Abstract
While the genomes of a number of Mycoplasma species have been fully determined, there has been limited characterization of which genes are essential. The surface protein (p47) identified by monoclonal antibody B3 is the basis for an enzyme-linked immunosorbent assay for serological detection of Mycoplasma gallisepticum infection and appears to be constitutively expressed. Its gene was cloned, and the DNA sequence was determined. Subsequent analysis of the p47 amino acid sequence and searches of DNA databases found homologous gene sequences in the genomes of M. pneumoniae and M. genitalium and identity with a gene family in Ureaplasma urealyticum and genes in M. agalactiae and M. fermentans. The proteins encoded by these genes were found to belong to a family of basic membrane proteins (BMP) that are found in a wide range of bacteria, including a number of pathogens. Several of the BMP family members, including p47, contain selective lipoprotein-associated motifs that are found in macrophage-activating lipoprotein 404 of M. fermentans and lipoprotein P48 of M. agalactiae. The p47 gene was predicted to encode a 59-kDa peptide, but affinity-purified p47 had a molecular mass of approximately 47 kDa, as determined by polyacrylamide gel analysis. Analysis of native and recombinant p47 by mass peptide fingerprinting revealed the absence of the carboxyl end of the protein encoded by the p47 gene in native p47, which would account for the difference seen in the predicted and measured molecular weights and indicated posttranslational cleavage of the lipoprotein at its carboxyl end. A DNA construct containing the p47 gene interrupted by the gene encoding tetracycline resistance was used to transform M. gallisepticum cells. A tetracycline-resistant mycoplasma clone, P2, contained the construct inserted within the genomic p47 gene, with crossovers occurring between 73 bp upstream and 304 bp downstream of the inserted tetracycline resistance gene. The absence of p47 protein in clone P2 was determined by the lack of reactivity with rabbit anti-p47 sera or monoclonal antibody B3 in Western blots of whole-cell proteins. There was no difference between the p47− mutant and wild-type M. gallisepticum in pathogenicity in chicken tracheal organ cultures. Thus, p47, although homologous to genes that occur in many prokaryotes, is not essential for growth in vitro or for attachment and the initial stages of pathogenesis in chickens.
Mycoplasmas are parasitic species that have evolved through reductive evolution from gram-positive bacteria and have developed complex relationships with their hosts (39, 40). In many of their mammalian hosts, they produce chronic infections causing respiratory, arthritic, and urogenital diseases. The absence of a cell wall has been associated with an increase in the number and functionality of lipoproteins in mycoplasmas and the development of strategies to evade or control the immunological environment of the host. A number of lipoproteins have been shown to be subject to antigenic and/or phase variation (4, 29, 30, 36, 37, 45, 49). There are also a small number of proteins that have been shown to be potent modulators of the immune system. These modulatory proteins include macrophage-activating lipoprotein 2 (MALP-2) of Mycoplasma fermentans (34), VlpA and VlpC of M. hyorhinis (35), and the superantigen, MAM, of M. arthritidis (6, 11). However, because of the limited availability of tools for genetic manipulation of mycoplasmas, there have been no studies specifically disrupting lipoprotein genes to establish whether they are essential or their roles in pathogenesis.
M. gallisepticum causes chronic respiratory disease in birds and also causes synovitis and occasionally encephalopathy in turkeys (27). There are at least 35 acylated proteins in M. gallisepticum, of which nearly half are immunoreactive with sera from infected chickens (26). The major membrane antigen of M. gallisepticum is VlhA (previously referred to as pMGA), a 67-kDa lipoprotein that is encoded by members of a multigene family and is subject to antigenic variation (31, 32). A second cell surface protein was first identified by monoclonal antibody (MAb) B3 (14). The MAb B3 epitope forms the basis of a blocking enzyme-linked immunosorbent assay used for serological identification of M. gallisepticum infection (13). The aims of this study were to identify the gene encoding the target for MAb B3, to develop systems with which to inactivate a specific gene (p47) of M. gallisepticum by homologous recombination, producing p47− mutant clones, and then to test whether a p47− mutant clone differs from the wild type in colonization of and pathogenicity in chicken tracheal organ cultures. The techniques and results will ultimately lead to further understanding of the pathogenesis of M. gallisepticum and other mycoplasmas.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. gallisepticum strains S6 and ts-11 (29, 48) were grown at 37 and 33°C, respectively, in modified Frey's medium (47), and the cells were harvested when the cultures reached the late logarithmic phase of growth. For colony growth, petri dishes containing modified Frey's medium (without phenol red or glucose) solidified with 0.8% Noble agar were inoculated with mycoplasma cells and the cells were grown at 37°C in an airtight container. For the growth and selection of tetracycline-resistant mycoplasma transformants, tetracycline was added to liquid or solid medium at a concentration of 4 μg/ml.
In experiments involving tracheal organ cultures, M. gallisepticum S6 and the transformant P2 were grown in modified Friis medium containing 18% horse serum instead of swine serum and 0.05% ampicillin instead of bacitracin and methicillin (21). The number of viable organisms was determined as previously described (43).
Affinity purification of protein.
An immunoaffinity column was constructed by using 5 mg of MAb B3 (14) coupled to 1 g of cyanogen bromide-activated Sepharose 4b (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions. Cells from an overnight culture of M. gallisepticum strain ts-11 were pelleted by centrifugation at 20,000 × g for 20 min at 4°C. The cells were then resuspended in phosphate-buffered saline (PBS). Centrifugation was repeated, and the cells were resuspended in 1/20 of the original volume of lysis buffer containing 0.01 M Tris-HCl (pH 7.4)-1% Triton X-100-0.1% phenylmethylsulfonyl fluoride and incubated for 2 h at 37°C. The cell lysate was centrifuged at 100,000 × g for 30 min at 4°C, and the supernatant was passed over the MAb B3 affinity column. Unbound protein was washed from the column with lysis buffer, and bound protein was eluted by addition of 0.5 M diethylamine (pH 11.5). Fractions containing bound protein were collected in a predetermined amount of glacial acetic acid, and the pH was stabilized by the addition of 2 M Tris-HCl (pH 7.4) and dialyzed against PBS overnight at 4°C.
Radioimmunoprecipitation.
Cells from an overnight culture of M. gallisepticum strain ts-11 were pelleted by centrifugation as described above, resuspended in 1/10 of the volume of broth medium, and metabolically radiolabeled by adding 2 mCi of [35S]methionine-cysteine (Amersham Pharmacia Biotech) and incubation for 18 h at 33°C. Radioimmunoprecipitation (RIP) was conducted as described previously (18), with minor alterations. Radiolabeled cells were collected by centrifugation, washed twice in PBS as described above, and solubilized in RIP lysis buffer (0.05 M Tris-HCl [pH 7.5], 1% Triton X-100, 0.6 M KCl, 1 mM phenylmethylsulfonyl fluoride), sonicated briefly, and centrifuged at 100,000 × g at 4°C for 30 min. Non-specifically reactive proteins were removed by incubation of 10 μg of a MAb of the same isotype as MAb B3 and then repeated addition and removal of aliquots of protein A Sepharose beads (Amersham Pharmacia Biotech). To equal aliquots of precleared antigen, 6 μg of MAb 71 (specific for VlhA, a 67-kDa protein of M. gallisepticum) or 10 μg of MAb B3 (with each MAb bound to 50 μl of protein A beads) was added and the mixtures were incubated for 30 min at 4°C with shaking. The beads were pelleted, washed twice with RIP assay buffer (0.05 M Tris-HCl [pH 8.0], 1 mM EDTA, 0.15 mM NaCl, 0.25% [wt/vol] bovine serum albumin, 1% Triton X-100) and twice with PBS, and finally resuspended in reducing sample buffer for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The samples and 14C-labeled molecular weight standards (Amersham Pharmacia Biotech) were separated in a 10% polyacrylamide gel. The gel was incubated in Enhance (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions, dried, and autoradiographed with Biomax film (Kodak).
Preparation of immune sera.
The affinity-purified 47-kDa protein (p47) was separated by SDS-PAGE and lightly stained with Coomassie brilliant blue (CBB) in water. The CBB-stained band containing p47 was excised from the gel, emulsified with Freund's complete adjuvant, and used to immunize New Zealand White rabbits in accordance with the method of Harlow and Lane (24). Three further boosters at 1-month intervals were given by using polyacrylamide gel slices containing p47 emulsified with Freund's incomplete adjuvant. Serum was obtained 10 days after each booster, and the titer was assessed by immunostaining.
Construction of expression library, cloning, DNA sequencing, and Southern blot analysis.
An expression library of M. gallisepticum strain ts-11 DNA cloned into expression vector pGEX1N was screened by using rabbit anti-p47 sera as described previously (19). Briefly, to identify the gene encoding p47, M. gallisepticum strain ts-11 DNA was partially digested with the restriction enzyme Sau3AI and ligated into the BamHI site of the pGEX1N expression vector and the mixture was used to transform Escherichia coli DH5α cells. The resultant recombinant colonies were screened for antigen production by using a 1/20 dilution of rabbit anti-p47 sera that had been adsorbed against E. coli colonies. A single immunoreactive clone (5b) was selected and found to contain a 34-bp Sau3AI insert. Oligonucleotide primer AZ (Table 1), which is identical to the insert of clone 5b, was synthesized, radiolabeled with [γ-32P]ATP (Amersham Pharmacia Biotech) and polynucleotide kinase (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions, and used to probe Southern blots of restriction endonuclease digests of M. gallisepticum ts-11 genomic DNA as described previously (32). A genomic library of BglII-digested strain ts-11 genomic DNA ligated to BamHI-digested pUC18 vector DNA in E. coli DH5α was screened with the radiolabeled oligonucleotide probe as previously described (32). A probe-reactive clone (t46) containing a 3.8-kbp insert was recovered, and the complete DNA sequence was determined by cycle sequencing with Big Dye Terminator chemistry (Applied Biosystems) with a combination of primer walking, subcloning, and utilization of the universal priming sites within the vector.
TABLE 1.
Oligonucleotides used in this study
| Oligo- nucleotide | Sequencea (5′-3′) |
|---|---|
| BQ | ggatccTCTAGTGGTGCTGCTTCTTCAC |
| BP | GTTCCAGGTCGTTTcCACACACCATTGTTAAC |
| BO | GTTAACAATGGTGTGTGgAAACGACCTGGAAC |
| BN | TGTTATCTGCTGCATTAGAGCTTACcCAAGAA |
| BL | TTGgGTAAGCTCTAATGCAG |
| BU | GTAGCTAAGTTATTcCATCCATATCCATAG |
| BT | CTATGGATATGGATGgAATAACTTAGCTAC |
| BG | gtcgacTTAGTTTAATTTCTTGAAGAC |
| BC | CTGTTGAGAAAGCGATTGTC |
| BD | CATATTCAGTGCTGTAGAAG |
| CB | gaattcCTAGTAGAGAAAACTGGAAA |
| BR | TAAGAAAAATGGAactagtATTTCAGCAAT |
| BS | ATTGCTGAAATactagtTCCATTTTTCTTA |
| CD | GAGTTGTATACCTCTATAGC |
| CE | GCATAGTCATTATATAATCCGAC |
| AZ | GATCAAACTACTCCAAGACAAGAAATGTCTGATC |
| TetFor | GAAAagatctGGAGTAATTGGAAG |
| TetRev | actagtCCATATTTATATAACAACTT |
Lowercase indicates nuclotide modification to produce a restriction endonuclease cleavage site or alter the UGA tryptophan codon to UGG.
Expression of p47 in E. coli.
To overcome the truncation of expression products in E. coli at mycoplasma tryptophan codon UGA, the first three UGA codons of the p47 gene were mutagenized to UGG by overlap extension PCR. Briefly, separate PCRs were conducted with 5 ng of clone t46 DNA as the template for each of the oligonucleotide primer pairs BQ-BP, BO-BN, BL-BU, and BT-BG (Table 1) in a reaction volume of 50 μl containing 5 μl of 10× reaction buffer, 10 μM each deoxynucleoside triphosphate, 2 μl of 50 mM MgSO4, 12.5 μM each primer, and 1.25 U of Platinum Taq thermo-polymerase (Life Technologies Inc.). PCRs were performed in a thermocycler (Hybaid) under the following conditions: 94°C for 4 min, followed by 35 cycles of 94°C for 20 s, 55°C for 20 s, and 68°C for 40 s, with a final extension at 68°C for 4 min. The PCR products were gel purified with a QIAquick gel extraction kit (Qiagen), and approximately equimolar amounts of the purified products, together with oligonucleotide primers BQ and BG (Table 1), were used in an overlap extension PCR under the same conditions as above, except that the extension time in each cycle was increased from 40 to 100 s. The resultant PCR product was purified with a QIAquick PCR purification kit (Qiagen) and cloned into the pGEM-T vector (Promega) in accordance with the manufacturers' instructions. The DNA sequence of the cloned PCR product was determined by cycle sequencing with Big Dye Terminator chemistry (Applied Biosystems) with the M13 universal priming sites of the vector and oligonucleotides BC, BD, BN, and BT (Table 1). The mutagenized p47 gene was subcloned into expression vector pGEX4T-1 (Amersham Pharmacia Biotech), and recombinants were selected in accordance with the manufacturer's instructions. Expression of recombinant p47 was induced in cultured E. coli by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 2 mM. The recombinant p47 protein was purified with a glutathione Sepharose column (Amersham Pharmacia Biotech), and the fusion partner glutathione S-transferase (GST) was removed by cleavage with thrombin (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions. The thrombin-cleaved recombinant p47 was tested for reactivity and purity by SDS-PAGE and Western blotting.
Northern blot analysis of p47 gene transcript.
The total RNA from a culture of M. gallisepticum strain ts-11 was extracted by the method of Chomczynski and Sacchi (9) as described previously (23). The RNA was separated by electrophoresis in a 1.2% agarose gel containing 0.22 M formaldehyde and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech). A product of 233 bp derived from the 3′ end of the p47 gene was amplified by PCR with oligonucleotide primers CB and BG (Table 1; see Fig. 3). The 233-bp DNA product and the full-length p47 gene were radiolabeled with [α-32P]dATP (Amersham Pharmacia Biotech) and a Random Primed DNA Labeling Kit (Amersham Pharmacia Biotech). The probes were hybridized with the membrane in Church's buffer (10) at 55°C overnight, washed three times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS for 10 min each time, and subjected to autoradiography with Biomax film (Kodak).
FIG. 3.
Pictorial representation of the cloned BglII DNA fragment encoding p47. The 3.894-kbp DNA sequence contained three ORFs, one complete ORF encoding p47 (ORF2) and two partial ORFs. The predicted leader sequences of ORF2 and -3 are boxed and hatched, and the acylated cysteine residue and the three amino acids immediately preceding each are also shown. The recognition sites for BglII, EcoRI, and HindIII are shown, together with the region identical to the sequence of oligonucleotide AZ (stippled box in ORF2). The positions of the UGA tryptophan codons are shown below ORF2 (p47). The positions for selected oligonucleotide primers from Table 1 are shown.
Peptide mass fingerprinting of recombinant and native p47.
Approximately 2 μg each of native and thrombin-cleaved recombinant p47 was separated by SDS-PAGE and visualized by staining with CBB. The p47 bands were excised from the acrylamide gel and vacuum dried.
Gel pieces were washed three times with 50% acetonitrile (AcN)-25 mM NH4HCO3 (pH 7.8) and dried. To the dried gel piece, 120 ng of trypsin (Promega) in 25 mM NH4HCO3 (pH 7.8) was added and the mixture was incubated at 37°C overnight. The resulting peptides were extracted from the gel with a 50% (vol/vol) AcN-1% (vol/vol) trifluoroacetic acid solution. A 1-μl aliquot was mixed with an equal volume of matrix (α-cyano-4-hydroxycinnamic acid at 8 mg/ml in 70% [vol/vol] AcN-1% [vol/vol] trifluoroacetic acid) and allowed to air dry.
Matrix-assisted laser desorption ionization-mass spectrometry was performed with a Micromass TofSpec 2E time-of-flight mass spectrometer. A nitrogen laser (337 nm) was used to irradiate the sample. The spectra were acquired in reflectron mode in the mass range of 600 to 3,500 Da. A near point calibration was applied, which will give a typical mass accuracy of around 100 ppm or less.
Production of a DNA construct for homologous recombination.
A DNA construct containing the p47 gene and the gene encoding tetracycline resistance was first prepared by amplifying the p47 gene in two regions so that an SpeI restriction site could be created approximately 400 bp from 5′ end of the coding sequence. This was accomplished with oligonucleotide primer pairs BQ-BS and BR-BG (Table 1) under PCR conditions similar to those described above. The full-length p47 gene containing an SpeI cleavage site was then regenerated by overlap extension PCR with primer pair BQ-BG (Table 1; see Fig. 3) as described above, and the PCR product was purified with the QIAquick PCR purification kit (Qiagen) in accordance with the manufacturer's protocols. The tetM gene of the vector pAM120 (22) was amplified with primers TetFor and TetRev (Table 1) under PCR conditions similar to those described above, and the product was cloned into the pGEM-T (Promega) vector in accordance with the manufacturer's protocols. Recombinant colonies were selected on Luria-Bertani agar containing 50 μg of ampicillin/ml and 20 μg of tetracycline/ml. The tetM gene was released from the pGEM-T plasmid by digestion with the restriction enzyme SpeI, by using both the SpeI restriction site engineered into the TetRev primer and that within the multicloning site of the vector, ligated to the SpeI-digested p47 PCR product, and cloned into the pGEM-T vector (Promega) as described above. Recombinants were selected, and the construct was verified by restriction endonuclease mapping. For large-scale purification of construct DNA, the Qiagen Plasmid Midi Kit was used in accordance with the manufacturer's protocols.
Transformation of M. gallisepticum.
M. gallisepticum strain S6 was transformed with polyethylene glycol as described by Minion and Kapke (33). Briefly, a broth culture of M. gallisepticum strain S6 was grown to the late log phase, harvested by centrifugation, washed twice in PBS, resuspended in 1/20 of the original volume of 0.1 M CaCl2, and incubated on ice for 45 min. Approximately 10 μg of the DNA construct in 0.1 M CaCl2 was added to the cells, followed by 2 ml of a 40% polyethylene glycol solution, the mixture was vortexed for 15 s, and 25 ml of PBS was added. The mycoplasma cells were pelleted by centrifugation, resuspended in warm mycoplasma broth medium, and incubated at 37°C for 1.5 h. The cells were inoculated onto mycoplasma agar medium containing 4 μg of tetracycline/ml and incubated at 37°C in a light-safe container. After 5 days, the agar plates were examined and colonies that had formed were selected and inoculated into mycoplasma broth medium containing 4 μg of tetracycline/ml. After growth at 37°C, each mycoplasma clone was examined by PCR for the presence of the tetM gene and cultures containing the tetM gene were further tested by colony blotting and Western blotting for the presence of p47 with specific antibodies. To examine the site of recombination, the DNA region encoding p47 in transformed clone P2 was amplified with oligonucleotide primer pair CD-CE and the resultant 5.1-kbp PCR product was cloned by insertion into the pGEM-T (Promega) vector and subjected to DNA sequencing.
Computer documentation of results.
Images for figures were captured and annotated with the Adobe Photoshop and Illustrator computer software packages.
Chicken embryo tracheal organ cultures.
The method of Cherry and Taylor-Robinson (8), with modifications, was used to prepare chicken embryo tracheal organ cultures. Twenty-day-old specific-pathogen-free chick embryos were removed from their shells and euthanized, and the tracheas were excised aseptically. After washing the tracheas with Eagle's basal medium, thin ring sections (about 0.5 mm) were prepared with a scalpel. The rings were placed individually into tubes containing 0.5 ml of medium composed of equal volumes of Eagle's medium and modified Friis medium. The tubes were placed into a roller drum (8 revolutions/h) and incubated at 37°C.
After 2 days, the rings were observed at 100× magnification with a Zeiss microscope. Rings not showing 100% ciliary motility were discarded.
Groups of rings with 100% ciliary motility (three rings in each group) were inoculated with 0.5 ml of different concentrations of either the M. gallisepticum strain S6 or the transformed P2 culture. The inoculated rings were checked for ciliary motility and color change of the medium every 4 to 5 h for 3 days.
Nucleotide sequence accession number.
The nucleotide sequence of the p47 gene of M. gallisepticum strain ts-11 has been submitted to the GenBank database and assigned accession number AY065985.
RESULTS
Immunoaffinity purification of a 47-kDa protein (p47).
M. gallisepticum strain ts-11 cellular proteins were subjected to immunoprecipitation with MAb B3 or 71 or to immunoaffinity chromatography with MAb B3. A single band of approximately 47 kDa (p47) was immunoprecipitated or affinity purified with MAb B3 (Fig. 1), while a band of approximately 67 kDa (VlhA) was immunoprecipitated with MAb 71 (Fig. 1).
FIG. 1.
Immunoprecipitation of p47 with MAb B3 and reactivity of rabbit antisera with p47. (A) M. gallisepticum strain ts-11 cell proteins radiolabeled with [35S]methionine-cysteine and immunoprecipitated with MAb B3 (lane 1) or 71 (lane 2). The radiolabeled immunoprecipitated proteins, together with 14C-labeled standards (Amersham Pharmacia Biotech), were separated by SDS-12.5% PAGE. (B) Affinity-purified protein from a MAb B3 Sepharose column together with low-molecular-weight standards (Bio-Rad) separated by SDS-12.5% PAGE and stained with CBB. (C) Western blot of whole-cell proteins of M. gallisepticum strains ts-11 and S6. Cellular proteins and prestained molecular weight markers (New England BioLabs) were separated by SDS-12.5% PAGE, Western transferred, and immunostained with rabbit anti-p47 sera.
Rabbit anti-p47 sera (raised against affinity-purified p47) reacted strongly in a Western blot with a band of approximately 47 kDa and faintly with a band of approximately 43 kDa in strain ts-11 whole-cell proteins and with a single band of approximately 47 kDa in strain S6 cellular proteins (Fig. 1).
Cloning of the gene encoding p47.
To clone the gene encoding p47, a library of M. gallisepticum strain ts-11 DNA ligated to expression vector pGEX1N was screened with rabbit anti-p47 sera. Immunoreactive clone 5b was found to contain a 34-bp mycoplasma DNA Sau3AI fragment. Oligonucleotide AZ (Table 1), with the same DNA sequence, was synthesized, radiolabeled, and hybridized to a Southern blot of M. gallisepticum strain ts-11 DNA digested with BglII, EcoRI, and HindIII. The radiolabeled oligonucleotide bound to single bands of 3.8, 4.6, and 1.3 kbp, respectively, in BglII-, EcoRI-, and HindIII-digested DNA (Fig. 2). A library of M. gallisepticum strain ts-11 DNA digested with BglII ligated into vector pUC18 was screened with the radiolabeled AZ oligonucleotide as a probe. Clone t46, containing a 3.8-kbp mycoplasma DNA insert, was recovered, and the DNA sequence of the insert was determined.
FIG. 2.
Southern blot analysis of M. gallisepticum strain ts-11 genomic DNA. M. gallisepticum strain ts-11 genomic DNA was digested with restriction endonucleases BglII (lane 1), EcoRI (lane 2), or HindIII (lane 3) and separated in a 0.8% agarose gel. HindIII-digested phage lambda DNA was used as a molecular weight marker. The separated DNA was transferred to a nylon membrane and probed with 32P-radiolabeled oligonucleotide AZ (Table 1).
Analysis of the DNA sequence revealed a 3.894-kbp insert containing one complete open reading frame (ORF) and two incomplete ORFs, with each ORF predicted to be transcribed in the same direction (Fig. 3). The nucleotide sequence of oligonucleotide AZ was present in ORF2. The putative p47 gene was 1.755 kbp in length and was immediately followed by a predicted transcriptional terminator at nucleotide positions 2163 to 2185. It would code for a peptide of 584 amino acids with a predicted molecular mass of 61.82 kDa and contained a 29-amino-acid leader sequence with a typical prokaryotic acylation consensus sequence (LASC). The acylation motif, together with observations that p47 partitioned into the hydrophobic phase following Triton X-114 fractionation (results not shown), suggested that p47 was a lipoprotein. Searches, with the BLASTX software available through the Australian National Genomics Information Service (38), of TrEMBL or SWISS-PROT databases (release numbers 17 and 39, respectively) revealed that the closest matches to the p47 peptide were M. pneumoniae MPN052 (SWISS-PROT accession no. Y040_MYCPN) and its homologue in M. genitalium MG040 (SWISS-PROT accession Y040_MYCGE). Similarity was also seen with Ureaplasma urealyticum genes UU226, UU480, and UU016 (TrEMBL accession no. Q9PQR5, Q9PQ10, and Q9PRC9, respectively) and, to a lesser extent, with M. agalactiae P48 (TrEMBL accession no. Q9X775) and the M. fermentans MALP-404 precursor (TrEMBL accession no. Q9R3N6). Searches of the Pfam database (version 6.6) (3) revealed that p47 contained protein domains with homology to the basic membrane protein (BMP) family. The BMP family includes BMP precursors A, B, C, and D of Borrelia burgdorferi, the TMPC precursor of Treponema pallidum, and the YUFN precursor and transcriptional activator protein MED precursor of Bacillus subtilis. A common theme among the members of the BMP family of proteins is that they are outer membrane proteins that produce an immune response when expressed by pathogenic strains in a host. The selective lipoprotein-associated (SLA) motif first identified in MALP-404 of M. fermentans by Calcutt et al. (5) and expanded and modified by Rosati et al. (44) following analysis of P48 of M. agalactiae may be found in several of the BMP family members. The two SLA motifs were found in p47, although the second SLA motif exhibited greater identity (Fig. 4, shaded regions in amino acid sequence).
FIG. 4.
Mass spectrometry of recombinant and native p47. Samples of recombinant and native p47 were subjected to digestion with trypsin. Resultant peptides were analyzed by mass spectrometry, and the spectrum of each digest is shown. The molecular weights of the peptides were compared to an in silico tryptic digest of p47. Peptides identified in both recombinant and native p47 are doubly underlined, and peptides identified in only recombinant p47 are singly underlined. Also shown are the SLA-1 (shaded, labeled 1) and SLA-2 (shaded, labeled 2) amino acid motifs predicted by Rosati et al. (44).
The ORF1 sequence of 229 bp (following the BglII restriction site) showed the highest level of identity (BlastP, p = 2e-10) with the recR gene (TrEMBL accession no. Q9PR56) of U. urealyticum, while ORF3 (truncated to 1,462 bp by the BglII restriction site) was a member of the vlhA gene family of M. gallisepticum, with the highest level of identity to the pMGA1.4 gene (TrEMBL accession no. Q49499).
Expression of the p47 gene.
To aid in the expression and cloning of the gene encoding p47, several nucleotide changes were made to the gene sequence by PCR overlap extension techniques. The gene was truncated by removing the leader peptide together with the acylation signal sequence (replacing the amino-terminal cysteine with serine) and converting the first three of four tryptophan UGA codons to UGG. The mutated gene, p47-mut, was cloned into the pGEX4T-1 vector, resulting in expression of a recombinant fusion protein. The GST-p47-mut fusion protein was purified over a glutathione Sepharose column (Fig. 5A, lane 1), resulting in two major bands of 88 and 74 kDa. Cleavage of the GST fusion partner from p47-mut by thrombin resulted in three major products (Fig. 5A, lane 2). The lowest band corresponded to the GST partner after thrombin cleavage, while the 62-kDa band corresponded to the full-length p47-mut gene product. The 48-kDa band product most probably corresponded to a premature truncation of the full-length protein as a result of translational termination at the fourth tryptophan UGA codon at amino acid position 518, which is predicted to yield a product 7.458 kDa smaller than full-length p47-mut (which has a predicted size of 59.2 kDa). Western blot analysis of the GST-p47-mut thrombin-digested products showed that rabbit anti-GST sera bound only the 26-kDa GST protein (Fig. 5B, lane 1). Rabbit anti-p47 sera reacted with the two main recombinant protein bands of 48 and 62 kDa and several minor bands lower than the 62-kDa band (Fig. 5B, lane 2) that probably resulted from degradation of the full-length product. MAb B3 reacted only with the upper band of 62 kDa (Fig. 5B, lane 3), suggesting that the epitope for the MAb resided close to, or carboxyl to, the fourth tryptophan codon (see below).
FIG. 5.
Expression, purification, and cleavage of recombinant p47 and Northern blot analysis of p47 gene transcript. Gene p47-mut was cloned and expressed with the pGEX4T-1 expression vector. The fusion product was purified over a glutathione Sepharose column and cleaved with thrombin. The fusion product and the cleaved fusion products, together with molecular weight standards (Novex) or prestained molecular weight standards (New England Biolabs), were separated by SDS-12.5% PAGE and either stained with CBB or Western transferred. Total RNA of M. gallisepticum strain ts-11 cells was extracted and, together with RNA standards (Promega), electrophoresed, blotted onto nylon membrane, and probed with the 233-bp region encoding the carboxyl-terminal end of p47. (A) CBB-stained gel of E. coli-expressed p47 protein following purification over a glutathione Sepharose column (lane 1) and after cleavage with thrombin (lane 2). (B) GST-p47 products immunostained with rabbit anti-GST sera (lane 1), rabbit anti-p47 sera (lane 2), or MAb B3 (lane 3). (C) Ethidium bromide-stained total RNA of M. gallisepticum strain ts-11 cells (lane 1). Northern blot hybridized with DNA from the carboxyl region of the p47 gene. The probe bound a 1.98-kb band (arrowed, lane 2).
Full-length RNA transcript of the p47 gene.
To determine if the full length of the p47 gene was transcribed, total RNA of M. gallisepticum strain ts-11 cells (Fig. 5C, lane 1) was electrophoretically separated and probed with radiolabeled DNA corresponding to the 3′ end or the full length of the p47 gene sequence. A band of approximately 1.98 kb hybridized with the 3′ end (Fig. 5C, lane 2) or the full-length (results not shown) gene probes. The size of this transcript was equivalent to that which would be predicted for the full-length monocistronic p47 gene transcript.
Peptide mass fingerprinting reveals that native p47 is truncated at the carboxyl-terminal end.
Recombinant and native p47 proteins were subjected to mass spectrometry following proteolytic cleavage with trypsin. The resultant mass spectra obtained were then used to search an in silico tryptic digest of p47. Analysis of the spectra revealed that most of the tryptic fragments within the amino-terminal half of p47 were present in both recombinant and native p47 (Fig. 4, double underlined). Three tryptic peptides were detected only in recombinant p47 (singly underlined). Even though native p47 did not contain the full TMVIMQYVNILAGTSTLLPAASRENWK sequence, it is possible that it contained some of the amino-terminal end of this peptide. Furthermore, since MAb B3 binds native p47, but only full-length recombinant p47, the epitope for MAb B3 may be close to or overlap the fourth encoded tryptophan, which is contained in this peptide. The other two tryptic peptides, which were detected only in recombinant p47, suggest that this region was absent from affinity-purified native p47 and that this was most likely responsible for the difference between the predicted molecular weight of the gene product and that determined by SDS-PAGE.
Transformation of M. gallisepticum with p47-tetM gene construct.
A DNA construct containing the p47 gene interrupted by the tetracycline resistance gene (Fig. 6A) was used to transform cells of M. gallisepticum strain S6. Of two tetracycline-resistant clones isolated, clone P2 was selected and its genomic DNA was Southern blotted and hybridized with radiolabeled tetM or p47 gene DNA. The tetM gene probe bound a single band of 3.2 kbp and two bands of 2.6 and 1.0 kbp, respectively, in BglII- and HindIII-digested P2 genomic DNA (Fig. 7A, part 1, lanes 2 and 4) but did not bind to similarly digested S6 DNA (Fig. 7A, part 1, lanes 1 and 3). Radiolabeled p47 DNA bound to a 3.0-kbp band and weakly to a 3.2-kbp band in BglII-digested P2 DNA (Fig. 7B, part 2, lane 2) and to bands of 2.6, 1.2, and 1.0 kbp in HindIII-digested P2 genomic DNA (Fig. 7B, part 2, lane 4). The p47 probe bound to a single band of 4.0 kbp in BglII-digested parental S6 DNA and to two bands of 1.2 and 1.3 kbp in HindIII-digested S6 genomic DNA (Fig. 7A, part 2, lanes 1 and 3). The predicted restriction endonuclease maps of the BglII DNA fragments containing the p47 gene of M. gallisepticum strain S6 and the P2 clone are shown in Fig. 7B and C.
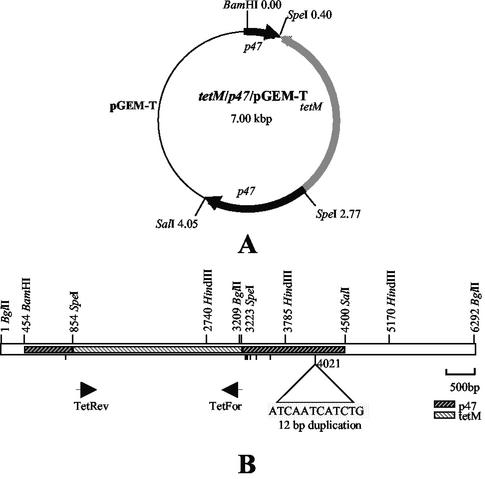
FIG. 6.
Schematic diagram of the p47-tetM gene construct in pGEM-T and clone C46. (A) Depiction of the vector pGEM-T containing the tetM gene ligated into the SpeI sites created in the p47 gene. (B) The location of the p47-tetM gene construct within the M. gallisepticum strain ts-11 3.894-kbp BglII fragment is depicted. The cleavage sites for BamHI, BglII, HindIII, SalI, and SpeI and the locations of oligonucleotides TetFor and TetRev are shown. The positions of nucleotides present in the C46 DNA sequence, but absent from the S6 p47 gene sequence, are indicated by vertical lines.
FIG. 7.
Southern blot analysis of DNA from transformed clone P2 of M. gallisepticum strain S6 and predicted restriction map of insert. The genomic DNA of M. gallisepticum strain S6 (lanes 1 and 3 of each part) and clone P2 (lanes 2 and 4 of each part) was digested with BglII or HindIII. The digested DNA and HindIII-digested lambda phage DNA (used as a molecular weight marker) were separated in a 0.8% agarose gel. The DNA fragments were transferred to a nylon membrane and probed with 32P-radiolabeled tetM gene DNA (panel A, part 1) or p47 gene DNA (panel A, part 2), and the predicted restriction maps based on hybridization and DNA sequencing are shown in panels B and C.
Oligonucleotides CD and CE were used to PCR amplify the region surrounding and including the p47 gene in transformant P2 (Fig. 6B). A PCR product of approximately 5 kbp was amplified, and subsequent restriction endonuclease digestion with SpeI and other enzymes showed it to contain an insert similar to that of the tetM gene (results not shown). The PCR product was cloned into the pGEM-T vector, and the resultant clone, C46, was subjected to DNA sequencing. The gene encoding p47 in M. gallisepticum strain S6 was also amplified by PCR, and the DNA sequence was determined. The DNA sequences of C46, the M. gallisepticum S6 p47 gene, and the original pGEM-T construct were aligned and analyzed with ClustalW (46). The alignment revealed that the tetM gene from clone C46 was located in the same position as in the p47-tetM construct. The BamHI and SalI cleavage sites (introduced by the BQ and BG oligonucleotide primers, respectively) in the p47-tetM pGEM-T construct were absent in the C46 DNA sequence. There was a 12-bp duplication in the C46 DNA sequence that was also present in the S6 p47 gene sequence but not in the p47 sequence from M. gallisepticum ts-11 used to create the pGEM-T construct (Fig. 8). There were several single nucleotide differences between the S6 and ts-11 p47 gene sequences in both the 5′ and 3′ regions adjacent to the SpeI restriction sites (Fig. 8). The nucleotide sequence of the C46 p47 gene sequences corresponded to the ts-11 p47 gene sequences between the 73rd nucleotide 5′ and the 304th nucleotide 3′ to the SpeI restriction sites in clone C46 DNA. This suggests that the region between nucleotide positions 377 and 796 was involved in the homologous recombination event.
FIG. 8.
Western blot analysis of M. gallisepticum (MG) strain S6 and transformant clone P2. (A) M. gallisepticum strain S6 and transformant clone P2 were grown by initially placing running drops of an actively growing broth culture on agar medium. After colony formation, nitrocellulose blots were taken and strips perpendicular to the running drop containing each culture were cut from the blots and immunostained with MAb 86 (lane 1) or B3 (lane 2). (B) Total cell proteins of M. gallisepticum strain S6 (lane 1) and transformant clone P2 (lane 2) and molecular weight standards (Novex) separated by SDS-12.5% PAGE and immunostained with rabbit anti-p47.
M. gallisepticum strain S6 and transformant P2 were grown in broth and subcultured onto agar by placing a single drop of culture on the agar and allowing it to run down the surface. Nitrocellulose colony blots were taken after 6 days of growth, and strips containing colonies of each culture were immunostained with either MAb B3 or 86 (which binds to the cell surface-exposed VlhA molecule). Both strain S6 and transformant P2 bound MAb 86 (Fig. 8A, lane 1) while strain S6, but not transformant P2, bound MAb B3 (Fig. 8A, lane 2). Western blots of whole-cell proteins from either transformant P2 or M. gallisepticum strain S6 immunostained with rabbit anti-p47 sera showed that only M. gallisepticum strain S6, and not clone P2, expressed detectable p47 (Fig. 8B, lanes 1 and 2).
Pathogenicity in chicken embryo tracheal organ cultures.
The number of CFU of each strain used to inoculate tracheal organ cultures was determined for M. gallisepticum strain S6 and clone P2 and are shown in Table 2. The medium color changed from yellow to a light rose 4 to 5 h before onset of ciliostasis, after which it became pink. Four tubes containing three tracheal rings each were used as uninoculated controls; the rings showed 100% motility after 5 days of incubation. There was no significant difference (t test, two sample, equal variance) in the time taken for ciliostasis to occur in tracheal rings inoculated with either M. gallisepticum strain S6 or clone P2 at any of the three inoculum concentrations tested (Table 2).
TABLE 2.
Time taken for ciliostasis to occur in tracheal rings inoculated with different concentrations of two M. gallisepticum strains
| Organism (inoculum size [CFU/ml]) | Log dilution | No. of CFU | Time (h) to 100% ciliostasis |
|---|---|---|---|
| M. gallisepticum S6 (2 × 108) | −4 | 1 × 104 | 28.3 ± 1.2a |
| −6 | 1 × 102 | 46 ± 0 | |
| −8 | 1 | 48 ± 2.3 | |
| Clone P2 (5 × 108) | −4 | 2.5 × 104 | 36 ± 8.6 |
| −6 | 2.5 × 102 | 41 ± 8.6 | |
| −8 | 2.5 | 46 ± 0 |
Mean time calculated for three tracheal rings inoculated with the same inoculum and concentration.
DISCUSSION
Past mycoplasma research has focused on phenotypic differences or altered capabilities to identify proteins, and ultimately genes, involved in the pathogenesis of these organisms. This has been exemplified by studies of attachment of M. pneumoniae to host cells. With advancement in genetic techniques and tools to transform mycoplasmas, it is now possible to produce specific mutants with which to investigate specific determinants involved in pathogenesis. The results of this study show, for the first time, transformation of the avian pathogen M. gallisepticum by homologous recombination to specifically inactivate a gene and produce a transformant with an altered phenotype.
The cell surface protein of M. gallisepticum recognized by MAb B3 was chosen as the candidate gene for mutation (14). By the techniques of immunoprecipitation and immunoaffinity chromatography, the target for MAb B3 of M. gallisepticum strain ts-11 was identified as a 47-kDa protein (p47). Previous studies with MAb B3 detected a 56-kDa protein in M. gallisepticum strain 1226 by Western blotting, although we were unable to replicate these results in this study (results not shown). In contrast, rabbit anti-p47 sera reacted in a Western blot of M. gallisepticum strain 1226 with a 47-kDa protein (results not shown), supporting our present finding that the target for MAb B3 is p47. Interestingly, immunoaffinity chromatography of whole cells of M. gallisepticum strain 1226 with MAb B3 purified a fraction containing proteins of 43, 45, 47, 56, and 64 kDa (12). Immunization of chickens with this fraction induced protection against virulent M. gallisepticum strain 1226. Whether protection resulted from immunization against one specific protein or the combination of proteins in the fraction was not determined. Vaccination of chickens with recombinant p47, followed by challenge studies, would determine if the protective antigen in the previous study was p47, although this experiment is outside the scope of this paper.
The translated p47 gene is predicted to be a lipoprotein, as it contains a signal peptide followed by an acylation signal sequence directing attachment of lipid moieties to the sulfhydryl group of the cysteine residue. Processing of the prelipoprotein by signal peptidase II would result in removal of the signal peptide to form a mature protein with a predicted molecular size of 59 kDa. A discrepancy in the predicted size of the p47 gene product and that of MAb B3 affinity-purified p47 may result from anomalous migration of the protein within the gel or transcriptional or posttranslational modification. It is unlikely to result from truncation of the gene transcript, as Northern blot analysis showed that a single band of 1.98 kb hybridized with both the full-length p47 gene and a 233-bp region at the 3′ end. Mass spectrometry data suggested that two tryptic peptides from the carboxyl end of the full-length p47 gene product were absent in native p47, indicating that posttranslational cleavage was involved in the production of native p47. Interestingly, there has been a report of cleavage of the p47 ortholog MPN052 (15) in M. pneumoniae by Regula et al. (42), where two fragments, one with a molecular mass of 22 kDa (amino acid positions 317 to 450 of MPN052) and another with a molecular mass of 6 kDa (amino acid positions 598 to 648 of MPN052) were identified by a combination of two-dimensional gel electrophoresis and mass spectrometry. In a later study, Regula et al. (41) reported that the 22-kDa peptide of MPN052 (amino acid positions 317 to 450) was found in the Triton X-100-insoluble fraction of M. pneumoniae.
M. fermentans MALP-404 shares amino acid sequence identity with p47. Within MALP-404 is an amino acid sequence motif that has been termed the SLA motif (5) and identified in other bacterial lipoproteins or in putative proteins. The SLA motif of Calcutt et al. (5) was found in P48 of M. agalactiae and expanded upon by Rosati et al. (44). Subsequent searches of protein databases by Rosati et al. (44) identified a second SLA motif in P48 that was present in peptides of a number of unrelated bacterial species. The peptides MALP-404, P48, MPN052, MG040, and P47 of M. hyorhinis (5, 44) have been identified as belonging to a family of BMPs (16). Classification of these proteins into a family of BMPs may give some insight into the function of these lipoproteins, and perhaps what is known about BMPs in mycoplasma may provide avenues for investigation of other bacterial species. With respect to one of the malp gene products, MALP-2, which has been shown to be a potent activator of macrophages, it is interesting to speculate about whether p47 may play a role in modulation of the host's immune response.
Previous studies have used chicken tracheal organ cultures as an in vitro model system with which to study the attachment and pathogenesis of M. gallisepticum (1, 2, 8, 28). Levisohn et al. (28) showed that inoculum size is approximately correlated with the time it takes for ciliostasis to occur, while Abdul-Wahab et al. (1) found that ciliostasis is independent of multiplication of the organism and thus unlikely to be caused by small changes in medium pH due to acid production by the organism. Ciliostasis observed in our experimental system was presumed to be directly related to the attachment and interaction of the organism with the tracheal rings, although the possibility cannot be ruled out that factors affecting ciliostasis other than those tested in this study may be involved. The results of the present study show that there is no difference in ciliostasis between M. gallisepticum strain S6 and clone P2, suggesting that p47 plays no role in the initial interactions between M. gallisepticum and its host. This does not preclude a role for p47 in vivo, where a more dynamic interaction would occur, but such investigations are outside the scope of this paper.
There has been one report of homologous recombination occurring in M. gallisepticum (7) and a limited number of reports of its use to specifically inactivate mycoplasma genes in other species. Cao et al. (7) showed integration of randomly cloned M. gallisepticum genomic DNA into the genome, although detailed analysis of the crossover event was not done. In another study, adherence-related protein MG218 of M. genitalium (17) was disrupted and inactivated, while in another, the recA gene of Acholeplasma laidlawii (20) was inactivated. More pertinent to this study is the report that the p47 homologue of M. genitalium, MG040, was disrupted and most likely inactivated by random transposon mutagenesis (25). However, studies were not performed to confirm this inactivation. It appears that p47 is not essential for the growth of M. gallisepticum in vitro, and this study has also shown that absence of p47 does not impair the abilities of the organism to attach to and produce ciliostasis in chicken embryo tracheal organ cultures, compared with those of the parental strain.
This study has identified and cloned the gene encoding p47, the target for MAb B3. The p47 gene was disrupted by homologous recombination, extinguishing the target for MAb B3. Through the use of this methodology, it will be possible to specifically disrupt genes in M. gallisepticum and other pathogenic mycoplasmas and determine what effect or consequence they have in the organism's survival in vitro and in vivo and to establish their role in the pathogenesis of disease.
Acknowledgments
This work was supported by funding from the Australian Research Council and Bioproperties Australia (Pty. Ltd.) to G.F.B. and the Rural Industries Research and Development Corporation to P.F.M.
REFERENCES
- 1.Abdul-Wahab, O. M., G. Ross, and J. M. Bradbury. 1996. Pathogenicity and cytadherence of Mycoplasma imitans in chicken and duck embryo tracheal organ cultures. Infect. Immun. 64:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avakian, A. P., and D. H. Ley. 1993. Inhibition of Mycoplasma gallisepticum growth and attachment to chick tracheal rings by antibodies to a 64-kilodalton membrane protein of M. gallisepticum. Avian Dis. 37:706-714. [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens, A., M. Heller, H. Kirchhoff, D. Yogev, and R. Rosengarten. 1994. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect. Immun. 62:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calcutt, M. J., M. F. Kim, A. B. Karpas, P. F. Muhlradt, and K. S. Wise. 1999. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans. Infect. Immun. 67:760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, G. W., B. C. Cole, and J. R. Ward. 1986. Differential effects of in vitro gold sodium thiomalate on the stimulation of human peripheral blood mononuclear cells by Mycoplasma arthritidis T cell mitogen, concanavalin A and phytohemagglutinin. J. Rheumatol. 13:52-57. [PubMed] [Google Scholar]
- 7.Cao, J., P. A. Kapke, and F. C. Minion. 1994. Transformation of Mycoplasma gallisepticum with Tn916, Tn4001, and integrative plasmid vectors. J. Bacteriol. 176:4459-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherry, J. D., and D. Taylor-Robinson. 1970. Large-quantity production of chicken embryo tracheal organ cultures and use in virus and mycoplasma studies. Appl. Microbiol. 19:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, B. C. 1991. The immunobiology of Mycoplasma arthritidis and its superantigen MAM. Curr. Top. Microbiol. Immunol. 174:107-119. [DOI] [PubMed] [Google Scholar]
- 12.Czifra, G., B. G. Sundquist, U. Hellman, and L. Stipkovits. 2000. Protective effect of two Mycoplasma gallisepticum protein fractions affinity purified with monoclonal antibodies. Avian Pathol. 29(4):343-351. [DOI] [PubMed] [Google Scholar]
- 13.Czifra, G., B. G. Sundquist, T. Tuboly, and L. Stipkovits. 1993. Evaluation of a monoclonal blocking enzyme linked immunosorbent assay for the detection of Mycoplasma gallisepticum-specific antibodies. Avian Dis. 37:680-688. [PubMed] [Google Scholar]
- 14.Czifra, G., T. Tuboly, B. G. Sundquist, and L. Stipkovits. 1993. Monoclonal antibodies to Mycoplasma gallisepticum membrane proteins. Avian Dis. 37:689-696. [PubMed] [Google Scholar]
- 15.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, S. B., M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, K. L., and K. S. Wise. 2002. Site-specific proteolysis of the MALP-404 lipoprotein determines the release of a soluble selective lipoprotein-associated motif-containing fragment and alteration of the surface phenotype of Mycoplasma fermentans. Infect. Immun. 70:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhandayuthapani, S., W. G. Rasmussen, and J. B. Baseman. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. USA 96:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drummer, H. E., M. J. Studdert, and B. S. Crabb. 1998. Equine herpesvirus-4 glycoprotein G is secreted as a disulphide-linked homodimer and is present as two homodimeric species in the virion. J. Gen. Virol. 79:1205-1213. [DOI] [PubMed] [Google Scholar]
- 19.Duffy, M. F., A. H. Noormohammadi, N. Bassegio, G. F. Browning, and P. F. Markham. 1998. Immunological and biochemical characterization of membrane proteins, p. 267-278. In R. Miles (ed.), Molecular biological methods for mycoplasmas. Humana Press, New York, N.Y. [DOI] [PubMed]
- 20.Dybvig, K., and A. Woodard. 1992. Construction of mutants of Acholeplasma laidlawii by insertional inactivation with a homologous DNA fragment. Plasmid 28:262-266. [DOI] [PubMed] [Google Scholar]
- 21.Friis, N. F. 1975. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare. Nord. Vet. Med. 27:337-339. [PubMed] [Google Scholar]
- 22.Gawron-Burke, C., and D. B. Clewell. 1984. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J. Bacteriol. 159:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glew, M. D., P. F. Markham, G. B. Browning, and I. D. Walker. 1995. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum strain S6. Microbiology 141:3005-3014. [DOI] [PubMed] [Google Scholar]
- 24.Harlow, E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 26.Jan, G., C. Fontenelle, M. Le Henaff, and H. Wroblewski. 1995. Acylation and immunological properties of Mycoplasma gallisepticum membrane proteins. Res. Microbiol. 146:739-750. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, F. T. W. 1990. Avian mycoplasmoses, p. 74-85. In F. T. W. Jordan (ed.), Poultry diseases, 3rd ed. W. B. Saunders, London, England.
- 28.Levisohn, S., M. J. Dykstra, M. Y. Lin, and S. H. Kleven. 1986. Comparison of in vivo and in vitro methods for pathogenicity evaluation for Mycoplasma gallisepticum in respiratory infection. Avian Pathol. 15:233-246. [DOI] [PubMed] [Google Scholar]
- 29.Markham, P. F., M. Glew, M. R. Brandon, I. D. Walker, and K. G. Whithear. 1992. Characterization of a major hemagglutinin protein from Mycoplasma gallisepticum. Infect. Immun. 60:3885-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markham, P. F., M. D. Glew, G. F. Browning, K. G. Whithear, and I. D. Walker. 1998. Expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect. Immun. 66:2845-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markham, P. F., M. D. Glew, J. Sykes, T. R. Bowden, T. D. Pollocks, G. F. Browning, K. G. Whithear, and I. D. Walker. 1994. The organisation of the multigene family which encodes the major cell surface protein, pMGA, of Mycoplasma gallisepticum. FEBS Lett. 352:347-352. [DOI] [PubMed] [Google Scholar]
- 32.Markham, P. F., M. D. Glew, K. G. Whithear, and I. D. Walker. 1993. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect. Immun. 61:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minion, F. C., and P. A. Kapke. 1998. Transformation of mycoplasmas. Methods Mol. Biol. 104:227-234. [DOI] [PubMed] [Google Scholar]
- 34.Muhlradt, P. F., M. Kiess, H. Meyer, R. Sussmuth, and G. Jung. 1997. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 185:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muhlradt, P. F., M. Kiess, H. Meyer, R. Sussmuth, and G. Jung. 1998. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect. Immun. 66:4804-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noormohammadi, A. H., P. F. Markham, M. F. Duffy, K. G. Whithear, and G. F. Browning. 1998. Multigene families encoding the major hemagglutinins in phylogenetically distinct mycoplasmas. Infect. Immun. 66:3470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noormohammadi, A. H., P. F. Markham, K. G. Whithear, I. D. Walker, V. A. Gurevich, D. H. Ley, and G. F. Browning. 1997. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect. Immun. 65:2542-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razin, S. 1999. Adherence of pathogenic mycoplasmas to host cells. Biosci. Rep. 19:367-372. [DOI] [PubMed] [Google Scholar]
- 40.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma ′cytoskeleton': the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045-1057. [DOI] [PubMed] [Google Scholar]
- 42.Regula, J. T., B. Ueberle, G. Boguth, A. Gorg, M. Schnolzer, R. Herrmann, and R. Frank. 2000. Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis 21:3765-3780. [DOI] [PubMed] [Google Scholar]
- 43.Rodwell, A. W., and R. F. Whitcomb. 1983. Methods for direct and indirect measurement of mycoplasma growth. Methods Mycoplasmol. 1:185-197. [Google Scholar]
- 44.Rosati, S., S. Pozzi, P. Robino, B. Montinaro, A. Conti, M. Fadda, and M. Pittau. 1999. P48 major surface antigen of Mycoplasma agalactiae is homologous to a malp product of Mycoplasma fermentans and belongs to a selected family of bacterial lipoproteins. Infect. Immun. 67:6213-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosengarten, R., and K. S. Wise. 1991. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency antigenic variation. J. Bacteriol. 173:4782-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whithear, K. G., D. D. Bowtell, E. Ghiocas, and K. L. Hughes. 1983. Evaluation and use of a micro broth dilution procedure for testing sensitivity of fermentative avian mycoplasmas to antibiotics. Avian Dis. 27:937-949. [PubMed] [Google Scholar]
- 48.Whithear, K. G., Soeripto, K. E. Harrigan, and E. Ghiocas. 1990. Safety of temperature sensitive mutant Mycoplasma gallisepticum vaccine. Aust. Vet. J. 67:159-165. [DOI] [PubMed] [Google Scholar]
- 49.Yogev, D., R. Rosengarten, R. Watson-McKown, and K. S. Wise. 1991. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 10:4069-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]