on the presence of Acinus-L or Acinus-S, respectively.
ASAP complex components colocalize in HeLa nuclei.
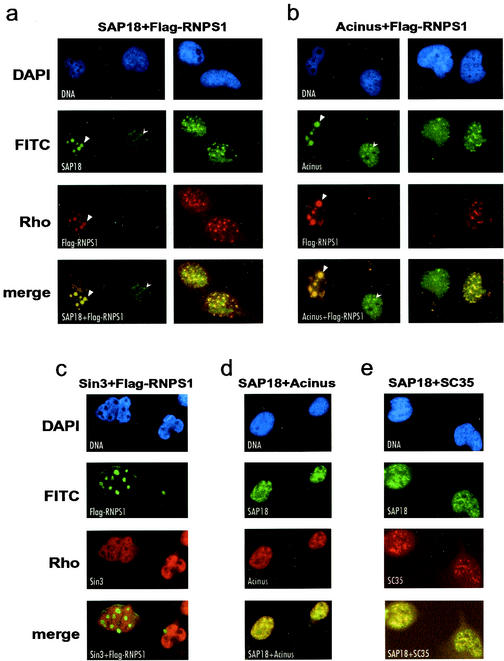
To determine the subcellular localization of the ASAP complex components, we performed indirect-immunofluorescence (IF) analyses with HeLa cells. Using antibodies against SAP18 or Acinus, we observed both generalized nucleoplasmic and punctuate nuclear staining with each antibody (Fig. 3a and b, left columns). Because we do not have an antibody against RNPS1 that functions in IF experiments, we transiently transfected HeLa cells with an expression construct for Flag-RNPS1 and detected the expressed protein with an antibody against the Flag tag. As observed previously (19), massive overexpression of Flag-RNPS1 in transiently transfected cells led to a reorganization of nuclear speckles and formation of so-called “mega-speckles,” which contained Flag-RNPS1 as detected by IF (Fig. 3a and b, left columns). These mega-speckles have been shown to occur in cells expressing high levels of RNPS1 and to coincide with nuclear interchromosomal granules (19) (compare also the 4′,6′-diamidino-2-phenylindole staining in Fig. 3) but do not colocalize with nucleoli as determined by IF analyses using antibodies against nucleolin and fibrillarin (data not shown). Importantly, in the transfected cells both SAP18 and Acinus colocalized with Flag-RNPS1 in the mega-speckles (Fig. 3a and b, left columns). Thus, this finding further stresses that the components are within a complex in vivo. In contrast, the nuclear staining obtained with an antibody against Sin3 is not reorganized by overexpression of Flag-RNPS1, and Sin3 does not colocalize with Flag-RNPS1 in the mega-speckles (Fig. 3c). It is noteworthy that transient expression of Flag-RNPS1 did not lead to the formation of mega-speckles in all transfected cells; rather, when Flag-RNPS1 was expressed at lower levels, RNPS1 was detected in smaller and more abundant sites in interphase nuclei, where it again colocalized with SAP18 and Acinus (Fig. 3a and b, right columns). We also performed IF experiments to determine whether SAP18 and Acinus colocalize in HeLa nuclei, using antibodies to SAP18 and a C-terminal region of Acinus that is present in all described nonapoptotic isoforms of Acinus ([C]; see Fig. 1a and Materials and Methods). As shown in Fig. 3d, the staining patterns for the two proteins clearly superimpose upon each other. Furthermore, it had previously been shown that RNPS1 partially colocalizes with the splicing factor SC35 in the mega-speckles (19). We therefore extended our IF analyses to examine whether other ASAP components colocalize with SC35. As shown in Fig. 3e, the IF stainings of SAP18 and SC35 in HeLa cells superimpose upon each other, indicating colocalization of ASAP with SC35. Taken together, the immunofluorescence data support the existence of protein complexes composed of SAP18, RNPS1, and Acinus in HeLa cells.
FIG. 3.
Localization of ASAP complex components in HeLa cells. (a) HeLa cells were transiently transfected with a Flag-RNPS1 expression construct. Overexpression of Flag-RNPS1 leads to formation of a few large mega-speckles (left panel, arrowheads) or more abundant and smaller speckles (right panel). Localization of Flag-RNPS1 and SAP18 was analyzed by indirect immunofluorescence with antibodies against the Flag tag and SAP18. SAP18 is present in multiple nuclear sites in untransfected cells (left panel, arrows) but colocalizes with Flag-RNPS1 in transfected cells. (b) HeLa cells were transfected and analyzed with antibodies against the Flag tag and Acinus as in panel a. Mega-speckles caused by overexpression of Flag-RNPS1 are indicated by arrowheads, and localization of Acinus in nuclear speckles in untransfected cells is indicated by arrows. Acinus colocalizes with Flag-RNPS1 in transfected cells. (c) HeLa cells transiently transfected with Flag-RNPS1 were analyzed with a fluorescein isothiocyanate-coupled monoclonal antibody against the Flag tag and by indirect immunofluorescence with an antibody against Sin3. Sin3 shows nuclear staining and does not colocalize with Flag-RNPS1 in the mega-speckles. (d) Untransfected HeLa cells were analyzed for localization of SAP18 and Acinus by indirect immunofluorescence with antibodies against SAP18 and the carboxyl terminus of Acinus, respectively. SAP18 and Acinus colocalize in nuclear speckles. (e) Untransfected HeLa cells were analyzed as in panel d with antibodies against SAP18 and SC35, respectively. SAP18 localizes to nuclear speckles containing SC35.
ASAP represses RNA processing in an in vitro splicing assay.
Several data point to a role of ASAP complex components in RNA processing (19, 25, 29, 36). To test directly whether the ASAP complex participates in pre-RNA splicing, we employed an in vitro splicing system that consists of a cytosolic S100 extract supplemented with the SR protein ASF/SF2. Figure 4 shows the results of such an assay using a simple splicing substrate (β-globin) with limiting (lanes 1 to 7) or saturating (lanes 8 to 14) amounts of ASF/SF2 (see Materials and Methods). As demonstrated previously by others (25), baculovirus-expressed RNPS1 stimulated splicing when ASF/SF2 was limiting (compare lanes 1 and 2) but had no effect on RNA processing when saturating amounts of ASF/SF2 were used (compare lanes 8 and 9). Addition of increasing amounts of bacterially expressed SAP18 had no effect on splicing activated by ASF/SF2 plus RNPS1 (lanes 5 to 7) or ASF/SF2 alone (lanes 12 to 14). In contrast, the addition of increasing amounts of the ASAP-L complex inhibited RNA processing mediated both by ASF/SF2 plus RNPS1 (lanes 3 and 4) and by saturating amounts of ASF/SF2 (lanes 10 and 11). All products of the splicing reaction were reduced, suggesting that ASAP-L inhibits RNA processing at an early stage. The addition of a later fraction of the Mono P column (Fig. 1c and d, fraction 48) had no effect under both conditions (data not shown). It is noteworthy that RNPS1-stimulated splicing was less sensitive to inhibition by ASAP-L than was RNA processing mediated by ASF/SF2 alone, suggesting that the ASAP-L complex acts in a pathway for splicing regulation that is distinct from the one that is regulated by RNPS1. Experiments performed with S100 extract complemented with SC35 instead of ASF/SF2 gave identical results (data not shown).
Finally, we analyzed the effects of ASAP-S as well as baculovirus-expressed Acinus-L and SAP18 on RNA processing. Figure 5 shows the results of an in vitro splicing assay employing saturating (lanes 11 to 20) or limiting (lanes 1 to 10) amounts of ASF/SF2 in the absence of recombinant RNPS1. While ASAP-S inhibited RNA processing activated by ASF/SF2 to an extent similar to that by ASAP-L (compare lanes 17 and 18 with 15 and 16), addition of recombinant Acinus-L alone led to a weak stimulation of splicing (lanes 12 to 14). Weak stimulation of splicing by Acinus-L could also be observed under limiting ASF/SF2 conditions (lanes 2 to 4), where neither ASAP isoform had any significant effect (lanes 5 to 8). Consistent with the results obtained using E. coli-expressed SAP18 (Fig. 4), SAP18 produced in the baculovirus system had no significant effect under any of the conditions employed (Fig. 5, lanes 9, 10, 19, and 20).
Taken together, these data suggest that the ASAP complexes function to repress splicing in vitro and that the ability of RNPS1 (and possibly Acinus-L) to activate splicing is overcome both when it is a component of ASAP and in the presence of the intact complex.
ASAP complexes accelerate cell death in HeLa cells and disassemble after induction of apoptosis.
Using an in vitro assay that employed permeabilized HeLa nuclei and an apoptotic extract obtained from bovine thymus, Acinus-p23 was originally isolated as an activity that mediated late-stage apoptotic chromatin condensation prior to DNA fragmentation. Furthermore, Acinus displayed effects on apoptosis in transient-transfection assays: while transfection of a sense construct of Acinus-L accelerated cell death after induction of apoptosis, transfection of an antisense construct led to a delay in cell death (30).
To analyze the influence of the ASAP complexes on HeLa cells in the absence of and after an apoptotic stimulus, HeLa cells were microinjected with highly purified ASAP-L and ASAP-S, respectively (Fig. 6a). In the absence of an apoptotic stimulus, cells microinjected with ASAP-L or ASAP-S showed no difference in viability from control cells injected with buffer only (data not shown). In contrast, after induction of apoptosis by staurosporine, cells microinjected with ASAP complexes showed a significant acceleration in cell death compared to the control cells or cells overexpressing caspase 8 (see Materials and Methods), indicating that ASAP is involved in the execution of apoptosis in vivo.
We next investigated whether the composition of ASAP complexes is changed during apoptosis. Figure 6b shows the results of IP experiments performed with extracts derived from HeLa cells which were either untreated or treated with staurosporine. Staurosporine-treated cells were apoptotic as judged by morphological criteria and the appearance of laddering of genomic DNA on agarose gels caused by oligonucleosomal DNA fragmentation (data not shown). An antibody against SAP18 specifically immunoprecipitated SAP18, RNPS1, and Acinus-L from an extract prepared from nonapoptotic cells (lane 5; see also Fig. 1e). In contrast, as expected due to cleavage of Acinus by caspases, Acinus-L could not be detected in precipitates obtained with an antibody against SAP18, when an extract derived from apoptotic cells is employed (lane 6). Most importantly, RNPS1, which is not proteolysed during apoptosis (compare lanes 2 and 3), did not coprecipitate with SAP18 from apoptotic cell extract (lane 6), indicating that RNPS1 is released from ASAP during apoptosis. Antibodies against SAP18 did not precipitate the unrelated protein ATRX from any extract analyzed (lanes 5 and 6). The results of the IP experiments strongly indicate that ASAP complexes disassemble during apoptosis.
DISCUSSION
Here we report the purification of a novel protein complex composed of the polypeptides SAP18, RNPS1, and Acinus. We chose the name ASAP complex because the subunit composition and our functional analyses suggest that the complex participates in both apoptosis and RNA splicing. The ASAP complexes exists in at least two isoforms, termed ASAP-L and ASAP-S, which are characterized by the presence of different isoforms of Acinus, and members of the ASAP complex have been identified in nuclear compartments thought to be involved in RNA processing (N. Saitoh and D. L. Spector, personal communication).
While SAP18 has not previously been implicated in pre-mRNA splicing, an involvement in RNA processing was suggested for both Acinus and RNPS1, which were purified as components of functional spliceosomes (29, 36). Furthermore, RNPS1 has been shown to localize to nuclear splicing factor compartments (19, 25) and was purified as a single-subunit protein that mediates the general activation of splicing that occurs in the presence of limited amounts of SR proteins in an in vitro splicing assay (25). In the context of the ASAP complex, this positive effect on RNA processing in vitro is suppressed (Fig. 4 and 5). Repression of RNA processing most likely occurs at an early stage of the splicing reaction. Therefore, ASAP inhibits splicing under conditions where RNPS1 alone can function with SR proteins to activate splicing, indicating that incorporation of RNPS1 into the ASAP complexes can serve to regulate the splicing activity of RNPS1. Modification of ASAP complex components might also modulate the activity of ASAP during RNA processing.
Several recent reports have presented evidence of a role for RNPS1 in downstream events of mRNA metabolism (16, 17, 20). In the current model, RNPS1 binds to RNA in the nucleus as part of a postsplicing complex deposited upstream of exon-exon junctions, which is thought to provide a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. RNPS1 in this exon junction complex travels with the RNA into the cytoplasm and communicates the position of exon-exon junctions for the mRNA surveillance machinery (20). It is unclear whether the fraction of RNPS1 incorporated in the ASAP complexes might play a similar role or if formation of the ASAP complex functions to sequester RNPS1, preventing it from functioning in this capacity.
Acinus had previously been implicated to function during apoptotic chromatin condensation (30). Microinjection analyses of the ASAP complexes clearly demonstrate that ASAP influences the progression of apoptotic cell death (Fig. 6a). Modulation of cell death by ASAP-L also occurred when apoptosis was induced by a combination of tumor necrosis factor α and cycloheximide (data not shown), suggesting that the effects caused by ASAP are at least partly direct. Various studies have provided evidence that SAP18 can associate with the Sin3-HDAC complex (4, 33). Considering the function of ASAP in execution of apoptosis and the potential role of Acinus in apoptotic chromatin condensation, these observations raise the exciting possibility that deacetylation of histones via recruitment of the Sin3-HDAC complex is involved in the process of chromatin condensation during apoptosis. However, we have not yet been able to demonstrate association of ASAP and Sin3-HDAC complexes. It is noteworthy that in the above-mentioned studies (4, 33), demonstration of interaction of SAP18 with the Sin3-HDAC complex required overexpression of SAP18, suggesting that just a small fraction of cellular SAP18 associates with the Sin3-HDAC complex. This is in agreement with our observations that although SAP18 is present in several multiprotein complexes of different sizes, the majority of SAP18 copurifies with RNPS1 and different Acinus isoforms (data not shown). To clarify a putative role of the Sin3-HDAC complex during apoptotic chromatin condensation, further experimentation is required.
Since the Acinus subunit of the ASAP complex is a target of proteolytic cleavage during apoptosis (30), it is possible that apoptotic stimuli lead to a regulation of the function of the complex in splicing through reorganization of the complex composition. Regulation of splicing has been shown to play an important role during apoptosis (3, 13). IP analysis of extracts derived from apoptotic cells strongly indicates that the complex disassembles during cell death (Fig. 6b). If activation of splicing by RNPS1 is repressed by incorporation into ASAP, this event would trigger release of the activation function of that fraction of RNPS1 in splicing, thereby modulating RNA processing of possibly selected target transcripts. Interestingly, a link between splicing and apoptosis is established not only by the Acinus protein but also by RNPS1. RNPS1 has been shown to interact with the 110-kDa isoform of the p34cdc2-related protein kinase PITSLRE (p110). p110 is processed during apoptosis into a smaller isoform(s) that does not interact with RNPS1 (19), and a recently identified cyclin, cyclin L, interacts with p110 (2, 5). The carboxyl terminus of this particular cyclin contains numerous arginine-serine-rich dipeptide repeats, characteristic of many splicing factors, and may target the p110/cyclin L complex to nuclear speckles. Antibodies against cyclin L inhibit RNA processing in an in vitro assay, and recombinant cyclin L stimulates splicing under suboptimal conditions (5). A smaller cyclin L protein is also made by alternative splicing, which retains the cyclin box region but no longer contains the RS domain. However, the caspase-processed p46 PITSLRE isoform interacts well in vivo only with the smaller form of cyclin L (J. H. Trembley, D. Hu, and V. J. Kidd, unpublished data). Further experimentation is required to decipher whether there is a functional interplay between the ASAP complex, PITSLRE isoforms, and cyclin L or L short forms during apoptosis.
In summary, we have identified a novel protein complex (ASAP complex) consisting of SAP18, RNPS1, and Acinus. Functional analyses suggest that ASAP is involved in both RNA processing and apoptosis, pointing to ASAP as a reasonable candidate for participation in the regulation of splicing during the execution of programmed cell death.
Acknowledgments
We thank Akila Mayeda for the generous gift of a baculovirus expressing RNPS1 and Douglas R. Higgs for the antibody against ATRX. We also thank members of the Reinberg laboratory, especially Andrei Kuzmichev, Erika Friedl, and Alejandro Vaquero, for helpful discussions. We thank Soumit Basu for initial work on the project, Simon Moshiach for help with the microinjection experiments, and Lois Wang for excellent technical assistance. We thank Anita Grewal for help with MS analysis.
This work was supported by National Cancer Institute core grants P30 CA08748 (to P.T.), R01 CA53370 (to E.W.), P30 CA21765 (to J.M.L. and V.J.K.), R37 GM48259 (to J.L.M.), grants GM44088 (to V.J.K.) and GM 37120 (to D.R.), a fellowship by the Deutsche Forschungsgemeinschaft (Schw 703/1-1; to C.S.), the American Lebanese Syrian Associated Charities (J.M.L. and V.J.K.), and the Howard Hughes Medical Institute (D.R. and E.W.).
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 2.Berke, J. D., V. Sgambato, P.-P. Zhu, B. Lavoie, M. Vincent, M. Krause, and S. E. Hyman. 2001. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32:277-287. [DOI] [PubMed] [Google Scholar]
- 3.Chalfant, C. E., K. Rathman, R. L. Pinkerman, R. E. Wood, L. M. Obeid, B. Ogretmen, and Y. A. Hannun. 2002. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J. Biol. Chem. 277:12587-12595. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, S. Y., and J. M. Bishop. 2002. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl. Acad. Sci. USA 99:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickinson, L. A., A. J. Edgar, J. Ehley, and J. Gottesfeld. 2002. Cyclin L: an RS domain protein involved in pre-mRNA splicing. J. Biol. Chem. 277:25465-25473. [DOI] [PubMed] [Google Scholar]
- 6.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 7.Erdjument-Bromage, H., M. Lui, L. Lacomis, A. Grewal, R. S. Annan, D. E. MacNulty, S. A. Carr, and P. Tempst. 1998. Micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. 826:167-181. [DOI] [PubMed] [Google Scholar]
- 8.Geromanos, S., G. Freckleton, and P. Tempst. 2000. Tuning of an electrospray ionization source for maximum peptide-ion transmission into a mass spectrometer. Anal. Chem. 72:777-790. [DOI] [PubMed] [Google Scholar]
- 9.Gibbons, R. J., D. J. Picketts, L. Villard, and D. R. Higgs. 1995. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome). Cell 80:837-845. [DOI] [PubMed] [Google Scholar]
- 10.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häcker, G. 2000. The morphology of apoptosis. Cell Tissue Res. 301:5-17. [DOI] [PubMed] [Google Scholar]
- 12.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 13.Jiang, Z.-H., and J. Y. Wu. 1999. Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med. 220:64-72. [DOI] [PubMed] [Google Scholar]
- 14.Kidd, V. J. 1998. Proteolytic activities that mediate apoptosis. Annu. Rev. Physiol. 60:533-573. [DOI] [PubMed] [Google Scholar]
- 15.Krämer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 16.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. L. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Hir, H., D. Gatfield, E. Izaurralde, and M. L. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 19.Loyer, P., J. Trembley, J. M. Lahti, and V. J. Kidd. 1998. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J. Cell Sci. 111:1495-1506. [DOI] [PubMed] [Google Scholar]
- 20.Lykke-Andersen, J., M.-D. Shu, and J. A. Steitz. 2000. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293:1836-1839. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 23.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 24.Mann, M., P. Højrup, and P. Roepstorff. 1993. Use of mass spectrometric molecular weight information to identify proteins in databases. Biol. Mass Spectrom. 22:338-345. [DOI] [PubMed] [Google Scholar]
- 25.Mayeda, A., J. Badolato, R. Kobayashi, M. Q. Zhang, E. M. Gardiner, and A. R. Krainer. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misteli, T. 2000. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci. 113:1841-1849. [DOI] [PubMed] [Google Scholar]
- 27.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 28.Prasad, J., K. Colwill, T. Pawson, and J. L. Manley. 1999. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 19:6991-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rappsilber, J., U. Ryder, A. I. Lamond, and M. Mann. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahara, S., M. Aoto, Y. Eguchi, N. Imamoto, Y. Yoneda, and Y. Tsujimoto. 1999. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature 401:168-173. [DOI] [PubMed] [Google Scholar]
- 31.Spector, D. L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9:265-315. [DOI] [PubMed] [Google Scholar]
- 32.Strasser, A., L. O'Connor, and V. M. Dixit. 2000. Apoptosis signaling. Annu. Rev. Biochem. 69:217-245. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1:1021-1031. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2000. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]