Abstract
The Escherichia coli ClpYQ (HslUV) is an ATP-dependent protease that consists of an ATPase large subunit with homology to other Clp family ATPases and a peptidase small subunit related to the proteasomal β-subunits of eukaryotes. Six identical subunits of both ClpY and ClpQ self-assemble into an oligomeric ring, and two rings of each subunit, two ClpQ rings surrounded by single ClpY rings, form a dumbbell shape complex. The ClpYQ protease degrades the cell division inhibitor, SulA, and a positive regulator of capsule transcription, RcsA, as well as RpoH, a heat shock sigma transcription factor. Using the yeast-two hybrid system, we explored the in vivo protein-protein interactions of the individual subunits of the ClpYQ protease involved in self-oligomerization, as well as in recognition of specific substrates. Interactions were detected with ClpQ/ClpQ, ClpQ/ClpY, and ClpY/SulA. No interactions were observed in experiments with ClpY/ClpY, ClpQ/RcsA, and ClpQ/SulA. However, ClpY, lacking domain I (ClpYΔI) was able to interact with itself and with intact ClpY. The C-terminal region of ClpY is important for interaction with other ClpY subunits. The previously defined PDZ-like domains at the C terminus of ClpY, including both D1 and D2, were determined to be indispensable for substrate binding. Various deletion and random point mutants of SulA were also made to verify significant interactions with ClpY. Thus, we demonstrated in vivo hetero- and homointeractions of ClpQ and ClpY molecules, as well as a direct association between ClpY and substrate SulA, thereby supporting previous in vitro biochemical findings.
In Escherichia coli, a number of ATP-dependent proteases have been identified and extensively studied at the genetic and biochemical levels. One family of proteases was designated Clp (for caseinolytic proteases) due to the capability of its members to degrade casein in vitro. Chief among the Clp family are the two-component proteases, ClpAP, ClpXP, and ClpYQ (also called HslUV). ClpA (13), ClpX (7), and ClpY (HslU) (19, 20) are the larger components that confer the ATPase activity and substrate specificity. ClpP (17) or ClpQ (HslV) (19, 20), the smaller subunit, contains only peptidase activity. In the presence of ATP, when ClpQ or ClpP is docked with its larger partner (ClpY or ClpA/ClpX), the resulting complex has proteolytic activity. To date, several E. coli proteins have been shown to be degraded in vivo by ClpAP, ClpXP, or ClpYQ (reviewed in reference 6).
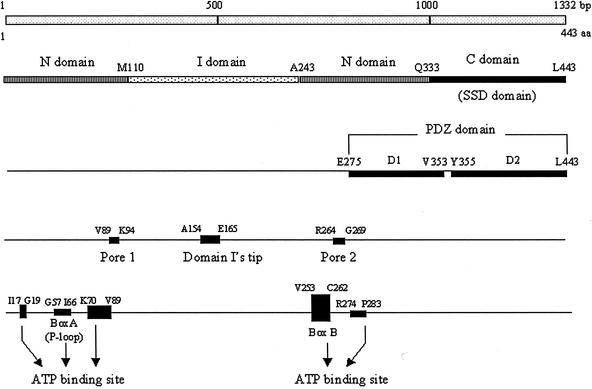
Based on amino acid sequence comparisons, ClpY is similar to ClpA and ClpX (3). In contrast, the smaller subunit, ClpQ, shares significant sequence similarity with only the proteasomal β-subunits (23). Only the structure of ClpYQ complex was recently resolved (1, 28). Structural analysis has determined that ClpY exists as a hexamer, as does ClpQ (14, 21). Twelve subunits of ClpQ and ClpY constitute the ClpYQ complex in which two tiers of hexagonal ClpQ subunits form a central barrel flanked at both ends by six subunits of ClpY (14, 21). The ClpY molecule has three domains defined from the crystal structure: the N-terminal (N; residues 2 to 109 and 244 to 332), I-intermediate (I; residues 110 to 243), and C-terminal (C; residues 333 to 443) domains (1) (Fig. 1). In terms of primary amino acids sequences, the N domains are, however, split into two parts; these include the ATP-binding sites conserved in the AAA domain, Walker boxA (P loop) and Walker boxB (Fig. 1). Domain I, with its flexible protruding teeth-like arrangement, recognizes and grasps protein substrates (1). The surfaces of the C-terminal domain (identical to a “sensor and substrate discrimination” [SSD] domain) of ClpY are primarily involved in intersubunit interaction (1), and its tail is important for the activation of ClpQ (24). The crystal structure also further defined functional motifs of ClpY, i.e., pore 1, pore 2, domain I's tip, and nucleotide binding pockets (see in Fig. 1) (28). The position of the pores, adjacent to ATP-binding sites, suggested that the cavities may allow the transfer of unfolded substrates for degradation and the tip, which is distal from the rest of the complex, probably functions in trapping substrates (28). Underlying this, ClpY lacking I domains failed in vitro to cleave one of its substrates, MBP-SulA, in the presence of the ClpQ (27). However, earlier biochemical data proposed that duo functional domains are localized in the C-terminal region of ClpY, i.e., PDZ-like domains, which contain ca. 100 amino acids with repeat motifs involved in protein-protein interactions (16). Accordingly, these include PDZ-like D1 (D1; residues 275 to 353) and PDZ-like D2 (D2; residues 355 to 443) domains that are also involved in substrate recognition (16) (Fig. 1). In similar fashion, the C domain, termed an SSD domain, of ClpY appeared to function in substrate binding (Fig. 1) (26).
FIG. 1.
Domains of the ClpY molecule and its functional motifs. The 443 aa (1,332 bp) that comprise ClpY were drawn to scale. The domains referred to in the text are indicated. The colored and shaded sticks show structures of domains, the N domains (residues 2 to 109 and 244 to 332), I domains (residues 110 to 243), and C (SSD) domains (residues 333 to 443). The PDZ-like domains are D1 (residues 275 to 353) and D2 (residues 355 to 443). The positions of pore 1 (residues 89 to 94), domain I's tip (residues 154 to 165), and pore 2 (residues 264 to 269) are indicated. The Walker boxA or P loop (residues 57 to 66) and boxB (residues 253 to 262) of the ATP-binding site are indicated and the dark box (residues 17 to 19, 57 to 66, and 70 to 89) are located in the nucleotide binding pocket.
To examine these discrepancies, we used the yeast two-hybrid system as a new approach to investigate both protease structure and substrate recognition and interaction. We demonstrate protein-protein interactions in vivo that implicate the oligomerization of ClpQ and ClpY in complex formation and substrate recognition. Our results suggest that the C-terminal regions of ClpY are specifically involved in protein association. Both I domains and PDZ domains are involved in substrate bindings. The critical amino acid residues of SulA for protein-protein interaction with ClpY were also determined.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strain Saccharomyces cerevisiae EGY48 [MATa his3 trp1 ura2 lexAop(x6)-leu2] was obtained from Clontech (Palo Alto, Calif.). E. coli strain XL1-Blue was purchased from Life Tech, Inc.(Rockville, Md.). The wild-type strain, E. coli MG1655 was a gift from Susan Gottesman. E. coli strain KC8 (leuB trpC hisB) was obtained from Clontech, as was the plasmid pGilda, a LexA DNA binding domain (BD) vector (5), and plasmid pACT2. Plasmid pB42AD, with a hemagglutinin (HA) epitope tag, a B42 polypeptide activation domain (AD) vector (8), was also purchased from Clontech. Plasmid pWF1 (clpQ+ clpY+) (29) and pGS165 containing a sulA gene have been described previously (10).
Media.
E.coli was grown in Luria broth. Yeast extract-peptone-dextrose and synthetic dropout (DO) minimal medium (SD) were purchased from Clontech for the growth of yeast. DO supplements contained specific mixtures of amino acids and nucleosides, as required, prepared according to the manufacturer's instructions. Glucose minimal medium was prepared as described earlier (18). Additional supplements were added at the following final concentrations: glucose, 2%; galactose, 2%; raffinose, 1%; ampicillin, 100 μg/ml; and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 80 μg/ml.
PCR and plasmid constructions.
The procedures used for PCR, as well as for cloning and restriction enzyme digestion, were performed as described by Sambrook et al. (22). Template DNAs were either plasmids carrying the appropriate genes or chromosomal DNA extracted from wild-type MG1655 by methods described elsewhere (22). Each PCR-generated DNA product had terminal restriction sites introduced by PCR primers. Genes encoding the polypeptides of interest were amplified by PCR with the primers listed in Table 1. Encoded proteins included ClpQ, ClpY, SulA, RcsA, RpoH, ClpYΔ337-443(ClpYΔC), ClpYΔ275-443 (ClpYΔPDZ), ClpYΔ110-197 (ClpYΔI), ClpYΔ1-335 (C), ClpYΔ1-274 (PDZ), ClpYΔN1-109+ΔC242-443 (I), ClpYΔN1-274+ΔC352-443 (D1), ClpYΔ110-197+ΔC337-443 (ClpYΔIΔC), SulA*Δ1-19 (SulA*ΔN19), SulA*Δ1-27 (SulA*ΔN27), SulA*Δ1-43 (SulA*ΔN43), SulA*Δ148-169 (SulA*ΔC148), SulA*Δ129-169 (SulA*ΔC129), and SulA*Δ1-43+Δ129-169 (SulA*ΔN43ΔC129). The resulting PCR DNA fragments carrying the above genes were, in most cases, each directly cloned into pGilda to create an in-frame fusion with DNA encoding the BD of LexA. Thus, the vector and the PCR fragments were digested with the appropriate enzyme (EcoRI and BamHI) and then ligated together. The recombinant plasmids were recovered after transformation into E. coli strain XL1-Blue by selecting for ampicillin resistance (Apr) and screening for the presence of an insert by restriction enzyme analyses.
TABLE 1.
Primers used in this study
| Gene | Primer
|
|
|---|---|---|
| Orientationa | Sequence | |
| clpQ | f | 5′-CCGCGGATCCTGACAACTATAGTAA-3′ |
| r | 5′-CCGGAATTCTTACGCTTTGTAGCTT-3′ | |
| clpY | f | 5′-CCCGAATTCATGTCTGAAATGACCCC-3′ |
| r | 5′-CCGCGGATCCTTATAGGATAAAACG-3′ | |
| sulA | f | 5′-CCGGAATTCATGTACACTTCAGGC-3′ |
| r | 5′-CGCGGATCCTTAATGATACAAATTAGAGTG-3′ | |
| rcsA | f | 5′-CCGGAATTCATGTCAACGATTATTATGGAT-3′ |
| r | 5′-CGCGGATCCTTAGCGCATGTTGACAA-3′ | |
| rpoH | f | 5′-CCCGAATTCTGAATGACTGACAAAATGC-3′ |
| r | 5′-TTTGGATCCCGGGTTACGCTTCAATGGCAGC-3′ | |
| clpYΔc | f | 5′-CCCGAATTCATGGCTGAAATGACCCC-3′ |
| r | 5′-CGCGGATCCTTACAGCGCCTGCAGTTCAAC-3′ | |
| clpYΔpdz | f | 5′-CCCGAATTCATGGCTGAAATGACCCC-3′ |
| r | 5′-CGCGGATCCTTAACGAGAAACATCCGGACC-3′ | |
| c | f | 5′-GGAGAATTCCTGCAAGGTCGTCTGCCA-3′ |
| r | 5′-CGTGGATCCTTATAGGATAAAACGGCTC-3′ | |
| pdz | f | 5′-CCGGAATTCGCGAGT CTTCCGGTC-3′ |
| r | 5′-CGTGGATCCTTATAGGATAAAACGGCTC-3′ | |
| i | f | 5′-CCGGAATTCATTCGCGATCTGACCG-3′ |
| r | 5′-CGCGGATCCTTAAGCGTCTTGCTTC-3′ | |
| d1 | f | 5′-CCGGAATTCATGGAAGGCGTTCAGCGTGAC-3′ |
| r | 5′-CGCGGATCCTTACACGGTGATAGAGGCATT-3′ | |
| sulA*ΔN1-19 | f | 5′-CCGGAATTCATTGCGCGCGTGTCTCT-3′ |
| r | 5′-CGCGGATCCTTAATGATACAAATTAGAGTG-3′ | |
| sulA*ΔN1-27 | f | 5′-CCGGAATTCATGAACACTACAGCCG-3′ |
| r | 5′-CGCGGATCCTTAATGATACAAATTAGAGTG-3′ | |
| sulA*ΔN1-43 | f | 5′-CCGGAATTCATGATGATGACGCAACTT-3′ |
| r | 5′-CGCGGATCCTTAATGATACAAATTAGAGTG-3′ | |
| sulA*ΔC149-169 | f | 5′-CCGGAATTCATGTACACTTCAGGC-3′ |
| r | 5′-CGCGGAATCCTTATACCGGACGCATAATAAAC-3′ | |
| sulA*ΔC125-169 | f | 5′-CCGGAATTCATGTACACTTCAGGC-3′ |
| r | 5′-CGCGGAATCCTTAAGCATGCTCTTCTTC-3′ | |
| sulA*ΔN1-43ΔC125-169 | f | 5′-CCGGAATTCATGATGATGACGCAACTT-3′ |
| r | 5′-CGCGGAATCCTTAAGCATGCTCTTCTTC-3′ | |
f, Forward; r, reverse.
To construct pGilda-clpQ, the clpQ gene was cloned into pACT2 (Clontech) after amplification with primers containing terminal BamHI and EcoRI restriction sites with pWF1 as the template. The clpQ was then transferred from the pACT2-clpQ plasmid into pAS2-1 (Clontech) at NcoI-EcoRI sites. Subsequently, the clpQ gene in pAS2-1-clpQ was inserted into the BamHI site of pGilda. The clpQ inserted in the correct orientation and in frame with DNA of the LexA-BD in pGilda was designated pGilda-clpQ.
To clone the clpYΔΙ gene into the pGilda, genes encoding an N-terminal region of the ClpY (amino acids [aa] 1 to 109) were PCR amplified with primers carrying EcoRI-PvuII sites (forward primer, 5′-CCCGAATTCATGGCTGAAATGACCCC-3′; reverse primer, 5′-ATAATCAGCTGTTTCACGGCGGCATCGGTCA-3′). The PCR products then replaced an EcoRI-PvuII fragment (aa 1 to 197 of ClpY) of pACT2-clpY, resulting in a construct (pACT2-clpYΔΙ) that carries a clpY deletion mutation that includes nucleotides coding for aa 1 to 109 and aa 198 to 443 joined in an in-frame fusion. The clpYΔΙ from pACT2-clpYΔΙ were subsequently subcloned into pGilda at EcoRI-BamHI sites, designated pGilda-clpYΔΙ. pGilda-clpYΔΙΔC was constructed by PCR amplification of the clpyΔΙΔC fragment, with pGilda-clpYΔΙ as a template and the same primer set used for clpYΔC, and subsequently cloned the PCR fragment into pGilda EcoRI-BamHI sites.
The DNA fragments of the above genes from the resulting pGilda derivative plasmids were each correspondingly transferred into pB42AD at EcoRI-XhoI sites. The resulting pB42AD derivative plasmids retain each gene in-frame with DNA encoding the B42 polypeptide, an artificial transcriptional AD of Gal4. The isolated plasmids were prepared after transformation into E. coli XL1-Blue. Fusion genes in the constructs were sequenced for verification to contain the expected gene fusion.
Yeast two-hybrid assays.
The yeast two-hybrid system (4), of which pGilda(BD), carrying a LexA-DNA BD, and pB42AD, carrying a B42-DNA AD, were used for the construction of fusion genes. The yeast strain EGY48[p8op-lacZ] was the reporter strain, which carries p8op-lacZ plasmids and lexAop(x6)-LEU2 integrated in the chromosome. The method of yeast two-hybrid assays, procedures for yeast transformation, and plasmid isolations were performed as described in the Clontech yeast protocol handbook.
The clpQ, clpY, and other gene fusions in BD or AD plasmids were cotransformed into EGY48[p8op-lacZ] and selected for histidine (His; for a selection of pGilda) and tryptophan (Trp; for a selection of pB42AD) on SD medium lacking His and Trp, respectively. The SD medium usually lacks uracil (Ura), which was omitted for the maintenance of plasmids p8op-lacZ. To assess expression of the reporters, the yeast strains, EGY48[p8op-lacZ] coexpressing BD and AD hybrid protein was overnight grown at 30°C on SD−Trp−His−Ura medium (i.e., medium lacking Trp, His, and Ura) supplemented with glucose, which would repress fusion gene expression. The overnight grown cells were pelleted, washed, and resuspended in an equal volume of distilled H2O, and one-tenth of the cells were inoculated into SD+Gal+Raf−Trp−His−Ura medium (Gal, galactose; Raf, raffinose) and grown to mid-log phase (i.e., an optical density at 600 nm of 0.5 to 0.8). At that point, galactose and raffinose were added for the induction of fusion genes expression. One-half of the mid-log-phase-grown cultures were collected, washed, and redissolved in an equal volume of H2O, which was prepared for LEU2 tests; the other half of cultures was otherwise saved for lacZ assays. The activation of LEU2 reporters was assessed by plating a 10-μl portion of washed yeast on medium lacking leucine (Leu; SD+Gal+Raf−Trp−His−Leu−Ura) and monitoring growth over 4 days. A 10-μl portion of the remaining yeast cultures was plated out on SD+X-Gal+Gal+Raf−Trp−His−Ura medium over 2 to 3 days to assess the activation of lacZ qualitatively, along with a β-galactosidase assay, by using the method described by Miller (18) to measure lacZ expression quantatively.
Western blot assays.
The overnight yeast cells expressing the hybrid fusion proteins in the repressing media were washed, and one-tenth was inoculated into 5 ml of inducing medium and grown to the mid-log phase (optical density at 600 nm of 0.5 to 0.8). The cells were then collected and washed with double-distilled H2O. After centrifugation, the pellet was treated with liquid nitrogen and resuspended in 400 μl of Laemmli loading buffer containing an equal amount of acid-washed glass beads (425 to 600 μm; Sigma). Equivalent amounts of protein extracts normalized for the original optical density were loaded, electrophoresed on sodium dodecyl sulfate-12% polyacrylamide gels, and electrotransferred onto 0.2-μm-pore-size nitrocellulose. Membranes were probed with monoclonal antibody to HA from Roach or with HA multiclonal antibody from Clontech, and the multiclonal antibody to LexA was from Clontech. The secondary antibody was either anti-mouse (for monoclonal antibody) or anti-rabbit (for multiclonal) antibody. The Western blots were developed by enhanced chemiluminescence.
Isolation of mutant SulA.
To isolate sulA mutations that resulted in SulA with either enhanced or inhibited interaction with ClpY, pGilda-sulA was subjected to hydroxylamine mutagenesis (18) and then transformed into XL-1 blue followed by selection for Apr colonies. About 10,000 Apr colonies were pooled together for plasmid extraction, and these mutagenized pGilda-sulA were transformed into yeast EGY48[p8op-lacZ] carrying pB42AD-clpY for His+ Trp+. The resulting yeast transformants were screened on X-Gal medium; both dark-blue colonies and white colonies were saved for further plasmid isolation. The extracted plasmids were subsequently transformed into KC-8 and selected for His+ on glucose minimal medium plus DO without the addition of His. Only plasmids carrying pGilda-sulA were recovered from His+ colonies. Clones expressing full-length SulA and that did not autoactivate the reporters when transformed back to EGY48[p8op-lacZ] were screened by immunoblotting with anti-LexA antibody (Clontech). The clones were then subjected to DNA sequence analysis to determine the mutations.
RESULTS
Several studies have shown that both ClpQ and ClpY hexamers undergo homo-oligomerization in vitro and that ClpQ interacts with ClpY to form an active protease complex (ClpYQ) for substrate degradation (14, 15, 19, 21). To ascertain whether these protein-protein interactions occur in vivo, we employed the yeast two-hybrid method. In the yeast two-hybrid, when the reporter host carries the Gal4 BD and Gal4 AD plasmids together, it cannot activate expression of the Gal4 targeting reporter genes since BD and AD are physically and functionally independent. Only when two test proteins, one fused into AD and the other fused into BD, interact with one another, can a functional GAL4 transcription activator be reconstituted, resulting in activation of the reporter expression.
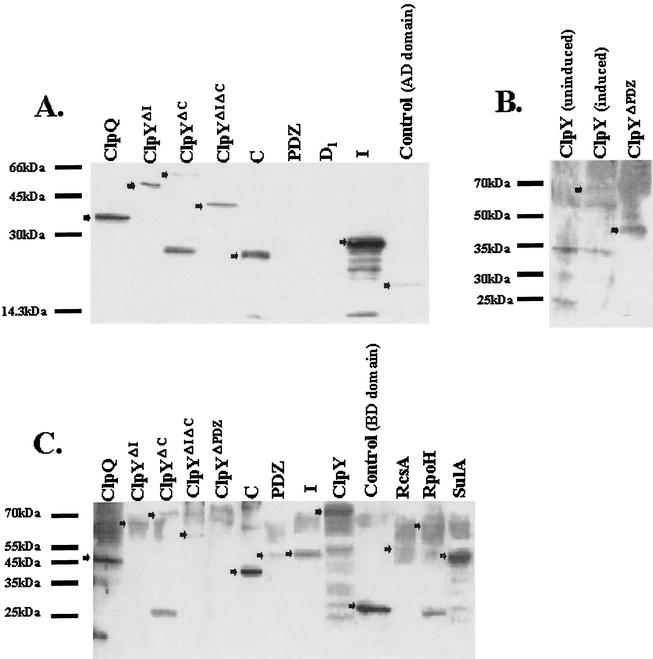
The clpQ, clpY, and other genes were initially cloned into pGilda(BD) and pB42AD vectors to construct BD or AD hybrid fusion genes as described in Materials and Methods. The yeast strain EGY48[p8op-lacZ], which carries leu2 and lacZ as reporter genes, was used as an indicator host. ClpQ, ClpY, or other gene products in a translational fusion with either the LexA-Gal4 DNA-BD of pGilda or with the B42 polypeptide, a Gal4 AD in pB42, were pairwise coexpressed by BD or AD chimeric plasmids in the yeast transformants. The expression of various fusion proteins was detected by Western blotting (Fig. 2). All AD fusion proteins (in pB42AD) were detected except for the PDZ and D1 domains (Fig. 2A and B). All of the BD fusion (in pGilda) proteins were detected except for ClpYΔPDZ (Fig. 2C). The X-Gal color changes and quantitative β-galactosidase assays were used to determine lacZ expressions and leu2 gene expression was monitored by growing yeast transformants on the SD selective medium depleted of Leu.
FIG. 2.
Western blotting of pB42AD and pGilda(BD) fusion proteins. In each condition, extracts were derived from EGY48[p8op-lacZ] carrying AD or BD plasmids expressing the hybrid fusion proteins. Each arrowhead indicates the corresponding fusion protein listed above each lane. The values to the left of each panel indicate the molecular masses (in kilodaltons) of the protein size standards. Analyses with anti-LexA (BD) monoclonal antibody (A), anti-LexA (BD) multiserum (B), and anti-HA (AD) multiserum (C) are shown.
ClpQ/ClpQ, ClpQ/ClpY, and ClpYΔI/ClpY interact in the yeast two-hybrid.
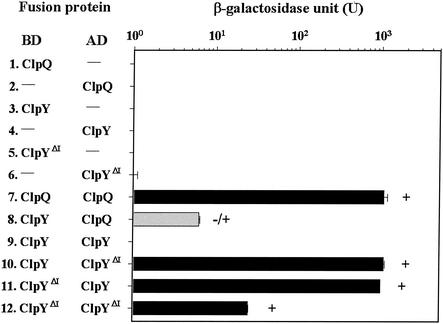
We first tested possible interactions in ClpQ/ClpQ, ClpQ/ClpY, and ClpY/ClpY in the two-hybrid assays. pGilda, pGilda-clpQ, pGilda-clpY, pB42, pB42-clpQ, or pB42-clpY were cotransformed in pairs into EGY48[p8op-lacZ], selecting for His+ Trp+ colonies on SD selective medium without Ura. All of the transformed EGY48[p8op-lacZ] strains with various BD and AD hybrid proteins were analyzed for expression of leu2 and lacZ reporter genes. However, the analysis of the two-hybrid assays revealed the orientation dependence of the induction of reporter gene expression since no interaction between ClpQ and ClpY was detected in the BD-ClpQ/AD-ClpY transformed yeast cells.
The controls, LexA and B42 hybrid proteins from strain EGY48[p8op-lacZ] carrying ClpQ or ClpY alone, showed no growth on SD medium lacking Leu; on X-Gal plates, the colonies were white and expressed low β-galactosidase activity (Fig. 3, lines 1 to 4). In contrast, ClpQ showed a strong interaction with itself when coexpressed in yeast, as indicated by rapid cell growth on Leu-selective medium and dark blue colony color on X-Gal plates, along with high-level β-galactosidase expression (Fig. 3, line 7). ClpQ and ClpY appeared to have a weak interaction, as demonstrated by the slow growth of yeast cells coexpressing the two proteins, the light blue colony color on X-Gal plates, and the low-level expression of β-galactosidase (Fig. 3, line 8). An interaction between ClpY and ClpY was not detected in the two-hybrid assays since cells expressing ClpY from two different fusion plasmids showed phenotypes indistinguishable from the controls (Fig. 3, compare line 9 to lines 3 and 4). No interaction was found between two ClpY molecules.
FIG. 3.
Expression of lacZ and LEU2 in strain EGY48[p8op-lacZ] carrying different pairs of the ClpQ, ClpY, or ClpYΔI fusion protein. The BD vector is pGilda, and the AD vector is pB42AD. Each lane represents the pairwise encoded fusion proteins from the BD or AD plasmid carrying fusion genes as indicated. A dash indicates that no proteins were fused to the BD or AD domain. In the column, each bar represents an average of the β-galactosidase levels with the standard deviation value in Miller units (18). Colony color was evaluated on Gal+Raf−Ura−His−Trp plates containing X-Gal over 2 days. The solid bar represents blue colonies; the shaded bar represents the light blue colonies. LEU2 expressions were tested on Gal+Raf−His−Trp−Leu plates over 4 days. +, Rapid growth of yeast cells in the selective medium as a result of elevated expression of the LEU2 gene; +/−, flash growth.
Since domain I of ClpY extends out from the rest of the molecule (1), we hypothesized that domain I might interfere with ClpY self-interaction in the two-hybrid assays. Subsequently , clpYΔI was constructed as in-frame fusion genes in BD and AD plasmids. These two fusion plasmids were each cotransformed with another set of plasmids either carrying AD and/or BD alone or carrying AD and/or BD clpY or clpYΔI fusion genes. Transformants expressing only one hybrid protein were selected as controls. All of the controls showed basal level expression of the reporters (Fig. 3, lines 3 to 6). Cells bearing ClpYΔI coexpressed with either ClpY or ClpYΔI were examined for subunit interactions. Compared to the controls, ClpYΔI appeared to interact with ClpY in both AD and BD orientations, as indicated by the high level of β-galactosidase activity, the dark blue colony color on X-Gal plates, and rapid growth on Leu-selective medium (Fig. 3, lines 10 and 11). ClpYΔI displayed also an interaction with itself (Fig. 3, line 12). These results suggest that I domains are not required for ClpY intersubunit interactions.
C-terminal region of the ClpY required for ClpY-ClpY interactions.
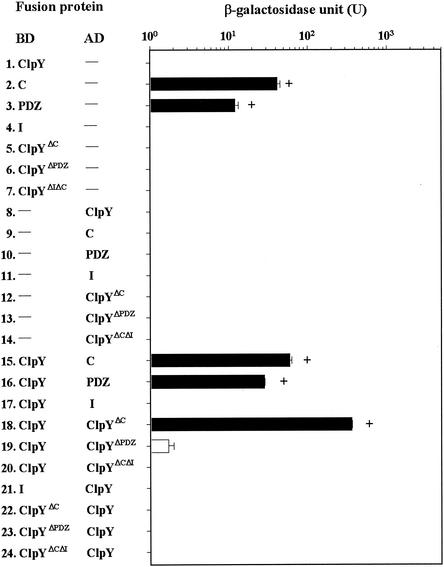
ClpY fragments instead of full-length ClpY were used in two-hybrid assays to determine the region of ClpY responsible for intersubunit interactions. It also has been proposed based on the crystal structure analysis that ClpY oligomerizes via the C-terminal domains of the molecule (1). To examine this possibility, BD or AD fused with C, ClpYΔC, and other ClpY deletion proteins were individually coexpressed with ClpY in the yeast cells as before. No autoactivated reporter gene expression was found in the controls (see Fig. 4, line 1 and lines 4 to 14) with the exception of a pGilda(BD) plasmid encoding a hybrid protein carrying C or PDZ domains, which led to false-positive results (Fig. 4, lines 2 and 3). Therefore, yeast cells bearing BD-C or BD-PDZ fusion proteins were excluded from the assays. We found that C domains alone interact with ClpY, as determined by the detection of all positive signals (Fig. 4, line 15). Unexpectedly, ClpY with deleted C domains still showed an interaction with ClpY (Fig. 4, line 18). However, although ClpYΔC was exchanged into an AD, the resulting AD-ClpYΔC hybrid proteins did not interact with ClpY (Fig. 4, line 22). The ClpY with more extensive deletion at the C-terminal ends, ClpYΔPDZ (see Fig. 1), was constructed in both BD and AD plasmids; the resulting hybrid plasmids were subsequently cotransformed with clpY in the reporter yeast. The ClpYΔPDZ did not interact with ClpY (Fig. 4, lines 19 and 23). As expected, PDZ domains, which include C domains, were also shown to interact with ClpY (Fig. 4, line 16). We also examined whether I domains were involved in ClpY self-association, and neither AD and BD I fusion proteins interacted with ClpY (Fig. 4, lines 17 and 21). The ClpYΔIΔC, in addition, was constructed and coexpressed with ClpY in both orientations; no interaction was found (Fig. 4, lines 20 and 24). We also used ClpYΔI substituted for ClpY to examine an interaction between each pair of molecules. ClpYΔI, although coexpressed with other ClpY deletion mutants used above, behaved as did ClpY (data not shown). ClpYΔC, ClpYΔPDZ, and ClpYΔIΔC were also examined one by one for self-association, and the results were negative (data not shown). All of these results taken together indicate that the C-terminal region of ClpY is necessary for an intersubunit interaction of ClpY with itself.
FIG. 4.
Expression of lacZ and LEU2 in strain EGY48[p8op-lacZ] tested for interactions between wild-type ClpY and ClpY deletion mutants. All of the signals shown in the diagram are as described in the Fig. 3 legend.
ClpY interacts with SulA in vivo, and the PDZs D1 and D2 (also C), together, are involved in an interaction.
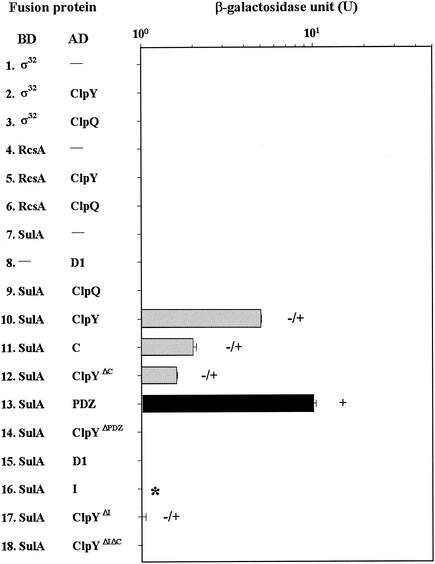
It was demonstrated previously that RcsA, RpoH, and SulA were regulated by ClpYQ protease (11, 12, 15, 25, 29) and that ClpY is a chaperone accessible for substrate binding in vitro (26). To determine whether the same accessibility is found in an in vivo yeast cell, strain EGY48[p8op-lacZ] was separately coexpressed as hybrid proteins (ClpQ-RcsA, ClpQ-RpoH, ClpQ-SulA, ClpY-RcsA, ClpY-RpoH, and ClpY-SulA) and then analyzed with respect to expression of the reporter. The controls, with one plasmid containing a hybrid fusion gene, showed low-level expression of β-galactosidase and did not grow on Leu-selective medium (Fig. 5, lines 1, 4, and 7). As expected, the full-length ClpY interacted with SulA, as shown in assays in which cells coexpressing ClpY and SulA possessed low but significant levels of β-galactosidase activity and grew on Leu-selective media (Fig. 5, line 10). However, there was no interaction detected between ClpY and RcsA and RpoH in that the colony phenotypes of the test and control cells were similar (Fig. 5, lines 1 and 2 and lines 4 and 5). Also, ClpQ alone did not appear to associate with SulA, nor did RcsA or RpoH in the two-hybrid assays, since all of the pairwise transformed cells showed low expression of leu2 and lacZ (Fig. 5, lines 3, 6, and 9). The exchange of the BD or AD for all of the above fusion genes resulted in only the BD-SulA/AD-ClpY pair showing a positive interaction. However, their β-galactosidase levels were lower than the results of cells with BD-ClpY/AD-SulA (data not shown).
FIG. 5.
Expression of lacZ and LEU2 in strain EGY48[p8op-lacZ] tested for interactions of SulA, RcsA, or RpoH with either subunit of ClpY or ClpQ. The SulA subunit was also tested for its interaction with ClpY derivative mutants. The signals shown in the diagram are as described in the Fig. 3 legend. ✽, blue color on X-Gal plates.
To define a region of the ClpY involved in SulA bindings, a series of ClpY deletion mutants, expressed from pB42 plasmids, was also tested for their interactions with SulA. Their pairwise transformants were again analyzed by monitoring expression of the reporter. Cells with PDZ-like domains, including domains D1 and D2 when coexpressed with SulA, demonstrated high-level β-galactosidase activity and a Leu+ phenotype comparable to those of cells that expressed full-length ClpY with SulA (Fig. 5, line 13). ClpY lacking PDZ-like domains showed a complete loss of binding affinity with SulA (Fig. 5, line 14). These results indicated that PDZs were necessary and sufficient for interaction with SulA. However, yeast cells containing a single PDZ-like domain, D1 or D2 (which overlaps with the C domain) alone, showed much less β-galactosidase activity (Fig. 5, compare lines 11 and lines 15 to 13). Hence, the PDZ-like domains, D1 and D2, are both present and have a higher substrate-binding affinity.
Based on X-ray crystal analyses, domain I of ClpY functions in substrate recognition. To test this, we also examined whether domain I alone was able to interact with SulA in a two-hybrid assay. No interaction was observed between I domains with SulA (Fig. 5, line 16), but samples were blue after an overnight induction on X-Gal plates. Transformants with the ClpYΔΙ/SulA pair showed low-level β-galactosidase activity (Fig. 5, compare lines 17 to 10), suggesting that the lack of domain I has an effect on the binding affinity of ClpY with SulA. In addition, ClpYΔC showed a lower affinity with SulA than did ClpY (Fig. 5, compare lines 12 to 10). These results also suggest that both the I and PDZ domains are involved in substrate binding.
Mutations in sulA can affect protein-protein interaction, and the N and C termini of SulA are dispensable for an interaction with ClpY.
To further characterize important residues in SulA that either reduce or enhance interaction with ClpY, cells containing plasmids carrying the sulA gene (pGilda-sulA) were randomly mutagenized by using hydroxylamine. When EGY48[p8op-lacZ] containing pB42AD-clpY was cotransformed with pGilda-sulA, the resulting colonies were blue on X-Gal medium. Screening of a pool of plasmids carrying an NH2OH-mutagenized sulA gene yielded transformants that were either dark blue or white on X-Gal medium. These colonies were then further analyzed, and the mutations in the sulA gene were identified by DNA sequence analysis of the subcloned gene.
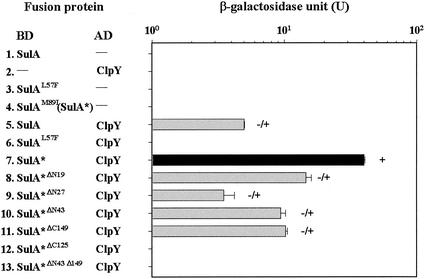
Sequencing of isolated clones revealed that four of the mutants resulted from a single base pair substitution. Two SulA mutants, isolated as white, had an identical amino acid altered from Leu to phenylalanine at position 57 (L57F), whereas two other mutants, isolated as darker blue, had an isoleucine substituted for a methionine at residue 89 (M89I). As shown in Fig. 6, the SulAL57F did not interact with ClpY, and the SulAM89I mutant (designated SulA*) did interact with ClpY strongly (Fig. 6, lines 6 and 7). These two SulA mutants, when singularly expressed in yeast, showed a basal level expression of the reporter (Fig. 6, lines 3 and 4).
FIG. 6.
Expression of lacZ and LEU2 in strain EGY48[p8op-lacZ] tested for interactions between SulA derivative mutants and ClpY. The SulA includes 169 aa residues. The signals shown in the diagram are as described in the Fig. 3 legend.
Since the SulA* mutant renders a strong interaction with ClpY, we used the sulA* gene as a template to construct serial deletion mutations and to determine the region necessary for an interaction with ClpY. SulA* with the N-terminal and/or the C-terminal amino acids deleted was first constructed in pGilda and designated SulA*ΔN19, SulA*ΔN27, SulA*ΔN43, SulA*ΔC149, SulA*ΔC125, and SulA*ΔN43ΔC125 separately. The BD hybrid proteins of these deletion mutants and SulA* were individually coexpressed with AD-ClpY and tested for their mutual interactions. As shown in Fig. 6, the SulA* N-end deletion mutants SulA*ΔN19, SulA*ΔN27, and SulA*ΔN43 maintain sufficient interaction with ClpY, a finding similar to that achieved with the full-length SulA* (Fig. 6, lines 8, 9, and 10). SulA*ΔC149, with the last 20 aa deleted from the C-terminal end, was also demonstrated to interact with ClpY (Fig. 6, line 11). However, SulA*ΔC125, with 40 aa deleted from the C terminus, has no interaction with ClpY; nor does SulA*ΔN43ΔC125 (Fig. 6, lines 12 and 13). These results suggest that the internal region of SulA was necessary for interactions with ClpY and that the N-terminal amino acids residues of SulA were not necessary for the interactions.
DISCUSSION
In the present study we used the yeast two-hybrid system to investigate an in vivo protein-protein interaction of the E. coli ClpYQ protease and the involvement of homo- or heterosubunit associations in substrate specificity. This is the first example of using the yeast two-hybrid approach in bacterial Clp protease studies, and it represents a good model system for detecting certain weak protein-protein interactions. Our studies demonstrate that there are in vivo protein-protein interactions between ClpQ and ClpQ, as well as between ClpQ and ClpY. These in vivo molecular interactions in ClpQ-ClpQ and ClpY-ClpQ pairs support previous in vitro biochemical and structural observations that ClpQ oligomerizes with itself and with ClpY (14, 15, 19, 21). No interactions were detected between two ClpY molecules in the yeast two-hybrid assay. Steric hindrance may have prevented two ClpY molecules from interacting with one another due to the larger molecular weight, resulting in essentially no expression of the reporters. Consistent with this, ClpYΔI, a ClpY molecule lacking the I domain, was shown to interact with ClpY. Thus, our data suggested that the I domain in ClpY may physically interfere with an intersubunit interaction of ClpY in two-hybrid assays. Additionally, ClpYΔI displayed an interaction with itself. Therefore, removing I domain from one of two ClpY molecules apparently facilitates ClpY subunit interaction. Deletion of both I domains allows interaction between two ClpYΔI at possibly lower intrinsic affinities. Our results also indicated that I domains are not essential for a direct contact between two ClpY molecules but may be necessary for proper configuration of ClpY in homo-oligomerization. The C-terminal regions of ClpY, in the present study, were found to be required for intersubunit interactions. All of these findings are consistent with the predicted structure of ClpY oligomers from crystal analyses, which indicated that domain I is distal to the rest ClpY molecule and not directly involved in intersubunit interaction. Likewise, the C-terminal ends are juxtaposed in adjacent ClpY monomers and are probably inaccessible for substrate interaction in a ClpY oligomer.
It is known that the ClpYQ complex regulates two proteins, SulA and RcsA (12, 15, 25, 29), as well as σ32 (RpoH), in combination with other ATP-dependent proteases (11). It has also been shown that overexpressed ClpY acting as a chaperone promotes SulA activities leading to bacterial death (25). ClpY appears to be responsible for recognizing and transferring substrates into an inner catalytic core in which ClpQ degrades the substrates (1, 28). The results presented here indicate that in vivo it is ClpY, not ClpQ, that accounts for the interaction of the ClpQ-ClpY complex with the substrate, SulA. No interactions were observed between ClpY and RpoH or between ClpY and RcsA. In the case of RcsA and RpoH, the lack of detectable interaction with ClpY in the yeast two-hybrid assay could reflect a transient interaction between the two molecules. Accordingly, SulA and ClpY also showed a weak interaction.
Our results demonstrated that ClpY without PDZ domains loses its binding affinity for SulA. However, PDZ domains in yeast two-hybrid analyses were able to interact with SulA in a fashion analogous to the full-length ClpY. Thus, our data support previous biochemical findings that PDZ domains are involved in substrate binding. X-ray crystal analyses indicated that I domains are more likely to be responsible for substrate recognition (1, 28). With the yeast two-hybrid assay, no interaction was detected between the domain I and SulA, since the β-galactosidase levels were not increased when cells were assayed on day 1. Based on the conformation of domain I, it is not surprising that interaction with SulA would not occur. Interestingly, colonies containing SulA and I domain hybrid fusion proteins were routinely blue after an overnight induction on X-Gal plates (see Fig. 5, line 16). This observation was contradicted by the results of the β-galactosidase assays, and the reason for this is unclear.
The crystal structure analyses have proposed two states of I domains: an exposed state for substrate binding and a buried state for substrate release (28). Our data suggest that the conformation of domain I may lead to lower substrate-binding affinities, whereas the conformational organization in the PDZ domains may result in higher substrate-binding affinities. Both domains are likely to be important for substrate binding, since domain I may function in grasping the substrate and the PDZ domain may be required for proper positioning of the substrate in the inner surfaces of the cylinder. More studies are needed to further characterize the function of both regions.
Through thorough genetic screening, we have repeatedly isolated SulA mutants L57F and M89I, which were shown to affect SulA interactive strength with ClpY, suggesting that an interactive part is located in the middle of the SulA molecule. To interpret the effect of mutations on SulA structure, the computer programs GCG and Pepplot were used to predict SulA secondary structure, antigenic domains, and a profile of charged residues. The SulA mutants, L57F and M89I, contained in two single amino acid substitutions located in highly antigenic and hydrophilic regions, probably corresponding to molecule surfaces. It is reasonable that residues involved protein-protein interactions would be positioned on an exterior layer of the SulA molecule. Consistent with secondary structure predictions, the M89I substitution is positioned in a junction between a turn and an α-helix. The two single amino acids are highly conserved, i.e., L57 (100%) and M89 (60%), based on a comparison of five different bacterial SulAs. We plan to further analyze these two SulA mutant proteins to distinguish an active state (folded) or an inactive state (possibly unfolded) with regard to their functional activities in E. coli. In addition, we demonstrated that the N terminus of the SulA* molecule is not essential for interactions with ClpY in yeast two-hybrid analyses. However, extensive C-terminal deletions in the internal region of SulA* completely eradicated interactions with ClpY, suggesting that the internal region of SulA plays an important role in interaction with ClpY.
Our results also indicated that the ClpYQ recognizes SulA at positions different from the sites for Lon, an ATP-dependent and a major protease of SulA. It was demonstrated that the C-terminal 20 residues of SulA are responsible for recognition by and complex formation with Lon (9). The middle region in SulA, on the other hand, is necessary for degradation by Lon. Since E. coli Lon has higher affinities for SulA (2, 29), more studies are needed to test whether these dissimilarities are related to different affinities.
Using the yeast two-hybrid system, we demonstrated in vivo homo- and heteroassociation of the ClpYQ subunits and specific substrates of the protease. Only pGilda and pB42 pair plasmids used in the present study successfully demonstrated the applications. Since the ClpY works as a chaperone, ClpY also binds unfolded proteins in vivo. In our studies, although we were searching for substrates from the E. coli genomic library, we found that ClpY also interacts with several small peptides; however, from our preliminary data, the specificities of these small peptides recognized by the ClpY are still inconclusive. However, according to the chaperone function of ClpY, this also indicates that the partial fragments of ClpY, although they interacted with intact ClpY, are involved as substrates rather than domains in specific oligomerization. However, initially selected ClpY mutants defective in ClpY self-association were mapped in the C-terminal domains in support of our earlier findings that the C domains are necessary for ClpY self-oligomerization. Independently, Seong et al. (24) recently demonstrated that deletion of the C-terminal 10 residues of ClpY prevented the formation of ClpY hexamer. We are now searching for ClpYQ substrates and are analyzing different point mutants of the ClpY and ClpQ subunits to determine the role of specific amino acid residues in subunit interaction, as well as in substrate recognition.
Acknowledgments
We thank Susan Gottesman for the gift of MG1655 and comments for the manuscript. We also thank Brenda Collins for critical reading of the manuscript, and Chia-Yin Lee for technical assistance. We thank Lan Feng Fan for assistance with the illustrations.
This work was supported by grants 89-2313-B-002-035 and 89-2313-B-002-172 from the National Science Council of Taiwan.
REFERENCES
- 1.Bochtler, M., C. Hartmann, H. K. Song, G. P. Bourenkov, H. D. Bartunik, and R. Huber. 2000. The structures of HslU and the ATP-dependent protease HslU-HslV. Nature 403:800-805. [DOI] [PubMed] [Google Scholar]
- 2.Canceill, D., E. Dervyn, and O. Huisman. 1990. Proteolysis and modulation of the activity of the cell division inhibitor SulA in Escherichia coli lon mutants. J. Bacteriol. 172:7297-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuang, S.-E., I. G. P. B. Burland, D. L. Daniels, and F. R. Blattner. 1993. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene 134:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Fields, S., and O. K. Song. 1993. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 5.Gimeno, R. E., P. Espenshade, and C. A. Kaiser. 1996. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol. Biol. Cell 7:1815-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman, S., W. P. Clark, V. de Crecy-Lagard, and M. R. Maurizi. 1993. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 268:22618-22626. [PubMed] [Google Scholar]
- 8.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi, a human G1 phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 9.Higashitani, A., Y. Ishii, Y. Kato, and K. Horiuchi. 1997. Functional dissection of a cell-division inhibitor, SulA, of Escherichia coli and its negative regulation by Lon. Mol. Gen. Genet. 254:351-357. [DOI] [PubMed] [Google Scholar]
- 10.Jubete, Y., M. R. Maurizi, and S. Gottesman. 1996. Role of the heat shock protein DnaJ in the Lon-dependent degradation of naturally unstable proteins. J. Biol. Chem. 271:30798-30803. [DOI] [PubMed] [Google Scholar]
- 11.Kanemori, M., K. Nishihara, H. Yanagi, and T. Yura. 1997. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J. Bacteriol. 179:7219-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanemori, M., H. Yanagi, and T. Yura. 1999. The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J. Bacteriol. 181:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama-Fujimura, Y., S. Gottesman, and M. R. Maurizi. 1987. A multiple-component ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262:4477-4485. [PubMed] [Google Scholar]
- 14.Kessel, M., W.-F. Wu, S. Gottesman, E. Kocsis, A. C. Steven, and M. R. Maurizi. 1996. Six-fold rotational symmetry of ClpQ, the E. coli homolog of the 20S proteasome, and its ATP-dependent activator, ClpY. FEBS Lett. 398:274-278. [DOI] [PubMed] [Google Scholar]
- 15.Khattar, M. M. 1997. Overexpression of the hslVU operon suppresses SOS-mediated inhibition of cell division in Escherichia coli. FEBS Lett. 414:402-404. [DOI] [PubMed] [Google Scholar]
- 16.Levchenko, I., C. K. Smith, N. P. Walsh, R. T. Sauer, and T. A. Baker. 1997. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell 91:939-947. [DOI] [PubMed] [Google Scholar]
- 17.Maurizi, M. R., W. P. Clark, Y. Katayama, S. Rudikoff, J. Pumphrey, B. Bowers, and S. Gottesman. 1990. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265:12536-12545. [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Missiakas, D., F. Schwager, J. M. Betton, C. Georgopoulos, and S. Raina. 1996. Identification and characterization of HslV HslU(ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 15:6899-6909. [PMC free article] [PubMed] [Google Scholar]
- 20.Rohrwild, M., O. Coux, H.-C. Huang, R. P. Moerschell, S. J. Yoo, J. H. Seol, C. H. Chung, and A. L. Goldberg. 1996. HslV-HslU: a novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. USA 93:5808-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohrwild, M., G. Pfeifer, U. Santarius, S. A. Muller, H.-C. Huang, A. Engel, W. Baumeister, and A. L. Goldberg. 1997. The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat. Struct. Biol. 4:133-139. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Seemuller, E., A. Lupas, D. Stock, J. Lowe, R. Huber, and W. Baumeister. 1995. Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268:579-582. [DOI] [PubMed] [Google Scholar]
- 24.Seong, I. S., M. S. Kang, M. K. Choi, J. W. Lee, O. J. Koh, J. Wang, S. H. Eom, and C. H. Chung. 2002. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J. Biol. Chem. 277:25976-25982. [DOI] [PubMed] [Google Scholar]
- 25.Seong, I. S., J. Y. Oh, S. J. Yoo, J. H. Seol, and C. H. Chung. 1999. ATP-dependent degradation of SulA, a cell division inhibitor, by the HslVU protease in Escherichia coli. FEBS Lett. 456:211-214. [DOI] [PubMed] [Google Scholar]
- 26.Smith, C. K., T. A. Baker, and R. T. Sauer. 1999. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci. USA 96:6678-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, H. K., C. Hartmann, R. Ramachandran, M. Bochtler, R. Behrendt, L. Moroder, and R. Huber. 2000. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA 97:14103-14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, J., J. J. Song, M. C. Franklin, S. Kamtekar, Y. J. Im, S. H. Rho, I. S. Seong, C. S. Lee, C. H. Chung, and S. H. Eom. 2001. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9:177-184. [DOI] [PubMed] [Google Scholar]
- 29.Wu, W. F., Y. N. Zhou, and S. Gottesman. 1999. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J. Bacteriol. 181:3681-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]