Abstract
The Epstein-Barr virus (EBV)-encoded lytic activator Zta is a bZIP protein that can stimulate nucleosomal histone acetyltransferase (HAT) activity of the CREB binding protein (CBP) in vitro. We now show that deletion of the CBP bromo- and C/H3 domains eliminates stimulation of nucleosomal HAT activity in vitro and transcriptional coactivation by Zta in transfected cells. In contrast, acetylation of free histones was not affected by the addition of Zta or by deletions in the bromo or C/H3 domain of CBP. Zta stimulated acetylation of oligonucleosomes assembled on supercoiled DNA and dinucleosomes assembled on linear DNA, but Zta-stimulated acetylation was significantly reduced for mononucleosomes. Western blotting and amino-terminal protein sequencing indicated that all lysine residues in the H3 and H4 amino-terminal tails were acetylated by CBP and enhanced by the addition of Zta. Histone acetylation was also dependent upon the Zta basic DNA binding domain, which could not be substituted with the homologous basic region of c-Fos, indicating specificity in the bZIP domain nucleosome binding function. Finally, we show that Zta and CBP colocalize to viral immediate-early promoters in vivo and that overexpression of Zta leads to a robust increase in H3 and H4 acetylation at various regions of the EBV genome in vivo. Furthermore, deletion of the CBP bromodomain reduced stable CBP-Zta complex formation and histone acetylation at Zta-responsive viral promoters in vivo. These results suggest that activator- and bromodomain-dependent targeting to oligonucleosomal chromatin is required for stable promoter-bound complex formation and transcription activity.
Chromatin modification is thought to be an early and integral step in the regulation of transcription and DNA replication (2, 18, 27, 28, 30, 44). Initiation sites for transcription and DNA replication are specified by sequence-specific DNA binding proteins which are subject to multiple levels of regulation through signaling pathways that respond to changes in intra- and extracellular conditions. Higher order chromatin structures associated with repressive heterochromatin are thought to prohibit sequence-specific transcription and DNA replication factors from binding to their recognition sites. Precisely how sequence-specific factors can access their sites in these regions of the genome, recruit chromatin modifying and remodeling activities, and initiate the processes of transcription and DNA replication remains poorly understood (50). Previous work, however, has suggested that a subset of bZIP proteins can stimulate chromatin modifying activity in the absence of sequence-specific recognition, suggesting that these proteins may function as early-acting factors in the chromatin remodeling process (9).
The Epstein-Barr virus (EBV) immediate-early protein Zta is a transcription and DNA replication factor that must disrupt chromatin-repressed latent viral genomes to initiate the lytic cycle transcription program (15, 39; reviewed in references 34 and 43). Zta is a member of the bZIP family with significant sequence similarity to the basic and zipper regions of the adipocyte differentiation factor C/EBP, the erythroid-specific factor NF-E2, and the protooncogene c-Fos. In latently infected B lymphocytes, the EBV genome exists as a nonintegrated nuclear episome with nucleosome phasing indistinguishable from that of cellular chromatin (32, 52). The majority of viral genes required for lytic replication are repressed but can be reactivated by overexpression of Zta or by treatment of cells with pleiotropic agents, such as sodium butyrate, trichostatin A, calcium ionophores, or phorbol esters (34, 43). It is thought that chromatin-based repression is one important component of maintaining the transcription silence of lytic cycle gene products during viral latency. Consistent with this hypothesis is the fact that transcriptional coactivators with histone acetylase activity can cooperate with Zta to stimulate lytic cycle gene expression (1, 55).
The CREB binding protein (CBP) and its close relative, p300, are transcriptional coactivator proteins with intrinsic histone acetyltransferase activity (3, 41; reviewed in references 6, 7, and 26). CBP and p300 are essential for early embryonic development and can costimulate transcription driven by numerous cellular and viral transcription factors (47, 53). The association of CBP and p300 with several activators can be regulated by phosphorylation, and it is thought that recruitment of CBP and/or p300 can be limiting for transcription initiation of several target genes (7, 25, 48). In this way, CBP and p300 are thought to integrate the complex network of transcription signals. The inactivation of p300 and/or CBP by viral oncogenes, including SV40 T antigen, adenovirus E1A, and papilloma virus E7, may play an important role in cellular growth transformation. The near ubiquitous requirement for CBP and/or p300 in transcription regulation underscores the importance of coactivators and histone acetylation in coordinating gene expression. Despite the enormous number of transcription factor interactions with CBP, the precise mechanism of CBP coactivator function remains poorly understood, and it is especially unclear whether CBP can function to facilitate the initial recognition of sequence-specific factors to their cognate binding sites in promoter regulatory regions.
Zta activates transcription of numerous viral and cellular genes in the early stages of the EBV lytic reactivation cycle (10, 19, 33, 37). Zta exists as a homodimer that binds with high affinity to a large family of degenerate sequence recognition sites found in the upstream regulatory regions of responsive viral genes (36). In addition to activation of transcription, Zta also binds to several sites in the EBV origin of lytic DNA replication and recruits virally encoded lytic replication enzymes to the origin (24, 45, 46). Thus, Zta not only is a transcriptional activator but also participates directly in the process of DNA replication initiation. The amino-terminal transcriptional activation domain of Zta has been examined by several groups in some detail (11, 16, 20). It has been found that aromatic hydrophobic residues in the activation domain are essential for transcription activation and recruitment of CBP (16). The Zta transcriptional activation domain binds to the cysteine-histidine (C/H)-rich regions 1 and 3 of CBP, and this interaction is strictly dependent on the aromatic residues that are critical for transcription function (16, 55). It was also found that Zta can stimulate CBP histone acetyltransferase (HAT) activity directed toward small oligonucleosomes (SONs) in vitro (9). Stimulation of HAT activity was also dependent on the aromatic residues in the Zta activation domain, suggesting that a strong correlation exists between the activation of HAT activity and transcription. Stimulation of CBP HAT activity was found to be dependent on the Zta DNA binding domain as well, and this correlated with the ability of Zta to bind directly to oligonucleosomes. Similar concentrations of other bZIP or CBP binding proteins did not share this activity, with the exception of C/EBP and NF-E2, two bZIP proteins with high sequence similarity to the Zta basic DNA binding and zipper dimerization domains. These results suggested that a subset of bZIP proteins shared the ability to direct CBP HAT activity to nucleosomal DNA lacking an obvious sequence recognition site (9).
In this study, we further characterize the activation of CBP HAT activity by Zta and suggest that the mechanism of activation is more complex than the simple recruitment of enzyme to substrate. We found that the activation of EBV lytic transcription correlated with the stimulation of CBP HAT activity by Zta, and both required the CBP bromo- and C/H3 domains. We also found that the basic region of Zta is essential for targeting CBP to oligonucleosomes, which also correlated with transcription activation. Finally, chromatin immunoprecipitation (ChIP) assays revealed that stable promoter-bound CBP and histone acetylation are dependent on the CBP bromodomain. Based on these results, we propose that Zta initiates nucleosomal targeting of CBP HAT activity, which facilitates stable promoter binding and sequence-specific transcription activation.
MATERIALS AND METHODS
Cells and plasmids.
293 and D98/HR1 cells were passaged in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics. Raji and ZKO-293 cells were passaged in RPMI supplemented with 10% fetal bovine serum and antibiotics. EBV reactivation in Raji cells was induced with 100 ng of tetradecanoyl phorbol acetate (TPA) per ml and 1 μM ionomycin (Sigma). The BRLF1-luciferase plasmid contains nucleotides −178 to +28 relative to the transcription initiation site cloned into the NheI and HindIII sites of pGL3Basic (Promega). BHLF1-Luc has been described previously (16). CMV-CBP-ΔHAT (L1690K/C1691L), CMV-CBP-ΔBromo (Δ1101-1171), and CMV-CBP-ΔC/H3 (D1805-1890) were generated by site-directed mutagenesis of pCMV-CBP-FLAG (gift of M. G. Rosenfeld). Baculovirus expression plasmids for CBP-FL, CBP-ΔBromo, and CBP-ΔHAT were generated by subcloning the CBP gene from the cytomegalovirus (CMV) expression vector construct into the NotI site of pFASTBAC (Pharmingen). Zta wt and ZFZ were expressed in the pQE8 bacterial expression plasmid (Qiagen). ZFZ was derived from a mammalian expression vector provided by G. Miller and consists of Zta (nucleotides 1-171), Fos (nucleotides 137-162), and Zta (nucleotides 198-245) (22). Zta substitution mutants were generated by site-directed mutagenesis (Stratagene Quick Change) in pCDNA3 (Invitrogen) for mammalian cell expression and in the pQE8 (Qiagen) vector for expression in Escherichia coli. Saccharomyces cerevisiae NAP1 was expressed from plasmid pET28-yNAP1 (a gift from T. Tsukiyama). Mammalian expression vectors SRα (pcDLSRα296) and SRα-Zta have been described previously (55). 293 and D98/HR1 cells were transfected with Lipofectamine 2000 (Invitrogen), and luciferase assays were performed as described previously (16).
HAT assays and kinetic analysis.
Typical HAT assays contained 200 ng of histone substrate (SONs, dinucleosomes, mononucleosomes, or free histones), 0.25 μCi of [3H]acetyl coenzyme A (Amersham), and 20 ng of baculovirus-expressed CBP in a 30-μl reaction volume as described previously (9). For kinetic studies, baculovirus-derived CBP (20 ng) was mixed with 0.1, 0.2, 0.5, or 1.0 μg of purified HeLa SONs in a 15-μl reaction volume and incubated at 30°C for 1, 2, 5, or 10 min in the presence or absence of 200 ng of Zta. Histone acetylation was quantitated by phosphorimager analysis using a tritium-sensitive screen. Lineweaver-Burk analysis was used to calculate Vmax and Km.
Nucleosome assembly in vitro.
Nucleosomes were assembled in vitro by use of yeast NAP1 as an assembly protein as previously published (23), with modifications by T. Tsukiyama (personal communication). Yeast NAP1 was expressed and purified from E. coli by MonoS chromatography. Histone octamers were purified from HeLa nuclear pellets by hydroxyapatite chromatography to remove nucleosomal DNA (14). Chromatin was assembled in a reaction containing 0.1 mg of bovine serum albumin per ml, 0.75 mg of yNAP1, 0.3 mg of histone octamer, and 0.3 mg of pBKSII plasmid DNA in buffer ExB (10 mM HEPES [pH 7.6], 50 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, and 10% glycerol). Nucleosomes were assembled by incubation for 4 h at 30°C. Assembly was confirmed by a gel mobility shift assay (data not shown) and by a supercoiling assay with topoisomerase-relaxed plasmid DNA as described previously (23). Topoisomerase I was purchased from Topogen, Inc. Histone acetylation reactions were described previously (9). Mononucleosomes and dinucleosomes were assembled by high-salt dialysis and purified by glycerol gradient centrifugation as described previously (13).
ChIP.
ChIP was performed as described previously (42). EBV sequences were amplified with the following primers: BMRF1 sense, ACCTACATGACTAGCATCAAGCAA; BMRF1 antisense, GGCCTCCATAGTTTACAGACAGAA; BHLF1 sense, AGGTGTGTCATTTTAGCCCG; BHLF1 antisense, CCTACCTCTAGGCTCCACCC; Cp sense, AAAATTGAACCTTGTTGGCG; Cp antisense, ATGGTAAGAACCCTGCGATG; EBER sense, AGGTCAGACCACGAAAGGTC; EBER antisense, AGACAGACTACGTCACCGTG; BZLF1 sense, ATCTTCAGCAAAGATAGCAAAGGT; BZLF1 antisense, TAGAGTCCATGACAGAGGATTTGA; BRLF1 sense, TTCTGCCATGCAATACAAATAAAT; BRLF1 antisense, ATTTCCTCCAGAAGAGTGACATC; cellular gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sense, TCACCACCATGGAGAAGGCT); and GAPDH antisense, GCCATCCACAGTCTTCTGGG. Antibodies for acetylated histones were purchased from Upstate Biotechnologies, Inc. Hemagglutinin (HA) antibodies were obtained from Babco, Inc., and Boehringer. Flag M2 antibody was obtained from Sigma. CBP antibodies (A2) and normal rabbit immunoglobulin G (IgG) were purchased from Santa Cruz Biotechnologies. EBV-specific antibodies for Western blotting were monoclonal anti-Rta (Argene), anti-EA-D (Advanced Biotechnologies), and polyclonal rabbit anti-Zta.
RESULTS
CBP bromo- and C/H3 domains are required for transcriptional coactivation by Zta.
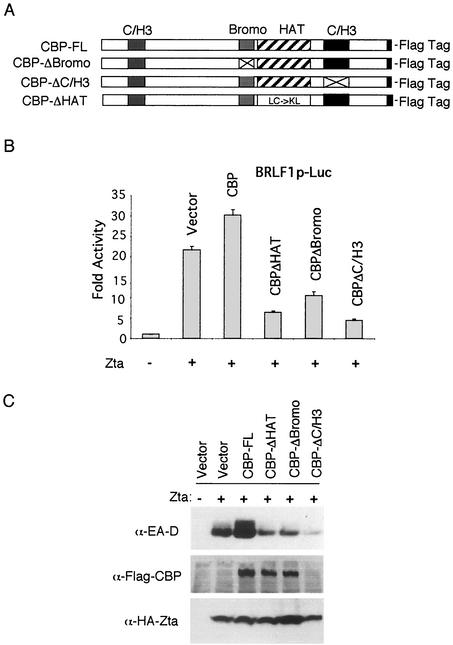
We have found that cotransfection of CBP with Zta can cooperatively stimulate transcription from several viral promoters in transient transfection assays (55). In a previous study, it was found that deletion of the CBP HAT domain compromised transcription activation of the BZLF1 promoter, indicating that HAT activity is required for the autoactivation of Zta gene transcription (9). We and others have subsequently found that the CBP HAT domain is particularly important for the coactivation function of the BRLF1 promoter (Fig. 1B) (1). BRLF1 encodes the Rta transactivator, which is required for full lytic cycle gene expression and is a critical early target of Zta transcription regulation. To further explore the mechanism of CBP coactivation, we tested whether deletion of additional subdomains of CBP affected transcription stimulation by Zta. We deleted the CBP bromodomain and E1A interacting region of the C/H3 domain. The bromodomain has been implicated in chromatin (38) and acetyl-lysine binding (17, 29, 31, 54), and the C/H3 domain is the target of numerous protein interactions, including contacts with Zta (55). Consistent with previous findings, full-length CBP modestly stimulated Zta activation of the BRLF1 promoter (Fig. 1B). Cotransfection of CBPΔHAT repressed Zta transcription activation of BRLF1, indicating that HAT activity is important for BRLF1 activation. We now show that deletion of the bromodomain (CBPΔBromo) and the E1A interacting region of the C/H3 domain (CBPΔC/H3) also inhibits transcription activation of BRLF1 by Zta (Fig. 1B). Since Zta can bind to the C/H1 domain of CBP, we suggest that these mutations in CBP function as dominant-negative inhibitors that interfere with Zta and native CBP associations. This suggests that native CBP requires the bromo- and C/H3 domains for coactivation of the BRLF1 promoter by Zta.
FIG. 1.
The CBP bromo- and C/H3 domains are required for transcriptional coactivation. (A) Schematic diagram of CBP mutants used for these studies. (B) The BRLF1-luciferase reporter plasmid was cotransfected with Zta and CBP deletion mutants in 293 cells and assayed at 48 h posttransfection for luciferase activity. CBP and CBP deletion mutants are indicated above each bar. (C) Zta and CBP deletion mutants were cotransfected into D98/HR1 cells latently infected with EBV and assayed for expression of EA-D viral antigen by Western blotting (top panel). Transfected Flag-tagged CBP derivatives were detected with anti-Flag antibody (middle panel). HA-Zta was detected with anti-HA antibody (lower panel). Transfected plasmids are indicated above each lane.
CBP full-length and internal deletion mutants were tested for the ability to stimulate viral reactivation from latently infected D98/HR1 cells. Viral reactivation was measured by Western blotting of early antigen D (EA-D). We found that CBPΔHAT and CBPΔBromo were modest inhibitors of Zta activation, while full-length CBP stimulated EA-D activation by Zta. Although CBPΔC/H3 was barely detectable by Western blotting, it had the strongest inhibitory effect on EA-D coactivation by Zta (Fig. 1C). These results are consistent with the findings shown in Fig. 1B and indicate that CBP HAT, bromo- and C/H3 domains are important for Zta coactivation of latent EBV genomes as well as of transiently transfected reporter genes.
The CBP bromo- and C/H3 domains are required for stimulation of nucleosomal HAT activity.
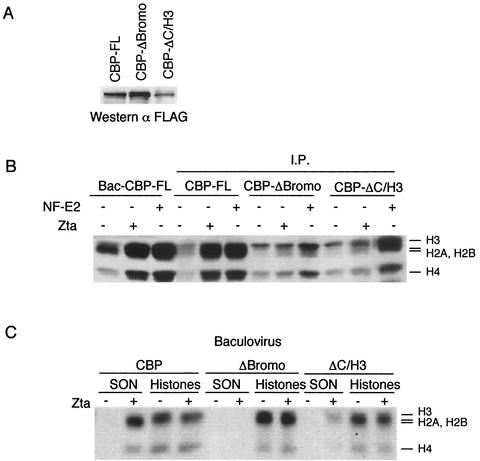
Previous work had indicated that Zta stimulated CBP HAT activity directed toward SONs. We have previously shown that HAT activity of immunoprecipitates of CBP expressed in 293 cells could be stimulated by the addition of recombinant Zta in vitro. We also showed that CBPΔHAT lacked HAT activity in immunoprecipitates and could not be further stimulated by the addition of Zta, indicating that the CBP HAT domain and not a CBP-associated HAT was required for HAT activity. We now tested whether the CBP bromodomain or the C/H3 domain was important for mediating Zta stimulation of CBP HAT activity (Fig. 2). Flag-tagged full-length (FL) CBP, CBP-ΔBromo, and CBP-ΔC/H3 were expressed in 293 cells and subjected to immunoprecipitation with anti-Flag antibody (Fig. 2A). As controls, we used baculovirus-expressed and -purified CBP-FL and the transcription factor NF-E2, which had also been shown to stimulated CBP HAT activity in vitro. Addition of Zta or NF-E2 to baculovirus CBP-FL stimulated HAT activity as expected (Fig. 2B). Similarly, Zta and NF-E2 stimulated HAT activity of immunoprecipitates of Flag-tagged CBP-FL. In contrast, neither Zta nor NF-E2 was capable of stimulating CBP-ΔBromo. Interestingly, we found that HAT activity of immunoprecipitates of CBP-ΔC/H3 was not stimulated by Zta and was stimulated at reduced levels by NF-E2. These results suggest that Zta and NF-E2 may have different mechanisms for stimulating CBP HAT activity, although both activators require the bromodomain.
FIG. 2.
The CBP bromodomain is required for stimulation of HAT activity by Zta. (A) Western blot of immunoprecipitated Flag-tagged CBP proteins used for HAT assays. (B) HAT assay showing the fluorogram of 3H-acetylated histones in reactions with either baculovirus-generated CBP (lanes 1 to 3) or immunoprecipitates (I.P.) of CBP-FL (lanes 4 to 6), CBP-ΔBromo (lanes 7 to 9), or CBP-ΔC/H3 (lanes 10 to 12). Zta or NF-E2 proteins were added (+) or not added (−) to HAT reactions as indicated above. (C) HAT assay with baculovirus-expressed and purified CBP or deletion mutant ΔBromo or ΔC/H3 in the presence (+) or absence (−) of Zta. SONs or free histones were used as substrates in HAT assays as indicated above each lane.
To confirm these results and rule out the possibility that additional cellular factors bound to CBP were influencing the properties of CBP deletion mutants, we assayed CBP deletion mutants that were expressed and purified from baculovirus (Fig. 2C). It was previously found that baculovirus-expressed CBP, but not CBPΔHAT, could be stimulated by Zta (9). As expected, Zta stimulated HAT activity of full-length CBP when SONs were used as substrates. In contrast, CBP-ΔBromo had robust HAT activity when assayed with free histones but had no detectable HAT activity with SONs in the presence or absence of Zta. Deletion of the C/H3 domain also reduced Zta-activated acetylation of SONs but had little effect on free histone acetylation. These results indicate that the CBP bromo- and C/H3 domains are required for Zta-stimulated nucleosome acetylation by CBP, suggesting that this activity correlates with transcription activation in vivo.
Stimulation of CBP HAT requires nucleosome assembly.
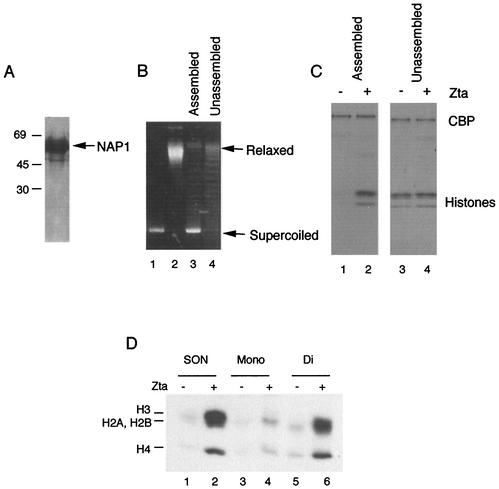
Histone octamers stripped of nucleosomal DNA by hydroxyapatite were not substrates for Zta activation, while SONs could be stimulated for acetylation (9). To better understand the substrate specificity of SONs, we determined whether in vitro-assembled nucleosomes on supercoiled plasmid DNA could reconstitute the substrate specificity for Zta and CBP. Yeast Nap1 functions as a histone chaperone that deposits histone octamers on supercoiled DNA resembling natural chromatin to some extent (23, 40). Nucleosomes were assembled in vitro using HeLa octamers, yeast NAP1, and supercoiled Bluescript plasmid DNA (Stratagene), which lacks any obvious high-affinity Zta binding sites. Nucleosomes were assembled in the presence of yNap1 (23), and the extent of the assembly reaction was determined by examining the change in superhelical density by use of a topoisomerase relaxation assay (Fig. 3B). We found that nucleosomes assembled in this manner could not be acetylated by CBP, suggesting that nucleosome assembly inhibits histone acetylation by CBP (Fig. 3C). However, the addition of Zta strongly stimulated acetylation of histones assembled in the presence of yNap1. In contrast, octamers incubated with DNA in the absence of yNap1 were not assembled (Fig. 3B), did not inhibit CBP acetylation (Fig. 3C), and were not further acetylated by the addition of Zta (Fig. 3C). These results indicate that Zta can facilitate the acetylation of nucleosomes assembled in vitro on supercoiled DNA. Inclusion of specific Zta-responsive element (ZRE) binding sites in the assembled templates did not significantly alter the stimulation of histone acetylation, suggesting that Zta interaction with nucleosomes was sufficient for this activation (data not shown). These results also indicate that nucleosome assembly inhibits CBP acetylation of histones and that Zta functions to derepress this inhibition.
FIG. 3.
In vitro reconstitution of Zta activation of CBP HAT activity. (A) Yeast NAP1 was expressed and purified from E. coli and examined by Coomassie staining of sodium dodecyl sulfate-polyacrylamide electrophoresis gels. (B) A supercoiling assay was used to indicate that nucleosomes were deposited on assembled templates. A supercoiled plasmid (lane 1) or topoisomerase I-relaxed template (lane 2) was used as an indicator of supercoiled DNA. Templates assembled with yNAP1 (lane 3) or without yNAP1 (lane 4) were compared in agarose gels containing chloramphenicol. (C) Plasmid DNA assembled with core histone and yNAP1 (lanes 1 and 2) or without yNAP1 (lanes 3 and 4) was assayed in HAT reactions with CBP in the absence (−) or presence (+) of Zta as indicated. (D) HAT assays reconstituted with recombinant CBP and 200 ng of SONs, dinucleosomes (Di), or mononucleosomes (Mono) in the presence (+) or absence (−) of 50 ng of Zta.
We also compared the ability of Zta to stimulate histone acetylation on defined-length templates that were assembled by high-salt dialysis and purified by use of a glycerol gradient (13). We found that equivalent concentrations of oligonucleosomes, dinucleosomes, and mononucleosomes were acetylated at similar levels in the absence of Zta (Fig. 3D). However, Zta-dependent HAT activity was significantly higher for SONs and dinucleosomes than for mononucleosomes. These results indicate that oligonucleosomal length is an important parameter for substrate targeting by Zta.
Zta stimulates acetylation of all lysine residues in histones H3 and H4.
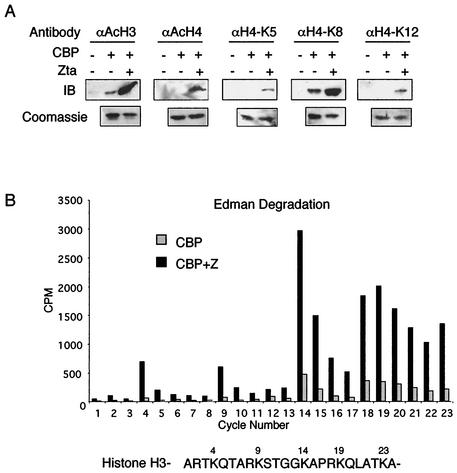
Specific lysine residues in histones H3 and H4 have been associated with particular functions in gene expression and chromatin assembly in various organisms and developmental models. To determine if Zta targets specific lysine residues for acetylation by CBP, we first utilized acetyl-lysine-specific antibodies in Western blots (Fig. 4A). SONs were assayed in vitro with recombinant CBP and Zta in the presence of unlabeled acetyl-coenzyme A and were subjected to Western blot analysis with various lysine-specific antibodies. Antibodies for the hyperacetylated forms of histone H3 and histone H4 showed significant stimulation of acetylated histones in the presence of Zta. We also found that antibodies specific for histone H4 K5 and K12 caused significantly increased acetylation after the addition of Zta. Histone H4 K8 acetylation was also stimulated by Zta, but to a lesser extent than other lysine residues. The acetylation of lysine residues on histone H3 was examined by Edman degradation of 3H-acetylated histones (Fig. 4B). In this analysis, we found that Zta stimulated acetylation of all lysines, including K4, K9, K14, K18, and K23. Interestingly, the greatest incorporation of the [3H]acetyl group per mole was at K14, K19, and K23, which are residues not typically thought to be subject to significant levels of acetylation. Edman degradation analysis of H4 lysines was not possible since the amino termini of H4 were blocked in HeLa cells. Nevertheless, these results indicate that Zta stimulates CBP acetylation of all lysine residues in the amino-terminal tails of histones H3 and H4 and may preferentially stimulate lysine residues at the C-terminal ends of the N-terminal tails.
FIG. 4.
Characterization of acetylated histone lysines stimulated by Zta. (A) Western blots of HAT reactions containing SONs, baculovirus CBP, and Zta as indicated. Acetylated histones were detected by Western blotting with antibodies specific for hyperacetylated H3, H4, or H4-acetyl K5, -acetyl K8, or -acetyl K12 as indicated (upper panel). A Coomassie stain of histone H3 is also shown (lower panel). (B) Histone H3 was assayed by N-terminal sequencing for incorporation of 3H. Histone H3 was isolated from sodium dodecyl sulfate-polyacrylamide electrophoresis gels from a reaction containing SONs, CBP, and [3H]acetyl-coenzyme A with or without Zta. 3H incorporation was measured in counts per minute per cycle.
The basic DNA binding domain of Zta is required for stimulation of HAT activity.
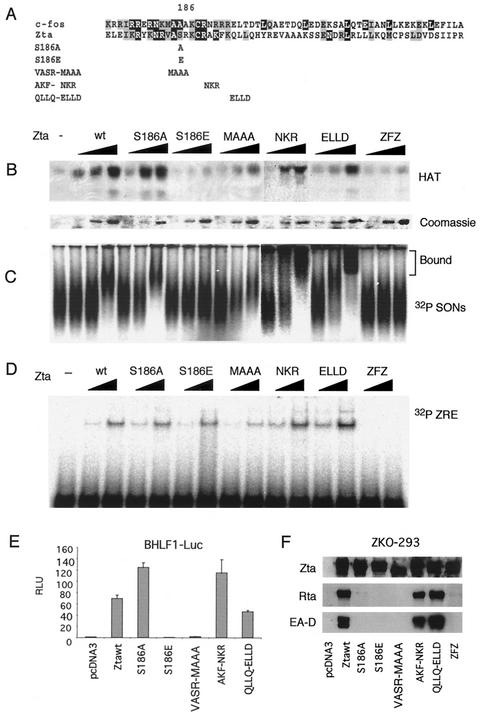
Our previous data suggested that the Zta bZIP domain provided a nucleosome binding function that is important for stimulation of CBP HAT activity (9). Substitution of the Zta basic region with the c-Fos basic region (ZFZ) has been shown by others to allow binding of Zta to AP-1 sites and stimulation of transcription from AP-1-responsive reporter genes to levels similar to those of wild-type Zta. ZFZ failed to activate lytic cycle gene expression since it could not bind to all ZRE sites. However, substitution of serine 186 with alanine resulted in the ability of Zta to bind ZRE sites and to activate EBV lytic promoters in transfected reporter plasmids but in its failure to activate chromatin-associated viral genes (21, 22). To determine if the Zta basic region conferred specific properties important for chromatin-specific reactivation function, we assayed several basic region substitution mutants for the ability to stimulate CBP nucleosomal HAT activity, bind nucleosomes and ZRE DNA, and stimulate transcription from plasmid or viral chromosomal targets (Fig. 5). Substitution mutants S186A, S186E, VASR-MAAA, AKF-NKR, QLLQ-ELLD, and ZFZ (Fig. 5A) were expressed and purified from E. coli, normalized by Coomassie staining, and assayed for the ability to stimulate acetylation of SONs by CBP (Fig. 5B). We found that S186E, MAAA, and ZFZ had significantly reduced stimulation of HAT activity. These same mutants were then assayed for nucleosome binding by an electrophoretic mobility shift assay (EMSA) with 32P-labeled SONs (Fig. 5C) (9). We found a perfect correlation between nucleosome binding and stimulation of HAT activity, suggesting that nucleosome binding is necessary for HAT stimulation by Zta. These substitution mutants were then assayed for binding to a ZRE from the BHLF1 promoter (Fig. 5D). We found that all of the substitution mutants, except for ZFZ, were capable of binding to this ZRE, indicating that nucleosomal binding was distinct from DNA binding. These same mutants were then assayed in transient transfection assays for activation of BHLF1 promoter fused to luciferase. Transcriptional activation of the transfected plasmid DNA correlated perfectly with the ability of Zta mutants to bind nucleosomes and stimulate CBP HAT activity (Fig. 5E). However, when these mutants were assayed for the ability to activate transcription from viral chromosomal genes, we found that S186A, in addition to S186E, MAAA, and ZFZ, was defective (Fig. 5F). This result is consistent with the findings of Francis et al. (21, 22), who showed that S186A is specifically defective for transcription of viral chromosome-associated genes. These findings with S186A suggest that stimulation of CBP HAT is not sufficient for transcriptional activation of viral chromosomal genes and that posttranslational modification of this residue may also be required for viral reactivation in vivo (4). Nevertheless, these data suggest that nucleosomal binding is a specific feature of the Zta basic region, is not found in the c-Fos basic region, and is essential for stimulation of CBP HAT activity.
FIG. 5.
The basic region of Zta is required for stimulation of HAT activity. (A) Substitution mutants of the Zta basic region. (B) HAT assay of SONs in the presence of recombinant CBP and substitution mutants of Zta as indicated above each panel. Coomassie staining of Zta substitution mutants purified from E. coli is also shown (lower panel). (C) Binding of Zta substitution mutants to 32P-labeled SONs measured by EMSA. (D) Binding of Zta substitution mutants to 32P-labeled ZRE measured by EMSA. (E) Transcription activation of the BHLF1-luciferase reporter plasmid in transient transfections. (F) Western blot of latently infected 293-ZKO cells transfected with Zta substitution mutants and assayed 48 h later by Western blotting with antibodies to Zta, Rta, or EA-D.
The mechanism by which Zta stimulates CBP HAT activity was further analyzed by use of Michaelis-Menten kinetics (Table 1). Purified baculovirus-expressed CBP was assayed in the presence or absence of Zta with SONs (ranging from 0.1 to 1 μg) to generate plots for 1 to 30 min of reaction time. Lineweaver-Burk analysis was used to calculate the Km and Vmax for three independent experiments. We found that the addition of Zta decreased the Km by an order of magnitude but had only an approximately twofold effect on Vmax. This suggests that the predominant mechanism of activation of Zta is to increase the effective local concentration of nucleosomes for HAT catalysis by CBP.
TABLE 1.
Michaelis-Menten kinetic analysis of CBP acetylation of oligonucleosomes in the presence or absence of Ztaa
| Expt no. |
Km (mM)
|
Vmax (min−1)
|
||
|---|---|---|---|---|
| − Zta | + Zta | − Zta | + Zta | |
| 1 | 3.2 × 10−3 | 3.2 × 10−4 | 1.2 × 10−3 | 2.0 × 10−3 |
| 2 | 3.2 × 10−3 | 1.2 × 10−5 | 3.9 × 10−4 | 7.4 × 10−4 |
| 3 | 3.0 × 10−3 | 5.3 × 10−5 | 4.5 × 10−4 | 9.4 × 10−4 |
Lineweaver-Burk plot analysis was used to generate quantities for Km and Vmax.
Zta stimulates histone acetylation in vivo.
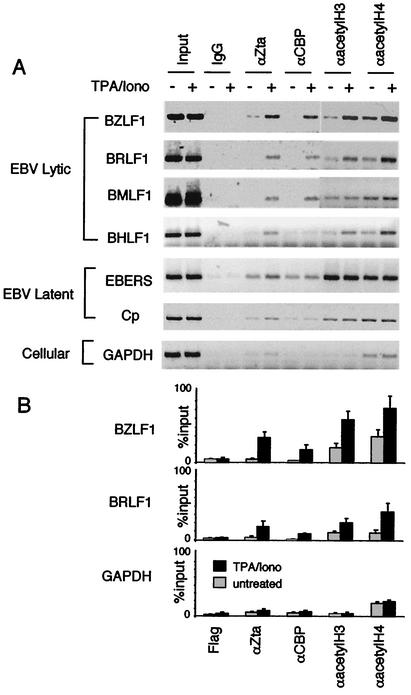
Latently infected Raji Burkitt lymphoma cells can be induced to enter the lytic cycle by treatment with TPA and ionomycin, which stimulates transcription of the BZLF1 promoter (49). This treatment has been shown by others to lead to hyperacetylation of the BRLF1 immediate-early promoters (8). However, in these studies it was not clear if TPA and ionomycin treatment involved Zta or CBP for gene activation and histone acetylation. We now show that TPA-ionomycin leads to an increased association of Zta and CBP with immediate-early promoters BZLF1 and BRLF1 in vivo by using a ChIP assay (Fig. 6). We also found that TPA-ionomycin leads to an increase in hyperacetylated histones H4 and H3 at the BZLF1 and BRLF1 promoter (Fig. 6). This result demonstrates that CBP functions in association with Zta at the immediate-early promoters during in vivo reactivation, and this correlates with an increase in histone acetylation.
FIG. 6.
Stimulation of CBP HAT activity in vivo. (A) Raji cells stimulated for reactivation with TPA-ionomycin were assayed by ChIP. Extracts from treated (+) or untreated (−) cells were immunoprecipitated with rabbit IgG (as a negative control) or antibodies specific for Zta, CBP, acetylated H3, or acetylated H4, as indicated. Immunoprecipitated DNA was assayed by PCR with primers specific for the promoter regions of EBV lytic genes (BZLF1, BRLF1, BMLF1, and BHLF1), EBV latent genes (EBERS and Cp), or the cellular GAPDH coding region. (B) Quantitation of three independent ChIP experiments is presented as percentages of input PCR product. All reactions were determined to be within the linear range of PCR (data not shown).
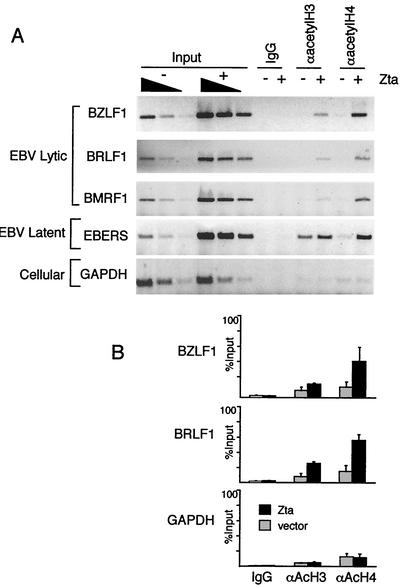
We have shown that Zta can stimulate CBP histone acetylation in vitro and transcription activation in vivo. To determine if Zta stimulates histone acetylation in vivo, we assayed histone acetylation by ChIP after transfection of latently infected D98/HR1 cells with a Zta expression plasmid (+) or control vector (−) (Fig. 7). Since Zta stimulates viral replication in these cell lines, input viral copy number was increased three- to ninefold after transfection with Zta (Fig. 7, input). ChIP revealed that histone H3 and H4 acetylation increased at EBV lytic promoters but not at a viral latent transcript (EBERS) or a cellular gene (glutaraldehyde-3-phosphate dehydrogenase [GAPDH]). Quantitation of the ChIP PCR products as percentages of input DNA revealed that lytic immediate-early promoters BZLF1 and BRLF1 were increased two- to fourfold with acetylated histones H3 and H4 (Fig. 7B). In contrast, Zta transfection did not stimulate histone acetylation at EBERS or GAPDH, and no PCR product was detectable with control IgG antibody (Fig. 7). These results indicate that Zta can stimulate acetylation of histones H3 and H4 at transcriptionally responsive lytic viral gene promoters.
FIG. 7.
Zta stimulates histone acetylation of viral lytic promoters in vivo. (A) D98/HR1 cells were transfected with Zta (+) or control expression vector (−) and fixed for ChIP assay at 48 h posttransfection. Chromatin immunoprecipitated with antibodies specific for hyperacetylated histone H3 or H4 was assayed for DNA by PCR with primers specific for EBV lytic promoters BZLF1, BRLF1, and BMRF1, EBV latent promoter EBERS, or cellular coding region GAPDH. (B) Quantitation of three independent experiments is presented as percentages of input PCR DNA product.
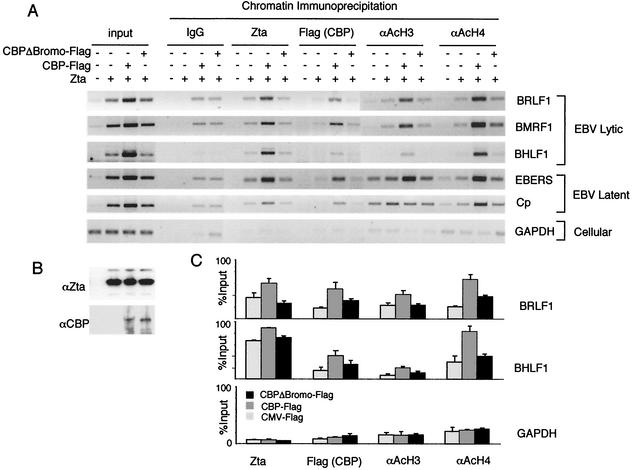
The ChIP assay was next used to determine if the bromodomain of CBP was important for histone acetylation at EBV-responsive promoters in vivo (Fig. 8). Zta was cotransfected with the CBP-Flag or CBP-ΔBromo-Flag expression vector into highly transfectable ZKO-293 cells. As expected, the addition of Zta increased the input copy number for EBV DNA. Interestingly, the addition of CBP, but not CBP-ΔBromo, further increased input DNA levels, suggesting that CBP stimulates DNA replication. ChIP assays with antibodies to Zta, Flag (CBP), and acetylated histones H3 and H4 revealed a striking increase at EBV lytic promoters (BRLF1, BMRF1, and BHLF1) relative to latent transcripts (EBERS and Cp) or cellular genes (GAPDH). By adjusting PCR levels as percentages of input DNA to normalize for replicating genomes, we found a consistent two- to fourfold increase of Zta, CBP, and acetylated histones at EBV lytic promoters. Importantly, this increase was dependent upon the bromodomain, since transfection of CBP-ΔBromo had no significant effect on histone acetylation or promoter association of Zta or CBP. CBP and CBP-ΔBromo were expressed at similar levels and did not affect Zta expression levels, as determined by Western blotting (Fig. 8B). These results indicate that the CBP bromodomain is required for the increase in histone acetylation at Zta-responsive promoters and for the stable association of Zta and CBP with promoter DNA in vivo.
FIG. 8.
CBP bromodomain is required for enhancement of histone acetylation and promoter binding of Zta and CBP. (A) ChIP assay of ZKO-293 cells transfected with Zta or control expression vector, CBP-Flag, or CBPΔBromo-Flag, as indicated above the lanes. ChIPs with rabbit IgG (negative control) or antibodies specific for Zta, Flag-tagged CBP, or acetylated histones H3 and H4 were assayed by PCR with primers for the genes indicated to the right. (B) Western blot of transfected cells indicating that equivalent levels of Zta and Flag (CBP transgenes) were expressed. (C) Quantitation of three independent ChIP experiments is presented as percentages of corresponding input DNA. Samples lacking Zta were not within the linear range in these reactions and therefore were not quantitated in these experiments.
DISCUSSION
Histone acetylation is thought to be a prerequisite for transcriptional activation of genes that may be silenced by heritable chromatin structures (5, 28). Transcriptional coactivators with intrinsic HAT activity can be recruited by sequence-specific DNA binding proteins to alter the local histone acetylation pattern and to stimulate transcription complex assembly (28, 51). The mechanism by which HAT complexes facilitate sequence-specific DNA binding of transcription factors remains poorly understood, since it is not clear how the initial sequence recognition occurs in higher-order chromatin structures that limit DNA accessibility. One simple mechanism that would overcome this problem is for initiating activators to recruit HAT complexes to nucleosome arrays found in repressed chromatin. Our data suggest that the EBV immediate-early bZIP protein Zta stimulates CBP acetylation of nucleosomal chromatin to enhance transcription complex assembly at viral lytic gene promoters.
Previously, it was shown that a class of bZIP proteins which included Zta, C/EBP-β, and NF-E2 was capable of stimulating CBP HAT activity directed towards oligonucleosomes (9). Here we further characterized the ability of Zta to stimulate CBP HAT activity. Most significantly, we found that the CBP bromo- and C/H3 domains were important for Zta activation of transcription and nucleosomal HAT activity. The bromodomain was required for both Zta and NF-E2 stimulation, but the C/H3 domain deletion had a greater effect on Zta activation, suggesting that Zta and NF-E2 may stimulate HAT activity by different mechanisms. Zta binds to the C/H1 and C/H3 domains of CBP with near equal affinity in vitro (55). Deletion of the C/H3 domain may reduce CBP binding to Zta, but it is unlikely to eliminate the strong interaction with C/H1 and therefore is unlikely to eliminate CBP-Zta association in vivo. We suggest that Zta binding to two domains of CBP provides allosteric enhancement of nucleosome-specific histone acetylation. Kinetic studies showed that Zta had a weak but reproducible effect on the Vmax of CBP HAT activity, suggesting that some stimulation of catalysis may exist (Table 1). However, the predominant effect of Zta was on the Km, which was reduced ∼10-fold in the presence of Zta. This suggests that Zta alters the substrate accessibility, either by recruitment of CBP to nucleosomes or by altering substrate-enzyme interactions to make acetylation more favorable. A similar observation was made with RpAp48, a histone binding factor that stimulates CBP HAT domain targeting to core histones and nucleosomes (56). Although simple recruitment of CBP to nucleosomes can stimulate nucleosome acetylation, we consider it more likely that Zta facilitates the accessibility of nucleosomal histone tails to the catalytic HAT domain of CBP. However, direct evidence for any conformational change in CBP or in the histone tails after the addition of Zta awaits further experimentation.
Bromodomains from several different proteins have been shown to interact with acetylated histones, and it has been proposed that the bromodomain four-helix bundles bind acetylated lysine (17, 31, 54). The p300 bromodomain, which is most closely related to the CBP bromodomain, was shown to be important for transcription initiation and responsiveness to estrogen receptor activation in vitro (35). The p300 bromodomain was also found to bind to chromatin and was essential for a stable, template-committed transcription complex (38). The bromodomains of Swi2 and GCN5 were also found to be required for anchoring of these chromatin-modifying complexes to acetylated chromatin templates (29). Binding to acetylated lysine may be important for the recognition of nucleosomal substrates, since we also observed that CBPΔBromo had a reduced ability to acetylate oligonucleosomes relative to free histones (Fig. 2). We suggest that the bromodomain facilitates processive targeting of multiple lysine residues in the amino-terminal tails of all four histones. Bromodomain association with acetylated lysines may help target adjacent lysine residues on neighboring histone tails. We hypothesize that Zta facilitates this process by interactions with the C/H1 and C/H3 domains of CBP as well as by providing essential interactions with nucleosomes.
Several lines of evidence suggest that the bZIP domain of Zta interacts with nucleosomes to facilitate histone acetylation by CBP. Zta bound HeLa-derived oligonucleosomes in vitro and stimulated histone acetylation of nucleosomes assembled on defined plasmid templates lacking known high-affinity ZRE sites (Fig. 3). Nucleosome assembly in vitro inhibited CBP acetylation of core histones, which could be reversed by the addition of Zta, suggesting that Zta altered the accessibility of histone tails in packed nucleosomes. Substitution of the Zta basic region with the c-Fos basic region eliminates the ability to target SONs for acetylation. Mutation of a single amino acid residue in the Zta bZIP domain from serine to glutamic acid inhibited nucleosome binding and HAT activation but did not eliminate ZRE DNA binding (Fig. 5). This suggests that S186 is important for nucleosome-specific binding and HAT activation. Interestingly, mutation of Zta serine 186 to alanine had no effect on HAT activity but eliminated transcription activation of viral chromosomal, but not plasmid-borne, genes. The complex behavior of S186 mutations suggests that additional events, like phosphorylation, may regulate the nucleosome binding and activation properties of Zta in vivo (4).
ChIP assays with TPA-ionomycin-induced Raji cells or Zta-transfected cells indicated that histone acetylation was increased at specific viral lytic promoters but not at viral latent transcripts or at cellular genes. Zta is known to stimulate transcription from these viral lytic genes (16), indicating that a good correlation exists between transcription activation and histone acetylation of Zta-responsive promoters. Interestingly, we found that cotransfection of wild-type CBP, but not CBP-ΔBromo, increased histone acetylation of Zta-responsive genes, as well as increased the promoter association of Zta and CBP themselves. Thus, transcription activation is correlated with increased occupancy of the activator-coactivator complex. This in vivo observation is consistent with recent biochemical studies showing that the bromodomain anchors coactivators to nucleosomal templates (29). The bromodomain-dependent increase in Zta binding to Zta-responsive genes suggests that CBP and Zta function in concert to select specific promoter sequences in nucleosome-repressed chromatin.
Higher-order chromatin structures can present a significant barrier to transcription initiation and DNA recognition by sequence-specific DNA binding proteins. Nucleosome binding and chromatin opening activity have been described for the “pioneering” proteins HNF3 and GATA4, which initiate liver-specific gene expression during development (12). Zta may also function as a pioneering protein with a different mechanism of action. Our data suggest that Zta recruits CBP to compact chromatin to induce transient histone acetylation, which in turn facilitates scanning for high-affinity Zta binding sites in compact chromatin. This pioneering function may be shared by other bZIP proteins, like C/EBP-β and NF-E2, that have a similar ability to target the nucleosomal HAT activity of CBP (16). The results presented here demonstrate the importance of the bromodomain as a nucleosome targeting element that may facilitate processive acetylation of histones in compact chromatin and promoter scanning by sequence-specific activators such as Zta. Finding the structural basis through which Zta facilitates nucleosome acetylation in a bromo- and C/H3 domain-dependent manner remains the work of future studies.
Acknowledgments
We thank W. Wunner and the Vector Expression Core at the Wistar Institute for baculovirus infections and acknowledge the technical support of Latasha Day.
This work was supported by a grant from NIH (CA-85678).
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J. Virol. 73:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annunziato, A. T., and J. C. Hansen. 2000. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 9:37-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, M., H. Mischak, S. Dammeier, W. Kolch, O. Gires, D. Pich, R. Zeidler, H. J. Delecluse, and W. Hammerschmidt. 1998. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J. Virol. 72:8105-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Blobel, G. 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 7.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C.-J., Z. Deng, A. Y. Kim, G. A. Blobel, and P. M. Lieberman. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevallier, G. A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi, T., and M. Carey. 1993. The ZEBRA activation domain: modular organization and mechanism of action. Mol. Cell. Biol. 13:7045-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo, L. A., F. R. Lin, I. Cuesta, M. Jarnik, and K. S. Zaret. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9:279-289. [DOI] [PubMed] [Google Scholar]
- 13.Cirillo, L. A., and K. S. Zaret. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4:961-969. [DOI] [PubMed] [Google Scholar]
- 14.Cote, J., R. T. Utley, and J. L. Workman. 1995. Analysis of transcription factor binding to nucleosomes. Methods Mol. Genet. 6:108-128. [DOI] [PubMed] [Google Scholar]
- 15.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, Z., C.-J. Chen, D. Zerby, H.-J. Delecluse, and P. M. Lieberman. 2001. Identification of acidic and aromatic residues in the Zta activation domain essential for Epstein-Barr virus reactivation. J. Virol. 75:10334-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 18.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemington, E. K., A. M. Borras, J. P. Lytle, and S. H. Speck. 1992. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J. Virol. 66:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francis, A., T. Ragoczy, L. Gradoville, L. Heston, A. El-Guindy, Y. Endo, and G. Miller. 1999. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of early lytic cycle genes and synergy with the Epstein-Barr virus R transactivator. J. Virol. 73:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francis, A. L., L. Gradoville, and G. Miller. 1997. Alteration of a single serine in the basic domain of Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii-Nakata, T., Y. Ishimi, A. Okuda, and A. Kikuchi. 1992. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem. 267:20980-20986. [PubMed] [Google Scholar]
- 24.Gao, Z., A. Krithivas, J. E. Finan, O. J. Semmes, S. Zhou, Y. Wang, and S. D. Hayward. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman, P. S., V. K. Tran, and R. H. Goodman. 1997. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog. Horm. Res. 52:103-119. [PubMed] [Google Scholar]
- 26.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 27.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 28.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 29.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 30.Howe, L., C. E. Brown, T. Lechner, and J. L. Workman. 1999. Histone acetyltransferase complexes and their link to transcription. Crit. Rev. Eukaryot. Gene Expr. 9:231-243. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, P. J., U. K. Binne, and P. J. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenney, S., J. Kamine, E. Holley-Guthrie, J.-C. Lin, E.-C. Mar, and J. Pagano. 1988. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J. Virol. 63:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 35.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, G. 1990. The switch between latency and replication of Epstein-Barr virus. J. Infect. Dis. 161:833-844. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa, T., M. Bulger, M. Muramatsu, and T. Ito. 2001. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J. Biol. Chem. 276:27384-27391. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 42.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 43.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 44.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 45.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schepers, A., D. Pich, J. Mankertz, and W. Hammerschmidt. 1993. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 67:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, Y., and C. Mello. 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12:943-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shikama, N., J. Lyon, and N. B. La Thangue. 1997. The p300/CBP family: integration signals with transcription factors and chromatin. Trends Cell Biol. 7:230-236. [DOI] [PubMed] [Google Scholar]
- 49.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 50.Thanos, D., and S. Lomavardes. 2002. Opening chromatin. Mol. Cell 9:209-211. [DOI] [PubMed] [Google Scholar]
- 51.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcription activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 52.Wensing, B., A. Stuher, P. Jenkins, M. Hollyoake, C. E. Karstegl, and P. J. Farrell. 2001. Variant chromatin structure of the OriP region of Epstein-Barr virus and regulation of EBER1 expression by upstream sequences and OriP. J. Virol. 75:6235-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 54.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 55.Zerby, D., C.-J. Chen, E. Poon, D. Lee, R. Shiekhattar, and P. M. Lieberman. 1999. The amino-terminal C/H1 domain of CREB binding protein mediates Zta transcription activation of latent Epstein-Barr virus. Mol. Cell. Biol. 19:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Q., N. Vo, and R. H. Goodman. 2000. Histone binding protein RpAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol. Cell. Biol. 20:4970-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]