Abstract
Although DNA polymerase η (Polη) and other Y family polymerases differ in sequence and function from classical DNA polymerases, they all share a similar right-handed architecture with the palm, fingers, and thumb domains. Here, we examine the role in Saccharomyces cerevisiae Polη of three conserved residues, tyrosine 64, arginine 67, and lysine 279, which come into close contact with the triphosphate moiety of the incoming nucleotide, in nucleotide incorporation. We find that mutational alteration of these residues reduces the efficiency of correct nucleotide incorporation very considerably. The high degree of conservation of these residues among the various Y family DNA polymerases suggests that these residues are also crucial for nucleotide incorporation in the other members of the family. Furthermore, we note that tyrosine 64 and arginine 67 are functionally equivalent to the deoxynucleotide triphosphate binding residues arginine 518 and histidine 506 in T7 DNA polymerase, respectively.
The Y family DNA polymerases (Pols) differ from others in their low fidelity and ability to replicate through DNA lesions. Among the eukaryotic Y family polymerases, Polη is unique in its proficient ability to replicate through UV-induced cyclobutane pyrimidine dimers and other DNA lesions (6, 8, 9, 18). Because of its role in the error-free bypass of cyclobutane pyrimidine dimers, inactivation of Polη in humans (7, 13) causes an increase in UV mutagenesis (17, 21) and results in the cancer-prone syndrome, the variant form of xeroderma pigmentosum. Polη misincorporates nucleotides with a frequency of ∼10−2 to 10−3 on undamaged DNA (9, 19), and pre-steady-state kinetic studies with yeast Polη have indicated that it discriminates poorly between the correct and incorrect nucleotides both at the initial nucleotide binding step and at the subsequent rate-limiting conformational change step (20).
The Y family DNA polymerases contain five conserved sequence motifs, designated I to V. The crystal structures of Saccharomyces cerevisiae Polη (16), as well as those of Dbh and Dpo4 from Sulfolobus solfataricus (12, 15, 22), reveal a polydactyl right-handed architecture, in which motifs I and III map to the palm domain, motif II is part of the fingers domain on the left side of the palm, motif IV forms a helix lying atop the palm domain on the right side, and motif V is part of the thumb domain. Motifs I and III carry the catalytic residues (e.g., Asp30, Asp155, and Glu156 in yeast Polη) and are structurally analogous to motifs A and C in classical DNA polymerases such as the Klenow fragment of Escherichia coli and T7 DNA polymerase (3, 4, 14). However, these structures reveal a fingers domain that is radically different from that in other polymerases; moreover, the active site of these polymerases remains quite open in the ternary complex with the duplex DNA and the incoming nucleotide. This raises the intriguing question of whether Y family polymerases utilize a mode for nucleotide binding that is different from that used by classical polymerases. Here, we have mutated conserved residues that come close to the incoming nucleotide and have measured steady-state kinetic parameters for nucleotide incorporation. These results reveal unexpected commonalities (and differences) in the basic mechanism by which the incoming nucleotide is bound by the Y family and classical DNA polymerases.
MATERIALS AND METHODS
Generation and purification of Polη mutant proteins.
To generate point mutations in the yeast Polη (1-513) protein, a 1.5-kb HindIII/XbaI DNA fragment containing the yeast RAD30 open reading frame from codons 1 to 513 was cloned into a derivative of YIpLac128, generating plasmid pBJ928. All mutations were generated by PCR amplification of pBJ928 by using the Quick Change kit (Stratagene). For the Y64F and Y64A mutations, the oligonucleotide pair LP194(5′-CCATTATTGCAGTCTCTTTTGCAGCAAGAAAGTATGGTATATC-3′) and LP195 (5′GA TATACCATACTTTCTTGCTGCAAAAGAGACTGCAATAATGG-3′) and the oligonucleotide pair LP196 (5′-CCATTA TTGCAGTCTCTGCTGCAGCAAGAAAGTATGG-3′) and LP197 (5′-CCATACTTTCT TGCTGCAGCAGAGACTGCAATAATGG-3′) were used, respectively. For the R67A and K279A mutations, the oligonucleotide pair LP198 (5′-GTCTCTTATGCAGCAGCAAAGTATGGTATA TCAAGG-3′) and LP199 (5′CCTTGATATACCATACTTTGCTGCTGCATAAGAGAC-3′), and the oligonucleotide pair LP202 (5′-GCTTCTAATTACAAAGCACCTGATGCCCAAACAATTGTG-3′) and LP203 (5′-CACAATTGTTTGGGCATCAGGTGCTTTGTAATTAGAAGC-3′) were used, respectively. The 512-bp EcoRI/BglII DNA fragments harboring either the Y64F, Y64A, or R67A mutations were then used to replace the corresponding wild-type regions in plasmid pBJ928. The 337-bp BglII/MscI DNA fragment harboring the K279A mutation was used to replace its corresponding fragment in plasmid pBJ928. All PCR-generated fragments were sequenced to confirm the presence of each mutation. Subsequently, each Polη mutant gene was cloned in frame with the glutathione S-transferase gene under the control of a galactose-inducible PGK promoter in plasmid pBJ842. Plasmids pR30.180 (Rad30 [1-513], wild type), pR30.195 (Rad30 [1-513], Y64F), pR30.197 (Rad30 [1-513], Y64A), pR30.198 (Rad30 [1-513], R67A), pR30.202 (Rad30 [1-513], K279A) were transformed into yeast strain BJ5464, and cells were grown and induced as described previously (9). Proteins were affinity purified from yeast by using glutathione Sepharose 4B (Amersham Pharmacia) as described previously (9), and non-glutathione S-transferase-tagged proteins were batch eluted from the column by treatment with Prescission protease (Amersham Pharmacia). Cleavage of proteins by Prescission protease leaves a seven-residue N-terminal nonrelated leader peptide attached to each Polη protein.
DNA polymerase assays.
The standard DNA polymerase reaction mixture (5 μl) contained 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol, 100 μg of bovine serum albumin/ml, 10% glycerol, 50 μM concentrations of deoxynucleotide (dGTP, dATP, dTTP, and dCTP), and 10 nM of 5′-32P-labeled oligonucleotide primer annealed to an oligonucleotide DNA template. For steady-state kinetic analyses, 0.05 nM yeast Polη was used. Reactions were carried out at 30°C for 5 min and terminated by the addition of 4 volumes of loading buffer (95% formamide, 0.05% cyanol blue, 0.05% bromophenol blue, and 20 mM EDTA) before resolving on 12% polyacrylamide gels containing 8 M urea. Gels were dried before autoradiography at −70°C. DNA primer-template substrates were composed of the oligodeoxynucleotide primer, 5′-CGACG ATGCT CCGGTACTCC AGTGT AGGCA-3′, annealed to one of the following four template (53-mer) oligodeoxynucleotides: template G, 5′-ATGCCTGCAC GAAGAGTTCC TAGTG CCTACACTGG AGTACCGGAG CATCGTCG; template A, 5′-ATGCCTGCAC GAAGA GTTCG CTATGCCTAC ACTGGAGTAC CGGAGCATCGTCG; template T, 5′-ATGCC TGCACGAAGA GTTCAGCTTGCCTAC ACTGGAGTACCGGAG CATCGTCG; template C, 5′-ATGCCTGCAC GAAGAGTTCT AGCTGCCTAC ACTGGAGTAC CGGAG CATCGTCG. The primer was 5′ 32P end labeled by using polynucleotide kinase (Roche Molecular Biochemicals) and [γ-32P]ATP (Amersham Pharmacia Biotech).
Steady-state kinetic analysis.
Steady-state kinetic analyses for deoxynucleotide incorporation were done as described previously (2). The standard DNA polymerase assay was used, except that only a single deoxynucleotide was included. Gel band intensities of the substrate and products of the deoxynucleotide incorporation reactions were quantitated by using a PhosphorImager and the ImageQuant software (Molecular Dynamics). The observed rate of deoxynucleotide incorporation, vobs, was determined by dividing the amount of product formed by the reaction time. We plotted vobs as a function of the deoxynucleotide triphosphate (dNTP) concentration and fit these data to the Michaelis-Menten equation describing a hyperbola: vobs = (Vmax × [dNTP])/(Km + [dNTP]). From the best-fit curve, the apparent Km and Vmax steady-state kinetics parameters were obtained for the incorporation of deoxynucleotides by the wild-type and mutant yeast Polη proteins and the efficiencies of nucleotide incorporation (Vmax/Km) were determined.
RESULTS
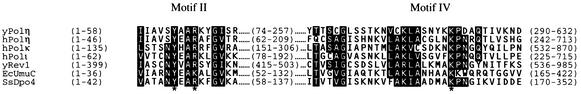
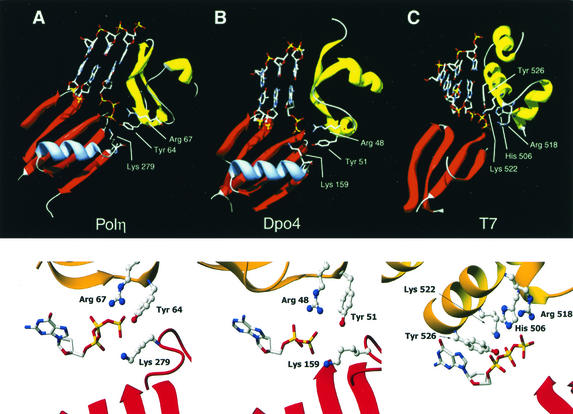
Figure 1 shows the alignment of motifs II and IV of several members of the Y family DNA polymerases. S. cerevisiae Polη has a tyrosine residue at position 64 and an arginine at position 67 in motif II and a lysine at position 279 in motif IV. All these residues are invariant among all the different Y family members, the only exception being the presence of a lysine residue in E. coli UmuC corresponding to arginine 67 in yeast Polη. The conservation of tyrosine 64 and arginine 67 is underscored by the inability to isolate biologically active mutations of the corresponding residues in human Polη in a random mutational analysis of motif II residues (5). Figure 2 shows these invariant residues as observed in the crystal structure of two Y family DNA polymerases, yeast Polη and the S. solfataricus Dpo4 protein. In the case of yeast Polη, based on the structural homology of its palm domain with that of T7 DNA polymerase, the positions of the DNA and the incoming nucleotide were modeled into the active site via the positions of the catalytic acidic residues (16), and in the case of Dpo4 protein, the DNA and incoming nucleotide were cocrystallized with the protein (12). In both cases, the conserved tyrosine, arginine, and lysine residues each come in close proximity to the triphosphate moiety of the incoming nucleotide. To determine the roles of these highly conserved tyrosine (Y), arginine (R), and lysine (K) residues in the Y family of DNA polymerases, we have mutated these residues in yeast Polη to alanine (A) and determined their effects on DNA synthesis and the efficiency of nucleotide incorporation. To determine whether the -OH group on tyrosine is critical for Polη function, we also made a mutation of tyrosine 64 to phenylalanine (F).
FIG. 1.
Alignment of motifs II and IV of Y family DNA polymerases. Highly conserved residues are highlighted. The asterisks indicate the positions of the conserved tyrosine and arginine residues in motif II and the conserved lysine in motif IV that were mutated in yeast Polη.
FIG. 2.
Contacts between the conserved residues and the incoming nucleotide. (A) Overview (top) and close-up view (bottom) of the positions of Tyr 64, Arg 67, and Lys 279 of yeast Polη in relation to the incoming deoxynucleotide, as predicted from the crystal structure. Tyr 64 and Arg 67 lie in the finger domain, and Lys 279 lies in a helix-turn above the palm domain. The DNA and nucleotide were modeled into the Polη structure by superposition of the Polη palm domain onto the equivalent domain in the T7 Pol/template-primer/ddGTP ternary complex. (B) Overview (top) and close-up view (bottom) of the homologous conserved Tyr, Arg, and Lys residues in the crystal structure of S. solfataricus Dpo4 protein with DNA and incoming dNTP. In this structure, the incoming ddATP was hydrolyzed to ddADP (12). (C) Overview (top) and close-up view (bottom) of amino acid residues in the T7 Pol/template-primer/ddGTP crystal structure that contact the incoming dNTP. In all cases, only the relevant regions of the fingers (yellow/gold) and palm (red) domains are shown.
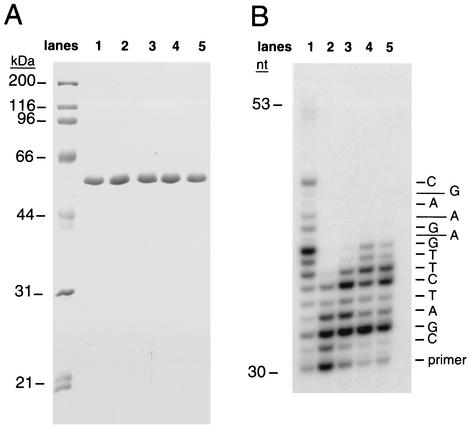
Figure 3 shows the purified wild-type and mutant yeast Polη proteins and their respective DNA synthesis activities. While all four mutants exhibit some DNA synthesis activity, it is reduced relative to that of the wild-type protein. The Y64A and Y64F mutations have the greatest effect, while the R67A and K279A mutations have a somewhat lesser effect on DNA synthesis activity.
FIG. 3.
Effects of mutations in the conserved Tyr 64, Arg 67, and Lys 279 residues in yeast Polη (1-513) on DNA synthesis. (A) Purified yeast Polη proteins. Lane 1, wild-type Polη; lane 2, Polη Y64F; lane 3, Polη Y64A; lane 4, Polη R67A; lane 5, Polη K279A. Molecular mass markers are shown on the left. (B) DNA synthesis by various yeast Polη proteins. Lane 1, wild-type Polη; lane 2, Polη Y64F; lane 3, Polη Y64A; lane 4, Polη R67A; lane 5, Polη K279A. Each protein (0.5 nM) was incubated with a DNA substrate (10 nM) and each of the four dNTPs (50 μM) for 5 min at 30°C. Template C was the substrate, and a portion of the sequence is shown on the right.
To determine the effects of these mutations on the efficiency of DNA synthesis, we analyzed the steady-state kinetic parameters for nucleotide incorporation for each mutant protein. As shown in Table 1, each of the mutations lowered the efficiency of nucleotide incorporation: for example, for the incorporation of C opposite template G, the Y64F mutant enzyme was 17-fold less efficient than the wild-type enzyme. In fact, the Y64A, R67A, and K279A mutant enzymes were all similarly reduced in their efficiency for the incorporation of C opposite template G. The incorporation of G opposite template C was reduced ∼20- to 60-fold for the various mutant enzymes. For the incorporation of T opposite template A, the Y64F and Y64A mutations both reduced the efficiency of nucleotide incorporation ∼140-fold compared with the 40- to 50-fold reductions seen for the R67A and K279A mutant enzymes. The incorporation of A opposite template T was reduced ∼70- to 80-fold for the Y64F and Y64A mutations, 100-fold for the R67A mutation, and ∼30-fold for the K279A mutation. Overall, the mutation of tyrosine 64 to either alanine or phenylalanine had the greatest effect and mutation of lysine 279 to alanine had the weakest effect. Furthermore, the reduction in the efficiency of nucleotide incorporation for all the mutant enzymes is somewhat base pair specific. For instance, for the formation of a G-C base pair, on average, there was an ∼25-fold reduction in efficiency of nucleotide incorporation by the mutant enzymes, whereas for A-T base pair formation, on average, the mutant proteins exhibited an ∼90-fold reduction in the efficiency of nucleotide incorporation. These results suggest that Watson-Crick hydrogen bonding makes a greater contribution to nucleotide incorporation by these mutant Polη proteins than by wild-type protein.
TABLE 1.
Steady-state kinetic parameters for correct nucleotide incorporation by yeast wild-type and mutant Polη proteins
| Polη protein | Template base | Incoming nucleotide | Vmax (nM/min) | Km (μM) | Vmax/Km | Relative efficiency | Fold reduction in efficiency |
|---|---|---|---|---|---|---|---|
| Wild type | G | dCTP | 0.9 ± 0.09 | 6.6 ± 1.2 | 0.14 | 1 | |
| Y64F | G | dCTP | 0.5 ± 0.01 | 59 ± 2.5 | 0.008 | 0.06 | 17 |
| Y64A | G | dCTP | 0.2 ± 0.004 | 20 ± 1.7 | 0.01 | 0.07 | 14 |
| R67A | G | dCTP | 0.5 ± 0.05 | 63 ± 13 | 0.008 | 0.06 | 17 |
| K279A | G | dCTP | 0.3 ± 0.01 | 32 ± 3.4 | 0.009 | 0.06 | 17 |
| Wild type | A | dTTP | 0.44 ± 0.04 | 7.9 ± 1.5 | 0.06 | 1 | |
| Y64F | A | dTTP | NDa | ≥250 | 0.0004b | 0.007 | 142 |
| Y64A | A | dTTP | ND | ≥250 | 0.0004b | 0.007 | 142 |
| R67A | A | dTTP | 0.3 ± 0.02 | 282 ± 26 | 0.001 | 0.02 | 50 |
| K279A | A | dTTP | 0.3 ± 0.03 | 196 ± 41 | 0.0015 | 0.025 | 40 |
| Wild type | T | dATP | 0.5 ± 0.06 | 8.8 ± 1.9 | 0.06 | 1 | |
| Y64F | T | dATP | 0.1 ± 0.01 | 109 ± 14 | 0.0009 | 0.015 | 67 |
| Y64A | T | dATP | 0.1 ± 0.008 | 143 ± 16 | 0.0007 | 0.012 | 83 |
| R67A | T | dATP | ND | ≥250 | 0.0006b | 0.01 | 100 |
| K279A | T | dATP | 0.2 ± 0.01 | 99 ± 13 | 0.002 | 0.03 | 33 |
| Wild type | C | dGTP | 0.8 ± 0.09 | 13.8 ± 2.6 | 0.06 | 1 | |
| Y64F | C | dGTP | 0.3 ± 0.003 | 148 ± 4 | 0.002 | 0.03 | 33 |
| Y64A | C | dGTP | 0.2 ± 0.004 | 170 ± 5 | 0.001 | 0.017 | 59 |
| R67A | C | dGTP | 0.3 ± 0.006 | 126 ± 6.3 | 0.002 | 0.03 | 33 |
| K279A | C | dGTP | 0.2 ± 0.02 | 70 ± 9 | 0.003 | 0.05 | 20 |
ND, not determined.
Since nucleotide incorporation remained linear throughout the dNTP concentration, the efficiency was determined from the slope of the line.
DISCUSSION
These studies provide evidence for a role of the Y64, R67, and K279 residues of yeast Polη in nucleotide incorporation. The high degree of conservation of Y64, R67, and K279 among the various Y family DNA polymerases strongly suggests that these residues are also crucial for nucleotide incorporation in the other members of this family. These residues are spread out in motifs II (Y64 and R67) and IV (K279), which map to the fingers and palm domains, respectively. In contrast, the conserved residues in E. coli Klenow and T7 DNA polymerase that bind dNTP are restricted solely to the fingers domain, namely to the O and N helices. In T7 DNA polymerase, for instance, the triphosphate binding residues R518, Y526, and K522 are present in the O helix while H506 is present in the N helix (Fig. 2) (4). Mutations of the analogous residues R754, F762, K758, and H734 in Klenow diminish the nucleotide incorporation ability of this polymerase (1, 10). The fingers domain of Y family DNA polymerases is radically different from that of classical DNA polymerases, being small and stubby and lacking the equivalent of O helix. Nonetheless, Y64 in Polη appears to be a functional equivalent of R518 in T7 DNA polymerase, where it is in a position to contact the γ-phosphate of the incoming nucleotide (Fig. 2). Thus, even though Y64 and R518 originate from different segments of their respective fingers domains, their terminal hydrogen bonding groups wind up in close proximity, the distance between the Y64 -OH group and R518 Nη2 atom being only ∼1.5 Å (based on an alignment of the structurally homologous palm domains). Interestingly, in the structure of Y family polymerase Dpo4 with DNA and incoming nucleotide (12), the incoming ddATP was unexpectedly hydrolyzed to ddADP, and therefore the structure did not yield information on the putative interactions between the conserved tyrosine and the γ-phosphate (Fig. 2). The kinetic data reported here support a direct role for this conserved tyrosine in dNTP binding for which, for instance, the Y64A and Y64F mutations show the lowest efficiency in nucleotide incorporation. The second conserved residue in Polη, R67, is in a position to contact the β-phosphate of the incoming nucleotide, in much the same way as H506 in T7 DNA polymerase (Fig. 2). The two residues structurally overlap, such that the distance between the R67 Nη2 atom and the His506 Nɛ2 atom is only 0.8 Å. Thus, in spite of very different fingers domains, two residues (Y64 and R67) in motif II of Polη play roles in dNTP binding that are functionally equivalent to those of the two conserved residues in motif B of Klenow (R754 and H734) and T7 DNA polymerase (R518 and H506). The third conserved residue, K279, stems from a helix- turn that is unique to Y family DNA polymerases. This helix- turn is situated above the central β-sheet of the palm domain, and it selectively positions the lysine for contacts to the γ-phosphate of dNTP, in addition to the contacts made by Y64 (Fig. 2). Conversely, T7 DNA polymerase contains two conserved residues, K522 and Y526, which contact the α- and β-phosphates of dNTP, respectively (4), and lack counterparts in Y family DNA polymerases (Fig. 2).
Overall, in spite of the very considerable sequence, structural, and functional differences between the Y family and classical DNA polymerases, the basic mechanisms of catalysis and to some extent dNTP binding have been preserved among them. The functional similarity of motifs II and B in dNTP binding reported here extends conservation between motifs I and III of Y family DNA polymerases and motifs A and C of classical DNA polymerases (3, 11). Motifs I/A and III/C form part of the palm domain that contains the invariant catalytic acidic residues in all these polymerases (4, 12, 16). Polη and other Y family polymerases (12, 16), however, differ from the classical high-fidelity DNA polymerases (4) in having a much more open active site, which makes less intimate contacts with the base of the incoming dNTP and the templating residue, and that likely accounts for their low fidelity and damage bypass ability.
Acknowledgments
This work was supported by NIH grants GM19261 (L.P.) and CA94006 (A.K.A. and L.P.).
REFERENCES
- 1.Astatke, M., N. D. F. Grindley, and C. M. Joyce. 1995. Deoxynucleoside triphosphate and pyrophosphate binding sites in the catalytically competent ternary complex for the polymerase reaction catalyzed by DNA polymerase I (Klenow fragment). J. Biol. Chem. 270:1945-1954. [DOI] [PubMed] [Google Scholar]
- 2.Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262:232-256. [DOI] [PubMed] [Google Scholar]
- 3.Delarue, M., O. Poch, N. Tordo, D. Moras, and P. Argos. 1990. An attempt to unify the structure of polymerases. Protein Eng. 3:461-467. [DOI] [PubMed] [Google Scholar]
- 4.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 391:251-258. [DOI] [PubMed] [Google Scholar]
- 5.Glick, E., K. L. Vigna, and L. A. Loeb. 2001. Mutations in human DNA polymerase η motif II alter bypass of DNA lesions. EMBO J. 20:7303-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haracska, L., S.-L. Yu, R. E. Johnson, L. Prakash, and S. Prakash. 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25:458-461. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 10.Kaushik, M., V. N. Pandey, and M. J. Modak. 1996. Significance of the O-helix residues of Escherichia coli DNA polymerase I in DNA synthesis: dynamics of the dNTP binding pocket. Biochemistry 35:7256-7266. [DOI] [PubMed] [Google Scholar]
- 11.Kondratick, C. M., M. T. Washington, S. Prakash, and L. Prakash. 2001. Acidic residues critical for the activity and biological function of yeast DNA polymerase η. Mol. Cell. Biol. 21:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling, H., F. Boudsocq, R. Woodgate, and W. Yang. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91-102. [DOI] [PubMed] [Google Scholar]
- 13.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 14.Ollis, D. L., P. Brick, R. Hamlin, N. G. Xuong, and T. Steitz. 1985. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313:762-766. [DOI] [PubMed] [Google Scholar]
- 15.Silvian, L. F., E. A. Toth, P. Pham, M. F. Goodman, and T. Ellenberger. 2001. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 8:984-989. [DOI] [PubMed] [Google Scholar]
- 16.Trincao, J., R. E. Johnson, C. R. Escalante, S. Prakash, L. Prakash, and A. K. Aggarwal. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8:417-426. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Y.-C., V. M. Maher, D. L. Mitchell, and J. J. McCormick. 1993. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell. Biol. 13:4276-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 274:36835-36838. [DOI] [PubMed] [Google Scholar]
- 20.Washington, M. T., L. Prakash, and S. Prakash. 2001. Yeast DNA polymerase η utilizes an induced fit mechanism of nucleotide incorporation. Cell 107:917-927. [DOI] [PubMed] [Google Scholar]
- 21.Waters, H. L., S. Seetharam, M. M. Seidman, and K. H. Kraemer. 1993. Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J. Investig. Dermatol. 101:744-748. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, B.-L., J. D. Pata, and T. A. Steitz. 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8:427-437. [DOI] [PubMed] [Google Scholar]