Abstract
The early events in filamentous bacteriophage infection of gram-negative bacteria are mediated by the gene 3 protein (g3p) of the virus. This protein has a sophisticated domain organization consisting of two N-terminal domains and one C-terminal domain, separated by flexible linkers. The molecular interactions between these domains and the known bacterial coreceptor protein (TolA) were studied using a biosensor technique, and we report here on interactions of the viral coat protein with TolA, as well as on interactions between the TolA molecules. We detected an interaction between the pilus binding second domain (N2) of protein 3 and the bacterial TolA. This novel interaction was found to depend on the periplasmatic domain of TolA (TolAII). Furthermore, extensive interaction was detected between TolA molecules, demonstrating that bacterial TolA has the ability to interact functionally with itself during phage infection. The kinetics of g3p binding to TolA is also different from that of bacteriocins, since both N-terminal domains of g3p were found to interact with TolA. The multiple roles for each of the separate g3p and TolA domains imply a delicate interaction network during the phage infection process and a model for the infection mechanism is hypothesized.
Filamentous bacteriophages are a large family of single-stranded DNA viruses that infect gram-negative bacteria (36). The current understanding of the infection process of filamentous bacteriophages comes mainly from studies of Ff phages (M13, fd, f1) infecting Escherichia coli carrying the F episome, coding for the F-pilus, which is the primary receptor of the host cell (24). The infection is specifically mediated by coat protein 3 (g3p) of the phage, which is located at one end of the phage particle and consists of 406 amino acid residues that form three functionally distinct domains linked by two flexible glycine-rich stretches. All three domains are involved in the infective process (38) but play different roles. The C terminus (CT) of g3p anchors the g3p in the phage coat by interacting with phage coat protein 6, while the two distal domains of g3p, N1 and N2, interact with each other to form a di-domain (N1L1N2) (22, 31). The binding of N2 to the tip of the bacterial F-pilus releases the N1 domain, which becomes free to interact with TolA (TolAIII), the C terminal of the membrane bound bacterial coreceptor (37). The F-pilus protrudes from the bacterial membrane and is believed to retract upon phage binding (5, 40), bringing the phage close to its coreceptor.
Penetration of the bacterial membrane and transfer of viral DNA are known to depend on the presence of the bacterial TolQRA membrane proteins (9, 10, 41). The Tol proteins of E. coli are involved in outer membrane stability and they are also required for the uptake of bactericidal group A colicins. The TolA protein consists of three domains. It is anchored in the inner membrane of the bacteria by its N-terminal domain (TolAI) through which it interacts with the membrane proteins TolQ and TolR. The second domain of TolA (TolAII) spans the periplasmic space through its extended α-helical conformation. Finally, the C-terminal domain of TolA (TolAIII) reaches the outer membrane of the bacteria and has been shown to interact with the N-terminal domain of group A colicins during colicin entry (1, 35), in addition to being the coreceptor for phage g3p during phage infection of bacteria. Recently, functional studies have shown TolA and colicins to be involved in multiple molecular interactions in the periplasm during the import of colicins (25).
Although some events of the phage infection are well characterized, it is still not known how the viral DNA gets translocated into the bacteria. In a recent study we showed that the first glycine-rich linker region of g3p (L1) had an active role in F-pilus-dependent infection (33). In another study, the CT of g3p was found to interact specifically with the two N-terminal domains (N1L1N2), leading to a stable compact conformation of g3p on the phage, which is expected to unravel during infection (8).
The aim of this study was to obtain a more complete understanding of the phage infection process through analyses of the molecular interactions between native proteins as well as separate domains of the g3p and TolA proteins. Using a biophysical approach for the measurements of protein interaction, we were able to determine the affinity constants for the interactions between the specified proteins involved in infection. Detection of a novel interaction between phage N-terminal domains and bacterial TolA, as well as interactions between domains of TolA, led us to propose a refined infection model.
MATERIALS AND METHODS
Strains.
The following E. coli strains were used for the cloning and expression of recombinant proteins and inhibition assays: Top10F′, TG1, XL1-Blue, MC1061, and C600 (for details on genetic markers see the work of Nilsson et al. [33] and http://omrf.ouhsc.edu/∼frank/hv_main.html).
Plasmids and primers.
The different g3p variants were amplified from wild-type (wt) VCSM13 (Stratagene), using specific oligonucleotides containing appropriate restriction sites. The construction of N1L1N2 has been described (32). For the construction of N1 and N2, see the work of Nilsson et al. (33).
The gene for TolA was amplified from E. coli Top10F′, using appropriate primers. The boundaries of TolA were based on a previous alignment (29). The tolA, tolAI, tolAII, and tolAIII genes were all cloned into the expression vector pPACIB9 (a generous gift from M. Duenas). The restriction sites used in cloning were 5′ EcoRI and 3′ BamHI. The vector pPACIB9 contains a trp promoter and a human interleukin-2 tag at the N terminus of subcloned inserts. The interleukin-2 tag promotes intracellular expression and allows for higher expression levels, without affecting the binding properties of the proteins fused to the tag (14). Two alanine and six histidine residues followed all constructs.
Production of pure recombinant proteins.
The N1L1N2 protein was produced as described earlier (32). The His-tagged recombinant forms of N1 and N2 were produced from XL1-Blue bacteria containing the expression vector with respective inserts. Cultures of single colonies were grown in 2×YT medium containing ampicillin (100 μg/ml) and tetracycline (10 μg/ml). The expression of proteins was induced at logarithmic growth phase (optical density reading at 600 nm = 0.4), by the addition of 0.25 mM isopropyl thiogalactoside (IPTG). After 4 h of expression, the cells were harvested by centrifugation. Proteins were purified from both supernatants and preparations of the periplasmic fraction of the cells (Qiagen, Ltd., Crawley, United Kingdom). The sample solutions were adsorbed onto nickel nitrilotriacetic acid agarose and were eluted stepwise using 80, 150, or 250 mM imidazole in 50 mM NaH2PO4 (pH 8.0), containing 300 mM NaCl. The samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [12.5 or 15% polyacrylamide]), followed by Western blotting and detection of purified protein, using an anti-tetra-his antibody from Qiagen (Qiagen Ltd.). After purification, the samples were run on either analytical or preparative gel filtration columns (Sepharose; Amersham Pharmacia). The samples were thus purified to more than 95%, as determined by SDS-PAGE and silver staining. Buffer changes and concentrations of samples were made, using Ultrafree centrifugal filters (molecular mass cutoff, 3.5 kDa) from (Millipore Corporation, Bedford, Mass.), as well as dialysis using Spectra/Por membrane tubing (Spectrum Laboratories, Inc., Rancho Dominguez, Calif.). Protein concentration was determined, using a bicinchoninic acid kit from Pierce (Perbio Science, Ltd., Cheshire, United Kingdom).
The pPACIB9 vector constructs containing the tolA, tolAI, tolAII, and tolAIII genes were transformed into C600 for expression. Cultures were grown to log phase in 2×YT medium supplemented with ampicillin (100 μg/ml) and tryptophan (100 μg/ml). Adding iodoacetic acid (final concentration, 10 μg/ml) to the culture induced expression. After 4 h of expression, the Qiagen protocol for cell lysis and solubilization with urea was followed, as was the refolding and purification protocol (23). After elution, the proteins were subjected to either analytical or preparative gel filtration, depending on sample homogeneity, as described above. The purified proteins were analyzed by SDS-12.5% PAGE and Western blotting, using both anti-tetra-His antibody and anti-TolA antibody (the latter antibody was a kind gift from R. E. Webster). The anti-TolA antibody is directed toward the second and third domains of TolA. The expression resulted in at least 95% pure protein, as determined by SDS-PAGE and Coomassie staining.
BIAcore analysis.
The CM5 sensor chip (research grade), amine coupling kit, containing N-hydroxysuccinimide (NHS), N-ethyl-N′′′-(3-diethylaminopropyl) carbodiimide (EDC), and 1 M ethanolamine hydrochloride (pH 8.5) for the BIAcore system, were from BIAcore AB (Uppsala, Sweden). Experiments were run at 25°C. Filtered and degassed HBS buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, 0.005% [vol/vol] surfactant P20 [polyoxyethylene sorbitan monolaurate] [Pharmacia Biosensor AB, Uppsala, Sweden] [pH 7.4]) was used as sample and running buffer for all experiments.
The CM5 chip was equilibrated with a 5-μl/min flow of HBS buffer and then activated by using standard amine coupling procedures. Equal volumes of NHS and EDC were mixed just before use, and 35 to 45 μl of the mixed solution was injected over the surface to activate the carboxymethylated dextran layer. For the capture of purified recombinant proteins, dilutions of each of the proteins to be coupled were made in 10 mM sodium acetate (pH 4 to 5, depending on protein) coupling solution, and injected across the activated surface. Excess reactive NHS esters were deactivated by injecting ethanolamine hydrochloride over the sensor chip and the surface was regenerated by 100 mM HCl (pH 1.0), followed by washing with HBS buffer to equilibrate. To verify that the specified proteins were indeed coupled to the chip surface, different control antibodies were used, i.e., polyclonal antibodies against TolA and the N2 domain of g3p (this does not give the amount of coupled protein). The latter antibody was generated by immunizing rabbits with purified N1L1N2 (Antibody AB, S Sandby, Sweden).
In order to reduce effects due to mass transport, rebinding, steric hindrance, and other transport-related artifacts, a low density of ligand is desirable on the sensor chip (34). Thus, the amount of protein coupled to the chip is recommended to be molecular weight/15 RU (the signal is expressed in resonance units [RU], and 1,000 RU is equivalent to a change in surface concentration of about 1 ng/mm2 [16]), by the manufacturer. MW/15 RU corresponds to 0.06 pmol of protein. In our experiments the amount of active protein (in picomoles) on the surfaces was determined by the obtained RU from the immobilization. The binding kinetics of the interactions between the selected proteins and their domains, to the immobilized counterparts, were then measured by passing series of each protein (from 50 μM down to picomolar concentrations, using at least four different dilutions per analysis) across the sensor chip, thus minimizing effects of mass transport (4). After each round of binding the surface is regenerated to remove all residual binders and make the surface ready for the next round of interaction. All proteins were analyzed for background binding to a reference surface with no protein attached. In no such case, could we detect any signal. Furthermore, irrelevant antibodies were used as cross reference for absence of binding to all coupled proteins.
Inhibition assay.
A culture of F-pilus- and TolQRA-expressing bacteria (Top10F′) was grown to log phase. The bacterial cells (100 μl) were preincubated with 10 μl of increasing concentrations of the inhibitory proteins at 37°C for 15 min and under moderate shaking (70 rpm). Thereafter, approximately 2,000 VCSM13 (Kanr) phage particles were added to the bacterial cells and allowed to infect at 37°C for 10 min. The samples were plated on 2×YT-agar plates, containing kanamycin, to detect infected cells. The plates were incubated overnight at 37°C. The level of inhibiting capacity for each of the protein preparations was determined by counting the CFU on each plate.
RESULTS
The phage infection mechanism is a dynamic process that involves a number of molecular interactions between phage and bacterial proteins. To investigate these interactions, we chose to produce pure proteins of all the N-terminal domains of phage minor coat protein g3p (N1L1N2, N1, and N2) and of the bacterial coreceptor protein TolA domains (TolA, TolAI, TolAII, and TolAIII). To measure the binding kinetics of the interactions between selected proteins, the surface plasmon resonance (SPR) technique was used. For each measurement, one of the two studied proteins must be coupled to the sensor chip surface (i.e., the ligand) while the other is passed over the chip (i.e., the analyte). In this real-time interaction analysis, all proteins (except TolAI) were used as ligand or analyte, respectively. This was done to reduce the risk of false-positive results due to the immobilization procedure. Analyzing the data generated from BIAcore sensorgrams, using an appropriate binding model (Langmuir 1:1; because the derivatized resonance curves gave straight lines [data not presented]) and global fitting, results in the generation of affinity constants (KD) from the association (ka) and dissociation (kd) rate constants. In general, the range of natural biological variation, in terms of KD values, is very broad; however, the range of affinity constants that can be determined by SPR technology is also very broad, ranging from millimolar to picomoloar concentrations (http://www.biacore.com/support/faq.lasso). There are reports on low-affinity interactions of biological relevance typically in the millmolar range, measured by SPR technology (39). Apart from positive and negative control antibodies and reference surface interaction determination, the binding pattern displayed by each of the investigated proteins revealed both specific binding to some proteins and nonbinding to other proteins, in all cases. Overall, the interactions characterized in this study can be divided into two categories: first, interactions between the phage protein domains and the domains of bacterial TolA, and second, interactions between the bacterial TolA domains alone.
Phage and TolA domain interactions.
The interactions of phage protein domains and the domains of the bacterial TolA were the first to be evaluated. Since we produced three phage protein variants and four bacterial protein variants there were 12 potential interactions that could be investigated. The mean value of the affinities for each of the six specific interactions detected is presented in Table 1 together with the standard deviations for the association and dissociation rate constants. First, we detected an interaction between TolAIII and N1. It has been established before that TolAIII interacts with the first N-terminal domain of g3p (N1) (37). The affinity between these two domains was measured to 1.9 and 1 μM, respectively, depending on which of them was used as the analyte (Table 1). This is the first affinity measurement between molecular TolAIII and N1, although soluble TolAIII has in vitro been shown to inhibit the interaction between N1 phage and TolAIII at concentrations down to 0.7 μM (37).
TABLE 1.
Affinities for interactions between N-terminal domains of phage g3p and bacterial TolA, measured by BIAcore analysis
| Interactiona | kac (M−1 s−1) | kdc (s−1) | KA (M−1) | KDb (μM) |
|---|---|---|---|---|
| N1L1N2-TolA | (2.5 ± 0.26) × 103 | (1.5 ± 0.4) × 10−3 | 1.7 × 106 | 0.6 |
| N1L1N2-TolAIII | (7.0 ± 2.3) × 103 | 0.164 ± 0.02 | 4.3 × 104 | 23 |
| N1-TolAIII | (123 ± 7) × 103 | 0.2 ± 0.005 | 5.4 × 105 | 1.9 |
| TolAIII-N1 | (676 ± 58) | (650 ± 0.3) × 10−6 | 1 × 106 | 1.0 |
| N2-TolA | (1.6 ± 0.09) × 103 | (6 ± 0.3) × 10−3 | 2.7 × 105 | 3.8 |
| TolA-N2 | (5.4 ± 0.1) × 104 | 0.3 ± 0.01 | 2.0 × 105 | 5 |
| N2-TolAII | (2.6 ± 0.19) × 105 | 0.3 ± 0.007 | 7.5 × 105 | 1.3 |
| TolAII-N2 | (2.7 ± 0.08) × 103 | (6.6 ± 0.4) × 10−3 | 4.1 × 105 | 2.4 |
| TolA-N1 | (1.7 ± 0.00009) × 105 | (24 ± 0.002) × 10−3 | 6.9 × 106 | 0.1 |
| N1-TolAI | ND | |||
| N1-TolAII | NI | |||
| N2-TolAI | NI | |||
| N2-TolAIII | NI | |||
| N1L1N2-TolAI | NI | |||
| N1L1N2-TolAII | NI |
The ligand is given first in each entry.
Abbreviations: ND, not fully determined due to conflicting results in the reverse setup; NI, no interaction detected.
Data are means ± standard deviations.
Since the TolAIII domain is part of the native TolA, the entire TolA should also be able to interact with N1. This was indeed confirmed by the setup in which TolA was used as ligand resulting in an apparent affinity of 0.1 μM (Table 1). However, in the reciprocal experiment we were unable to determine the association rate constant for TolA binding to N1. The observed difference in affinity between N1 and TolAIII (1 μM) compared to intact TolA (0.1 μM) was most probably not dependent on other interdomain (TolAI and TolAII) interactions. However, no conclusive result could be obtained for the interaction between N1 and TolAI, since the TolAI protein was not used as ligand in this study (Table 1).
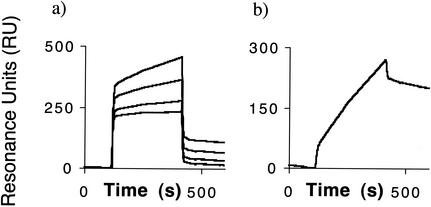
Furthermore, we then wanted to analyze whether TolA was able to interact with the more complex N1L1N2 domains. A specific and measurable interaction was evident between both TolA and TolAIII in relation to N1L1N2 (Table 1). Moreover, although N2 has never been reported to interact with TolA, we decided to investigate this possibility. Indeed, we detected a specific interaction between TolA and N2 as well (Fig. 1a and Table 1). This interaction was found to be dependent on the TolAII subdomain, since TolAII was the only individual domain of TolA that was able to bind N2, both as analyte and ligand (Table 1). Finally, if N1L1N2 was added to immobilized TolA or TolAIII no interaction was detected, contrasting the behavior of immobilized N1L1N2.
FIG. 1.
BIAcore sensorgrams. The increase in resonance (a) represents binding of bacterial TolA to the immobilized phage g3p second N-terminal domain (N2). After each round of binding the surface is regenerated to remove all residual binders and make the surface ready for the next round of interaction. This part of the sensorgram is removed from the figure. The KD (calculated by global fitting) for the interaction shown in panel a is 3.8 μM. (b) A control antibody directed towards N2 demonstrates specificity for the immobilized protein.
Interactions between domains of TolA.
The second category of interactions characterized by the BIAcore procedure consisted of those between bacterial TolA and its own domains. We found that native TolA was able to bind to itself, as did each of the separate domains TolAII and TolAIII at micromolar affinity (Table 2). Previous studies have shown that TolA forms a subcomplex in the inner membrane with the TolQ and TolR proteins, interacting with each other by their trans-membrane segments (12, 18, 26), but the binding kinetics of these interactions has not yet been characterized.
TABLE 2.
Affinities for interactions between homogenous domains of TolA and between different domains of TolA, measured by BIAcore analysis
| Interactiona | kab (M−1 s−1) | kdb (s−1) | KA (M−1) | KD (μM) |
|---|---|---|---|---|
| TolA-TolA | (3.4 ± 1) × 105 | 0.12 ± 0.008 | 2.8 × 106 | 0.4 |
| TolA-TolAI | (5.3 ± 1.1) × 105 | 0.12 ± 0.01 | 4.5 × 106 | 0.2 |
| TolA-TolAII | (2.3 ± 0.2) × 103 | (9.3 ± 0.4) × 10−3 | 2.5 × 105 | 4 |
| TolA-TolAIII | (7.0 ± 0.5) × 103 | (69 ± 3) × 10−3 | 1.0 × 105 | 9.9 |
| TolAII-TolAII | (7.4 ± 1) × 103 | (54 ± 4) × 10−3 | 1.4 × 105 | 7.2 |
| TolAIII-TolAI | (9.4 ± 0.9) × 103 | (5.7 ± 0.4) × 10−3 | 1.6 × 106 | 0.6 |
| TolAIII-TolAII | (1.5 ± 0.08) × 103 | (4.4 ± 0.1) × 10−3 | 3.4 × 105 | 2.9 |
| TolAIII-TolAIII | (5.1 ± 0.5) × 103 | (24 ± 1.3) × 10−3 | 2.1 × 105 | 4.7 |
The ligand is given first in each entry.
Data are means ± standard deviations.
As a result of the interactions detected between each of the separate domains of TolA, we asked whether there would be any cross talk between these domains. Interestingly, interactions between the C-terminal outer membrane domain (TolAIII) and each of the separate domains of TolA were detected (Table 2). The most intriguing of the TolAIII interactions was its ability to bind to TolAI, the inner membrane anchor of TolA. The periplasm spanning domain of TolA (TolAII) was able to interact with TolAIII and consequently also with the entire TolA molecule, again at micromolar affinity. However, no detectable affinity could be measured between TolAII and TolAI (the membrane anchor of TolA), despite the fact that TolAII has been seen to interact with outer membrane porins of E. coli (13). The observed interaction between the membrane domain TolAI with the complete TolA was a consequence of its affinity for TolAIII (Table 2).
Inhibitory capacity of selected proteins.
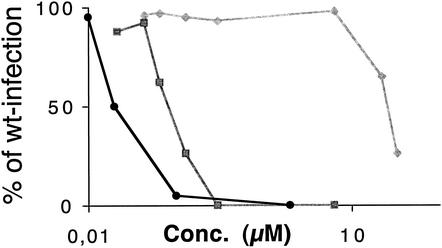
To determine the involvement of the selected proteins during normal phage infection, they were tested for their capacity to inhibit the wt phage infection of F+ TolA+ bacteria (Fig. 2). We show that the N1, N2, and N1L1N2 proteins were fully functional and capable of delaying wt phage infection. The N1 fragment showed inhibitory effect only when used at high concentrations, while N2 needed approximately 4 times the concentration of the N1L1N2 fragment to display the same inhibitory capacity (Fig. 2). The inhibitory effect of proteins N1, N2 and N1L1N2 found in this study correlates with previous reports (27, 33). The domains of the TolA protein, however, were not able to inhibit normal phage infection (data not shown). Previously, TolAIII molecules have been shown to inhibit phage binding to pilus in an enzyme immunoassay (37), although when added to phage particles TolAIII could not inhibit the actual bacterial infection (9).
FIG. 2.
The abilities of different proteins to inhibit wt phage infection of F-pilus-bearing bacteria were evaluated. All three tested phage protein domains functionally inhibited wt infection. The level of inhibition, expressed as a percentage of wt infection, varied with the protein concentration. Having both N-terminal domains of g3p (N1L1N2) present at the same time inhibited infection at the lowest concentration (circles), while N2 (squares) inhibited infection at about four times the inhibitory concentration of N1L1N2. N1 was not able to inhibit wt infection unless used at high concentrations (diamonds).
DISCUSSION
To study the specific molecular interactions taking place during phage infection in more depth, we chose to use pure recombinant proteins and fragments of proteins instead of constructing deletion mutants of phage and bacteria. This allowed us to study their interactions, using the BIAcore biosensor technique, and we show that this method is capable of detecting previously known as well as unknown molecular interactions between proteins involved in the infection pathway. Furthermore, it allows the determination of the apparent affinities between the selected proteins.
For the interaction between phage N1 and TolAIII, a domain within the bacterial coreceptor, we determined a KD of 1.9 μM. On the other hand, when TolAIII was added to immobilized N1L1N2, we noticed a 12-fold reduction in affinity between the phage N terminal and TolAIII (Table 1). However, this is in agreement with the fact that the TolAIII and N2 share the same binding site on N1 (37). We determined the affinity for the interaction between phage N1L1N2 and TolA (0.6 μM) to be within the same range as that of the apparent affinities reported between colicins and TolA (20, 35). Gokce et al. showed that the binding of TolA to immobilized colicin fusions had an affinity of around 0.9 μM. These authors also suggest that immobilization of the incomplete TolAII-III molecule results in the exposure of a second binding site for colicin N to TolA (KD1 = 0.9 μM, KD2 = 0.2 μM). Apart from the difference in binding mode to TolA, we also found that both N-terminal domains of phage g3p took part in binding to immobilized TolA, contrary to colicins, which only interact with TolA via their first N-terminal domain. Therefore, we argue that g3p interacts with TolA in a manner different from that of TolA-dependent colicins. Colicins, which are lethal to sensitive E. coli cells, use the Tol proteins of bacteria to pass the bacterial membrane. The domain structure of colicins is similar to that of g3p, and both the phage g3p N-terminal domain and the colicin N-terminal domain are prevented from binding with full affinity when the whole protein is in the native, compact state. Therefore, it has been suggested that colicins interact with Tol proteins in the same manner as phage g3p. The binding of both N1 and N2 to TolA, as presented here, and the involvement of different amino acids in the binding (20, 30), strongly suggest that these proteins interact differently with TolA. Furthermore, the more advanced structure of N1 compared to the colicin N-terminal domain is a clear indicator of a different mode of action towards TolA. A recent study (11), where a conformational change in TolAIII during binding of colicin A was demonstrated, also lends support to this notion. Taken together these data suggest divergent evolutionary pathways for phage coat proteins and colicins despite their topological similarities.
When studying the interaction between N1L1N2 and TolA/TolAIII we discovered a contrasting behavior between immobilized and soluble N1L1N2, resulting in a lack of interaction when N1L1N2 was used as an analyte. However, this lack of interaction between soluble N1L1N2 and TolA/TolAIII can be expected, since in the absence of F-pilus binding, N1L1N2 are engaged in extensive intraprotein interactions, as demonstrated by their crystal structure (31) and their ability to form a complex even in the absence of their covalent linker (37). This type of intraprotein interaction is probably disturbed when the protein is immobilized, resulting in a mix of available and nonavailable binding sites on the N1L1N2-coated chip, whereof the fraction of available binding sites contributes enough to detect a binding.
The fate of the second N-terminal domain of g3p (N2) after pilus binding has so far not been fully examined. After the process of pilus retraction, however, g3p presumably crosses the outer membrane by a yet-unknown mechanism and insert its CT in the inner membrane (2, 15). Whether or not N2 remains bound to pilin molecules after depolymerization remains to be elucidated. Here we report on the central domain of TolA (TolAII) having affinity for N2, the second N-terminal domain of g3p. An interaction with TolAII in the periplasm after the pilus has depolymerized would help g3p orient itself in the inner membrane, in a way that would facilitate its reuse and its possible role in the formation of a pore. However, it seems that the N2-TolAII interaction is not necessary for infection, as demonstrated by our previous finding that phage lacking N2 is capable of infecting pilus-bearing bacteria at almost the same frequency as wt phage (33). Also, the presence of the TolAII domain is not an absolute requirement for phage infection but contributes to the efficiency of TolA function in infection (9). The finding reported in this work, that TolAIII has affinity for TolAI, could explain why TolAII is not an absolute requirement for infection, as well as why infection is possible without the N2 domain.
To our knowledge TolA has not been shown to interact with itself. However, the interactions detected in this study suggest the possibility of colocalized TolA molecules forming subcomplexes. Also the cooperative effects of TolA molecules clustering together in the membrane could indicate a further enhancement of the capacity of TolA to mediate the infection. In order to weld the detected interactions with the known interactions of the current infection mechanism model we came up with one suggested model that incorporates all of the interactions detected in this study (Fig. 3). First, as it is known that there are three to five copies of g3p on the phage coat, the suggested clustering of TolA molecules would allow for all of the g3p molecules to be correctly oriented into the inner membrane by the cooperating TolA molecules. Second, our proposed clustering of TolA molecules might have an effect on the distance between the outer and inner membranes. TolA has been shown to locate near adhesion zones, where the outer and inner membranes meet (3, 12, 21). In this respect, our finding that the C-terminal domain of TolA (TolAIII) was able to bind to the N-terminal membrane anchor of TolA (TolAI) is of particular interest. This would allow the TolA to assume a more compact state of assembly, with its C terminus in close contact with the membrane anchor domain TolAI, thus bringing the outer and inner membrane of the bacteria closer together, to facilitate phage entry (Fig. 3). In E. coli the inner and outer membrane complexes are linked via TolAIII with Pal lipoprotein and TolB (6, 25), demonstrating the capability of TolAIII to interact with membrane associated proteins. However, it remains to be determined whether the interaction between TolAIII and TolAI is of functional significance in relation to the phage infection mechanism. Recently, it was proposed that multiple interactions between colicins and Tol proteins seem to be a general phenomenon (25). Also, it has recently been suggested that TolQ-TolR could function as a motor, energizing TolA to drive macromolecules through the cell envelope (6, 7, 17, 19). Our discovery of a novel interaction between the TolA protein and the N2 of phage g3p, as well as our indication of a binding capacity between TolA domains, fits well with these reports on TolA function. The notion that transient intermolecular interaction is a key feature of this membrane protein is thus strengthened.
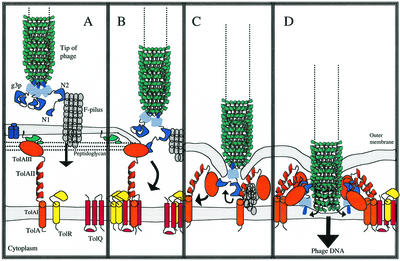
FIG. 3.
Model for the early events in the phage infection of E. coli. (A) The phage g3p second N-terminal domain (N2) interacts with the F-pilus on the outside of the bacteria. The outer membrane proteins OmpF (blue cylinders) and Pal lipoprotein (greenish) are also included, as there have been reports of TolA interacting with these proteins prior to infection (6, 28). (B) After F-pilus retraction, the first N-terminal domain of g3p (N1) binds to the C-terminal domain of bacterial TolA (TolAIII). (C) The retracting pilus brings phage g3p domains in closer contact with TolA domains. As a consequence, the TolA can assume a more compact state of assembly, thus bringing the outer and inner membranes of the bacteria closer together. At this stage, the central domain of TolA (TolAII) has the possibility to interact with the N2 domain of phage g3p. (D) The phage g3p is inserted into the inner membrane, and the cap of the phage head is opened to allow phage DNA to enter the bacteria. The data in Table 2 suggest that cooperating TolA molecules help achieve the insertion of g3p in the inner membrane.
In summary, we have shown that both the first and the second N-terminal domains of phage g3p contribute to the overall affinity of protein 3 for TolA. Also, we report on the extensive interaction between different domains of the TolA protein, suggesting that the TolA protein has the capability of interacting with itself in the bacterial membrane. Furthermore, based on the finding that both the N-terminal domains of g3p contribute to the overall affinity for TolA, we propose that g3p interacts with TolA in a manner completely different from that of TolA-dependent colicins.
Acknowledgments
This investigation was supported by The National Swedish Council for Engineering Sciences.
We thank Ann-Charlott Olsson for excellent laboratory assistance.
REFERENCES
- 1.Benedetti, H., C. Lazdunski, and R. Lloubes. 1991. Protein import into Escherichia coli: colicins A and E1 interact with a component of their translocation system. EMBO J. 10:1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke, J. D., P. Model, and N. D. Zinder. 1982. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 186:185-192. [DOI] [PubMed] [Google Scholar]
- 3.Bouveret, E., R. Derouiche, A. Rigal, R. Lloubes, C. Lazdunski, and H. Benedetti. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J. Biol. Chem. 270:11071-11077. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, D., S. J. Aylwin, D. J. Newman, and J. M. Burrin. 1999. Steroidogenic factor 1-DNA binding: a kinetic analysis using surface plasmon resonance. J. Mol. Endocrinol 22:241-249. [DOI] [PubMed] [Google Scholar]
- 5.Burke, J. M., C. P. Novotny, and P. Fives-Taylor. 1979. Defective F pili and other characteristics of Flac and Hfr Escherichia coli mutants resistant to bacteriophage R17. J. Bacteriol. 140:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubes. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol. Microbiol. 38:904-915. [DOI] [PubMed] [Google Scholar]
- 7.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 8.Chatellier, J., O. Hartley, A. D. Griffiths, A. R. Fersht, G. Winter, and L. Riechmann. 1999. Interdomain interactions within the gene 3 protein of filamentous phage. FEBS Lett. 463:371-374. [DOI] [PubMed] [Google Scholar]
- 9.Click, E. M., and R. E. Webster. 1997. Filamentous phage infection: required interactions with the TolA protein. J. Bacteriol. 179:6464-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Click, E. M., and R. E. Webster. 1998. The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deprez, C., L. Blanchard, F. Guerlesquin, M. Gavioli, J. P. Simorre, C. Lazdunski, D. Marion, and R. Lloubes. 2002. Macromolecular import into Escherichia coli: the TolA C-terminal domain changes conformation when interacting with the colicin A toxin. Biochemistry 41:2589-2598. [DOI] [PubMed] [Google Scholar]
- 12.Derouiche, R., H. Benedetti, J. C. Lazzaroni, C. Lazdunski, and R. Lloubes. 1995. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270:11078-11084. [DOI] [PubMed] [Google Scholar]
- 13.Derouiche, R., M. Gavioli, H. Benedetti, A. Prilipov, C. Lazdunski, and R. Lloubes. 1996. TolA central domain interacts with Escherichia coli porins. EMBO J. 15:6408-6415. [PMC free article] [PubMed] [Google Scholar]
- 14.Duenas, M., J. Vazquez, M. Ayala, E. Soderlind, M. Ohlin, L. Perez, C. A. Borrebaeck, and J. V. Gavilondo. 1994. Intra- and extracellular expression of an scFv antibody fragment in E. coli: effect of bacterial strains and pathway engineering using GroES/L chaperonins. BioTechniques 16:476-477, 480-483. [PubMed]
- 15.Endemann, H., and P. Model. 1995. Location of filamentous phage minor coat proteins in phage and in infected cells. J. Mol. Biol. 250:496-506. [DOI] [PubMed] [Google Scholar]
- 16.Fagerstam, L. G., A. Frostell-Karlsson, R. Karlsson, B. Persson, and I. Ronnberg. 1992. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J. Chromatogr. 597:397-410. [DOI] [PubMed] [Google Scholar]
- 17.Gaspar, J. A., J. A. Thomas, C. L. Marolda, and M. A. Valvano. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38:262-275. [DOI] [PubMed] [Google Scholar]
- 18.Germon, P., T. Clavel, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J. Bacteriol. 180:6433-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Germon, P., M. C. Ray, A. Vianney, and J. C. Lazzaroni. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183:4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gokce, I., E. M. Raggett, Q. Hong, R. Virden, A. Cooper, and J. H. Lakey. 2000. The TolA-recognition site of colicin N. ITC, SPR and stopped-flow fluorescence define a crucial 27-residue segment. J. Mol. Biol. 304:621-632. [DOI] [PubMed] [Google Scholar]
- 21.Guihard, G., P. Boulanger, H. Benedetti, R. Lloubes, M. Besnard, and L. Letellier. 1994. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J. Biol. Chem. 269:5874-5880. [PubMed] [Google Scholar]
- 22.Holliger, P., L. Riechmann, and R. L. Williams. 1999. Crystal structure of the two N-terminal domains of g3p from filamentous phage fd at 1.9 A: evidence for conformational lability. J. Mol. Biol. 288:649-657. [DOI] [PubMed] [Google Scholar]
- 23.Holzinger, A., K. S. Phillips, and T. E. Weaver. 1996. Single-step purification/solubilization of recombinant proteins: application to surfactant protein B. BioTechniques 20:804-806, 808. [DOI] [PubMed]
- 24.Jacobson, A. 1972. Role of F pili in the penetration of bacteriophage fl. J. Virol. 10:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Journet, L., E. Bouveret, A. Rigal, R. Lloubes, C. Lazdunski, and H. Benedetti. 2001. Import of colicins across the outer membrane of Escherichia coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42:331-344. [DOI] [PubMed] [Google Scholar]
- 26.Journet, L., A. Rigal, C. Lazdunski, and H. Benedetti. 1999. Role of TolR N-terminal, central, and C-terminal domains in dimerization and interaction with TolA and tolQ. J. Bacteriol. 181:4476-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krebber, C., S. Spada, D. Desplancq, A. Krebber, L. Ge, and A. Pluckthun. 1997. Selectively-infective phage (SIP): a mechanistic dissection of a novel in vivo selection for protein-ligand interactions. J. Mol. Biol. 268:607-618. [DOI] [PubMed] [Google Scholar]
- 28.Lazdunski, C. J., E. Bouveret, A. Rigal, L. Journet, R. Lloubes, and H. Benedetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levengood, S. K., W. F. Beyer, Jr., and R. E. Webster. 1991. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 88:5939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodawer. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Struct. Fold Des. 7:711-722. [DOI] [PubMed] [Google Scholar]
- 31.Lubkowski, J., F. Hennecke, A. Pluckthun, and A. Wlodawer. 1998. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat. Struct. Biol. 5:140-147. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson, N., F. Karlsson, J. Rakonjac, and C. A. Borrebaeck. 2002. Selective infection of E. coli as a function of a specific molecular interaction. J. Mol. Recognit. 15:27-32. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson, N., A. C. Malmborg, and C. A. Borrebaeck. 2000. The phage infection process: a functional role for the distal linker region of bacteriophage protein 3. J. Virol. 74:4229-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ober, R. J., and E. S. Ward. 1999. The influence of signal noise on the accuracy of kinetic constants measured by surface plasmon resonance experiments. Anal. Biochem. 273:49-59. [DOI] [PubMed] [Google Scholar]
- 35.Raggett, E. M., G. Bainbridge, L. J. Evans, A. Cooper, and J. H. Lakey. 1998. Discovery of critical Tol A-binding residues in the bactericidal toxin colicin N: a biophysical approach. Mol. Microbiol. 28:1335-1343. [DOI] [PubMed] [Google Scholar]
- 36.Rasched, I., and E. Oberer. 1986. Ff coliphages: structural and functional relationships. Microbiol. Rev. 50:401-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riechmann, L., and P. Holliger. 1997. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90:351-360. [DOI] [PubMed] [Google Scholar]
- 38.Stengele, I., P. Bross, X. Garces, J. Giray, and I. Rasched. 1990. Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J. Mol. Biol. 212:143-149. [DOI] [PubMed] [Google Scholar]
- 39.Strandh, M., B. Persson, H. Roos, and S. Ohlson. 1998. Studies of interactions with weak affinities and low-molecular-weight compounds using surface plasmon resonance technology. J. Mol. Recognit. 11:188-190. [DOI] [PubMed] [Google Scholar]
- 40.Sun, T. P., and R. E. Webster. 1986. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J. Bacteriol. 165:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster, R. E. 1991. The tol gene products and the import of macromolecules into Escherichia coli. Mol. Microbiol. 5:1005-1011. [DOI] [PubMed] [Google Scholar]