Abstract
The enzymes of the mandelate metabolic pathway permit Pseudomonas putida ATCC 12633 to utilize either or both enantiomers of mandelate as the sole carbon source. The genes encoding the mandelate pathway were found to lie on a single 10.5-kb restriction fragment. Part of that fragment was shown to contain the genes coding for mandelate racemase, mandelate dehydrogenase, and benzoylformate decarboxylase arranged in an operon. Here we report the sequencing of the remainder of the restriction fragment, which revealed three further open reading frames, denoted mdlX, mdlY, and mdlD. All were transcribed in the opposite direction from the genes of the mdlABC operon. Sequence alignments suggested that the open reading frames encoded a regulatory protein (mdlX), a member of the amidase signature family (mdlY), and an NAD(P)+-dependent dehydrogenase (mdlD). The mdlY and mdlD genes were isolated and expressed in Escherichia coli, and the purified gene products were characterized as a mandelamide hydrolase and an NAD(P)+-dependent benzaldehyde dehydrogenase, respectively.
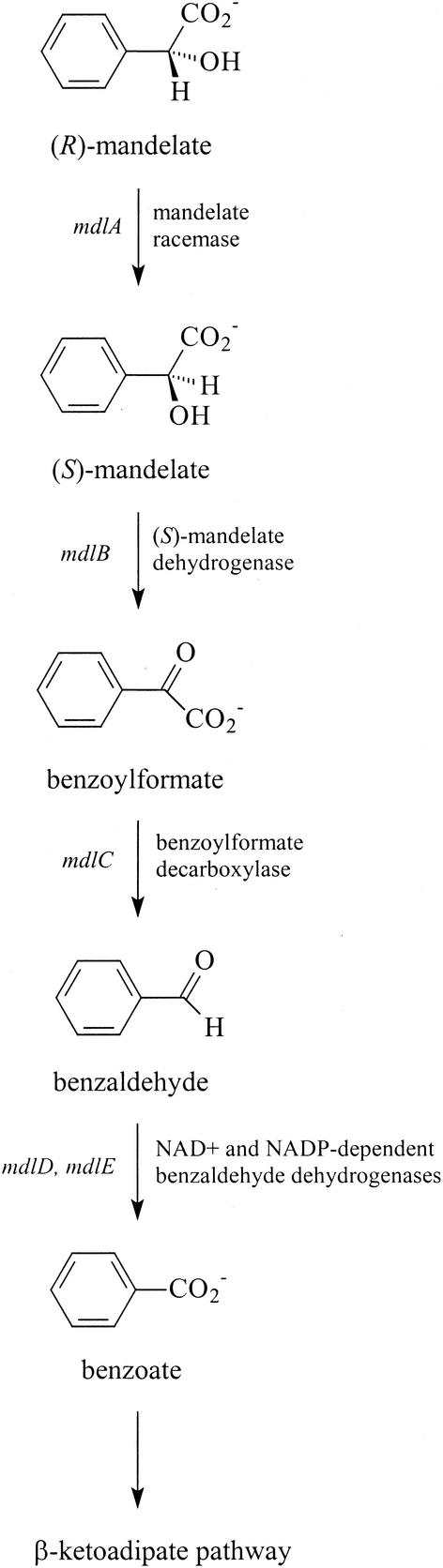
The mandelate metabolic pathway provides the means to allow a variety of pseudomonads, including Pseudomonas putida, to use one or both enantiomers of mandelic acid as the sole carbon source (7). Early studies suggested that the mandelate pathway of P. putida consists of five enzymes (8, 10) that facilitate the conversion of both (R)-mandelate and (S)-mandelate to benzoate, which is subsequently converted to acetyl coenzyme A (CoA) and succinyl-CoA by the enzymes of the β-ketoadipate pathway (Fig. 1). Genetic experiments indicated that the genes for the mandelate pathway were all closely clustered on the chromosome (36, 38), and, subsequently, it was reported that the mandelate pathway genes for P. putida ATCC 12633 resided on a single 10.5-kb restriction fragment (35). Sequencing of part of that fragment showed that three of the genes—those encoding mandelate racemase (mdlA), S-mandelate dehydrogenase (mdlB), and benzoylformate decarboxylase (mdlC)—were arranged in an operon (31, 35). Furthermore, it appeared that the genes for the benzaldehyde dehydrogenases (BADHs) must be located upstream of the mdlC gene (34) and must be transcribed independently.
FIG. 1.
Mandelate pathway in P. putida ATCC 12633 as described in reference 35.
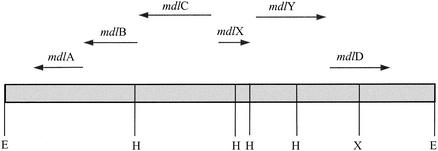
It has been suggested that the mandelate pathway is an excellent candidate for the study of enzyme functional evolution (30). Mandelate racemase, for example, proved to be sequentially and structurally homologous to muconate lactonizing enzyme (23). This observation ultimately led to the identification of the larger enolase superfamily (2, 3). The X-ray structures of S-mandelate dehydrogenase (22) and benzoylformate decarboxylase (9) are now available. In order to extend our studies of the mandelate pathway, it will be necessary to identify, isolate, and determine the three-dimensional structures of the other members of that pathway. We began that process by sequencing the remainder of the restriction fragment believed to contain the genes encoding the enzymes of the mandelate pathway (35). This has revealed three open reading frames, which we have denoted mdlX, mdlY, and mdlD (Fig. 2). Analysis of the deduced amino acid sequences suggests that these genes encode a transcriptional regulatory protein, an amidase, and a dehydrogenase, respectively. Finally, we have expressed the mdlY and mdlD genes in Escherichia coli, purified the gene products, and identified them as a mandelamide hydrolase (MAH) and an NAD(P)+-dependent BADH, respectively.
FIG. 2.
Schematic representation of the 10.5-kb EcoRI restriction fragment carrying the genes of the mandelate pathway of P. putida ATCC 12633. The arrows show the position and orientation of the genes, and relevant restriction sites are indicated: E, EcoRI; H, HindIII; and X, XbaI.
MATERIALS AND METHODS
Materials.
P. putida strains ATCC 12633 and ATCC 17453 were obtained from the American Type Culture Collection (Manassas, Va.). The plasmid pSCR4, which carries the mandelate pathway genes on a 10.5-kb EcoRI fragment, and P. putida (ATCC 17453) conjugated with pSCR4 were available from a previous study (35). Restriction enzymes, ligases, DNA polymerase, and other molecular biology reagents were obtained from New England Biolabs, Promega, Stratagene, or Novagen. PCR, mutagenesis, and sequencing primers were obtained from either the Biomolecular Resource Center (BRC, University of California, San Francisco) or the DNA Synthesis Core Facility (University of Michigan). Glycerol, dithiothreitol (DTT), KH2PO4, K2HPO4, potassium chloride, ammonium sulfate, and 2-(N-morpholino)ethanesulfonic acid (MES) were obtained from Fisher Scientific. Phenylmethylsulfonyl fluoride (PMSF), imidazole, magnesium chloride, reactive yellow 86, NAD+, NADP+, HEPES, and [(2-hydroxy-1,1-bis[hydroxymethyl]ethyl)amino]-1-propanesulfonic acid (TAPS) were obtained from Sigma. Dialysis tubing (12,000- to 14,000-molecular-weight cutoff) was from obtained from Spectrum Laboratory, while the Centriplus centrifugal concentration units were obtained from Amicon. Affi-Gel blue was obtained from Bio-Rad Laboratories.
Sequencing of mandelate pathway fragment.
The restriction endonucleases EcoRI, HindIII, and XbaI were used to digest pSCR4. The fragments were subcloned into pUC18, pUC19, or pTZ18R as required and were sequenced in both directions by both manual and automated sequencing methods. Sequence alignment was accomplished with the Beckman Microgenie analysis program (version 6.0), and open reading frames were identified with the GCG package of the Genetics Computer Group, Madison, Wis. The sequence was later confirmed by direct sequencing of pSCR4 with the appropriate primers.
Sequence analysis.
BLAST searches for screening homologous proteins (1) were made with BLASTp version 2.2.2 (http://www.ncbi.nlm.nih.gov/BLAST). Sequence alignments were carried out with the GCG package.
PCR amplification of mdlY and mdlD.
Using the sequence of the open reading frame, two primers were designed to amplify the mdlY gene. The forward primer, 5′-GGTAACATATGCGCCAC-3′, added a 5′ NdeI site (underlined), while the reverse primer, 5′-CCTAGCCGGATCCAGATAATT-3′ added the underlined 3′ BamHI site. A similar strategy was used to obtain the mdlD gene. The forward primer, 5′-CCACATATGAATTATCTGTCTCCGGC-3′, again added an NdeI site (underlined), but for mdlD, the reverse primer, 5′-CTGAATTCCAATAGCACAAGAAGCGCGC-3′, was designed to add the underlined EcoRI site. Amplification of both genes was achieved with Pfu DNA polymerase, using the plasmid pSCR4 as a template. The products were ligated into pET17b (Novagen) to generate the MAH and BADH expression vectors pET17mdlY and pET17mdlD, respectively. Both inserts were sequenced, and the fidelity of the amplification was checked by comparison with that of the original sequences.
Purification of MAH.
MAH was purified from E. coli strain BL21(DE3)pLysS, which had been transformed with pET17mdlY. A freshly transformed, single colony was selected and used to inoculate 50 ml of Luria broth containing 50 μg of ampicillin per ml and 25 μg of chloramphenicol per ml. This culture was grown overnight at 37°C and used to inoculate 1 liter of fresh medium. The fresh culture was grown at 37°C until the optical density at 600 nm reached 0.6 to 0.8. The cells were cooled to 28°C, and protein expression was induced by the addition of 0.4 mM isopropyl β-d-thiogalactopyranoside (IPTG). The cells were grown for an additional 6 h at 28°C prior to harvesting by centrifugation at 4,000 × g for 8 min at 4°C. The cell pellet was washed and resuspended in 50 mM phosphate buffer (pH 7.0) containing 1 mM EDTA. Cell lysis was achieved by sonication, and the lysate was clarified by centrifugation at 15,000 × g for 30 min at 4°C.
The clarified cell extract was subjected to ammonium sulfate fractionation. The protein fraction precipitating between 0 and 40% saturation with ammonium sulfate was collected by centrifugation and dissolved in 50 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA. Following exhaustive dialysis against the same buffer at 4°C, a small amount of precipitate was removed by centrifugation, and the solution was subjected to size-exclusion chromatography on a Sephacryl S-200 column, which had been equilibrated previously with 20 mM potassium phosphate buffer (pH 6.0)-1 mM EDTA (buffer A). Fractions containing MAH activity were pooled and concentrated with Centriplus centrifugal filter devices with a 30,000-molecular-weight cutoff. The concentrated enzyme was applied to a HiPrep Q anion-exchange column, using an Akta fast protein liquid chromatograph (Amersham Pharmacia Biotech). The column was washed with buffer A until there was no further elution of protein. MAH was eluted with buffer A containing 0.5 M NaCl, with activity eluting at about 250 mM NaCl. Fractions containing activity were pooled and concentrated as described above. In the final step, the concentrated enzyme solution was applied to HiTrap Q column (5 ml) equilibrated with 100 mM potassium phosphate buffer (pH 7.8)-1 mM EDTA. MAH eluted in the early fractions in a 0 to 0.5 M NaCl gradient in 100 mM potassium phosphate buffer (pH 7.8)-1 mM EDTA. These fractions were pooled, concentrated, and stored in aliquots at −20°C.
Assay of MAH.
MAH activity was determined at 30°C in 100 mM potassium phosphate buffer (pH 7.8) containing 1 mM EDTA. Both R-mandelamide and S-mandelamide were employed as substrates. The release of ammonia was monitored by the phenol-hypochlorite method of Krallmann-Wenzel (17). A 5-ml reaction volume was used, and at appropriate time intervals, 0.5-ml aliquots were withdrawn and added to 50 μl of a catalyst solution (a 3:1 mixture of 2 mM MnSO4 and acetone) and 1 ml of phenol reagent. Sodium hypochlorite solution containing 1.5 g of free chlorine (0.5 ml) per 100 ml was added, and the solution was vortexed. After a minimum of 30 min, the A636 was measured. The assay mixture, without enzyme, served as a blank. The amount of ammonia released was extrapolated from a calibration curve of ammonia in the range 0 to 100 nmol/ml. Assays were carried out in triplicate (at least), and kinetic parameters were calculated by fitting the data to the Michaelis-Menten equation by nonlinear regression analysis.
Preparation and assay of P. putida strains grown on glucose, mandelamide, and mandelate.
P. putida ATCC 12633, ATCC 117453, and ATCC 117453 conjugated with pSCR4 were grown at 30°C in M9 minimal media. Glucose, R,S-mandelamide, or R,S-mandelate was added as the sole carbon source to a final concentration of 20 mM. The cells were grown to late log phase and harvested by centrifugation at 4°C. The cell pellet was washed several times with water before being resuspended in 50 mM phosphate buffer (pH 7.0) containing 1 mM EDTA. Crude cell extracts were prepared by sonication followed by centrifugation at 15,000 × g for 30 min at 4°C. These extracts were assayed for MAH activity by the phenol-hypochlorite method described above.
Purification of BADH.
The mdlD gene was also expressed in E. coli strain BL21(DE3)pLysS in a manner identical to that described for MAH. Prior to sonication, the cell pellet was resuspended in 100 mM potassium phosphate buffer (pH 7.5) containing 100 mM KCl, 2 mM DTT, and 1 mM PMSF. Following ammonium sulfate fractionation, the pellet containing the 30 to 65% (NH4)2SO4 fraction was resuspended in a minimum volume of a buffer containing 25 mM imidazole (pH 7.2), 50 mM KCl, 2 mM DTT, and 10% glycerol (buffer B) and dialyzed against the same buffer overnight at 4°C. After centrifugation, the solution was loaded onto a DEAE-Sepharose anion-exchange column that had previously been equilibrated with buffer B and then was eluted with a linear gradient of 50 to 500 mM KCl in buffer B. The BADH activity eluted at ∼300 mM KCl, and fractions containing activity were pooled, desalted, concentrated, and exchanged into 10 mM MES buffer (pH 6.0) containing 5 mM KCl, 5 mM MgCl2, 2 mM DTT, and 10% glycerol (buffer D) by using Centriplus concentration units. The BADH was loaded onto a reactive yellow 86 column linked to an Affi-Gel blue column, both of which had been equilibrated with buffer D. After the columns had been washed thoroughly with buffer D, the reactive yellow 86 column was removed, and the Affi-Gel blue column was washed with a buffer (buffer E) containing 10 mM MES (pH 6.5), 50 mM KCl, 2 mM DTT, and 10% glycerol. BADH activity was then eluted with buffer E containing 1 mM NADP. Again fractions were collected, pooled, and concentrated before being applied to a Sephacryl S-200 column. Finally, highly purified BADH was eluted with 50 mM HEPES buffer (pH 7.5) containing 100 mM KCl, 2 mM DTT, and 2 mM NADP. After concentration, the purified enzyme was stored in aliquots at −20°C.
Assay of BADH.
BADH activity was monitored spectrophotometrically by observing the increase in A340 due to the reduction of either NAD+ or NADP+. Routine activity assays were carried out at 25°C with a reaction mixture containing TAPS buffer (pH 8.5) (100 mM), KCl (100 mM), DTT (1 mM), NAD+ or NADP+ (1 mM), and benzaldehyde (1 mM) in a 1-ml final volume. Reaction rates were determined with a molar extinction coefficient of 6220 M−1 for NADH or NADPH at 340 nm. For the determination of Km values for benzaldehyde, NAD+ or NADP+ was held at 1 mM, and the benzaldehyde concentration was varied as required. Kinetic data (triplicates) were again fitted to the Michaelis-Menten equation.
Nucleotide sequence accession number.
The nucleotide sequences determined in this work have been deposited under GenBank accession no. AY143338.
RESULTS AND DISCUSSION
Sequencing of the region immediately upstream from mdlC revealed an open reading frame of 699 bp, located 151 bp from mdlC and oriented in the opposite direction from the genes of the mdlCBA operon. The open reading frame of 233 residues was denoted mdlX, and S1 nuclease mapping experiments revealed the presence of a divergent promoter between it and mdlCBA (34). The full-length sequence was used for a BLAST search (1) of the nonredundant protein database at the National Center for Biotechnology Information. The results suggested that the mdlX gene product had 60% sequence similarity (over 222 residues) to a transcriptional regulatory protein from Bradyrhizobium japonicum (gi:27381525). In addition the C-terminal 60 residues could be aligned (85 to 95%) with families of transcriptional regulators that bind DNA by using a helix-turn-helix motif. We have yet to express the gene product for mdlX, and at present, its role remains unclear.
Sequencing of mdlY and identification of its gene product as an MAH.
A second open reading frame was found 107 bp downstream of mdlX. This was denoted mdlY, comprised 1,521 bp, and encoded a protein of 507 amino acids the function of which was, at this point, unknown. A BLAST search using the full-length sequence identified a number of bacterial amidases, including several indoleacetamide hydrolases, that were highly homologous (∼50% similar) to the unknown protein. In particular, the amino acid sequence contained the amidase signature sequence, a sequence rich in serine and glycine residues (6, 21). This homology, together with the location of the mdlY gene within the mandelate pathway gene cluster, suggested that mdlY might encode a protein that could hydrolyze mandelamide to mandelic acid. This is not unreasonable, because Hegeman, in his initial studies of the enzymes of the mandelate pathway, observed that R,S-mandelamide supported the growth of P. putida ATCC 12633 (10). Furthermore, Laverack and Clarke (18) were able to select for “mandelamidase” mutants of P. putida ATCC 12633 by alternating between mandelamide and isobutyramide growth media.
Figure 3 shows the results of a recent BLAST search. These data indicate that there are now at least 11 enzymes that contain the amidase signature sequence and show homology to MAH. These enzymes arise from organisms as diverse as Agrobacterium rhizogenes, Streptomyces coelicolor, Rhodococcus sp., Caulobacter crescentus, Saccharomyces cerevisiae, Aspergillus ustus, Flavobacterium sp., B. japonicum, chickens, Drosophila, rats, and humans. While the homology ranges from 50% to less than 20% sequence identity, the figure clearly demonstrates the presence of a GGSSXG motif, as well as an invariant serine residue located 21 amino acids toward the C terminus. Intriguingly, it may also be seen that, with the exception of glutamyl-tRNA amidotransferase subunit A (GATA), the group also contains the GXSXG motif of the serine proteases (5). This may not be entirely surprising, because proteases are also amidases, but to date, there is no evidence to link the evolution of the serine proteases to that of the amidase superfamily. Indeed, the evidence suggests that the mechanisms of the two families are quite distinct. Of the amidase signature family, only fatty acid amide hydrolase (FAAH) has been subjected to any detailed mechanistic studies (24-27). These indicated that the FAAH operated by a nonconventional mechanism using a Ser241 as a nucleophile while Lys142 operated as an acid/base catalyst (25). Lately, kinetic (16) and structural (32) studies of malonamidase E2 from B. japonicum have confirmed the novelty of the catalytic machinery of this superfamily.
FIG. 3.
Multiple sequence alignment of the region encompassing the amidase signature sequence (boldface) of enzymes identified as having sequence homology to MHA. The enzymes are in decreasing order of similarity to MAH and were identified by a BLAST search (1) of the nonredundant protein database. The top-scoring example of each enzyme was used for this alignment. MAH, MAH from P. putida; IAAH, indoleacetamide hydrolase from A. rhizogenes (gi:1170450), GATA, glutamyl-tRNA amidotransferase subunit A from S. coelicolor (gi:6225417); ESAM, enantiomer-specific amidase from Rhodococcus (gi:98837); AMID, amidase from R. erythropolis (gi:113712); PYRA, pyrazinamidase/nicotinamidase from Caulobacter crescentus (gi:13424188); UREA, urea amidolyase from S. cerevisiae (gi:6319685); ACET, acetamidase from Aspergillus ustus (gi:13569683); 6AHH, 6-aminohexanoate-cyclic-dimer hydrolase from Flavobacterium sp. strain K172 (gi:129000); MAE2, malonamidase E2 from B. japonicum (gi:17939381); FAAH, FAAH from Homo sapiens (gi:6225310); VD3H, vitamin D3 hydroxylase-associated protein from Gallus gallus (gi:2492838).
Cloning, expression, purification, and characterization of MAH.
Given the lack of detailed mechanistic information about this widely distributed class of enzymes, it was incumbent upon us to confirm that the mdlY gene product was, in fact, an MAH and a member of the amidase family.
Pfu DNA polymerase and primers containing an NdeI site at the initiation codon and a BamHI site downstream of the termination codon were used to amplify the mdlY gene. Using the newly constructed restriction sites, the amplified gene was placed in the pET17b expression vector, which uses the inducible T7 promoter. Excellent expression of MAH in E. coli BL21(DE3)pLysS was observed when induction with 0.4 mM IPTG was carried out at temperatures lower than 30°C. However, it was noted that induction at 37°C led to little or no protein expression, and for that reason, induction of MAH expression was routinely carried out at 28°C.
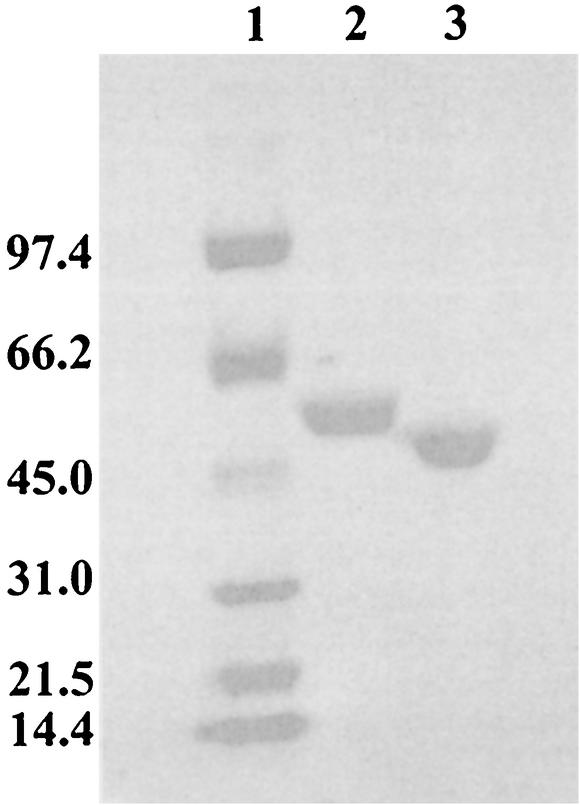
The purification of MAH was achieved in three steps, and the enzyme was greater than 95% homogenous as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 4). The molecular mass of the protein, determined by electrospray mass spectrometry, was 53,820 ± 4 Da, which was consistent with that calculated from the deduced amino acid sequence (53, 815 Da). Sedimentation equilibrium experiments provided an experimental molecular mass of 52.6 kDa, which is consistent with the mass spectral data and suggests that MAH exists as a monomer in solution (data not shown).
FIG. 4.
SDS-PAGE gel (4 to 20% polyacrylamide) showing MAH (lane 2) and BADH (lane 3) purified after expression in E. coli. Molecular mass standards (kilodaltons) are shown in lane 1.
The majority of assays used to determine the activity of amidases are discontinuous assays. These include the radiochemical method employed for FAAH (25) and hydroxamate-based assays, which, in effect, monitor the rate of transfer of the acyl group to hydroxylamine (4). Conversely, the release of ammonia can be monitored by reaction with indophenol (13). None of these methods proved satisfactory in our preliminary trials, so an assay based on the phenol-hypochlorite method of Krallmann-Wenzel (17) was developed. The method monitors the release of ammonia, and the reaction is monitored by the increase in A636. The rate of ammonia release was found to depend on both enzyme and substrate concentration and demonstrated typical Michaelis-Menten saturation behavior. Both R-mandelamide and S-mandelamide proved to be excellent substrates for MAH, with Km values in the low micromolar range (Table 1). Interestingly, the kcat/Km values for both enantiomers were almost identical (Table 1), suggesting that MAH was not enantioselective. In contrast, the amidase from Rhodococcus erythropolis (12) was shown to convert R,S-arylamides to the corresponding S-acids with enantiomeric excesses above 99%.
TABLE 1.
Kinetic parameters for MAHa
| Substrate | Km (μM) | kcat (min−1) | kcat/Km (min−1 M−1) |
|---|---|---|---|
| R-Mandelamide | 34.2 ± 1.9 | (9.64 ± 0.54) × 102 | 2.82 × 105 |
| S-Mandelamide | 19.8 ± 3.8 | (4.64 ± 0.82) × 102 | 2.34 × 105 |
Experiments were carried out at 30°C and pH 7.8 as described in Materials and Methods. The data are the mean ± standard deviation of at least three individual experiments.
Not unexpectedly, given that it can hydrolyze both enantiomers of mandelamide, MAH was readily able to hydrolyze phenylacetamide. It was also able to hydrolyze 3-phenylpropionamide, albeit considerably less efficiently, while benzamide was not a substrate (data not shown). MAH proved susceptible to reagents such as 3,4-dichloroisocoumarin and PMSF, both of which would be expected to modify serine residues (15). This would suggest that MAH, like FAAH (27) and malonamidase E2 (16), also uses a serine nucleophile for catalysis. Further experiments, designed to elucidate the catalytic mechanism of MAH, are now under way.
Although it was not unreasonable to propose that the mdlY gene product would be an MAH, conclusive evidence for the physiological role of MAH requires the preparation of a gene knockout. This, presumably, would confer a mandelamide-negative phenotype. In the absence of such a knockout, we sought alternative evidence. It has been shown that the mandelate racemase, mandelate dehydrogenase, benzoylformate decarboxylase and BADH of P. putida ATCC 12633 can all be fully induced by R,S-mandelate and benzoylformate (10). Potentially, if MAH was indeed a member of the mandelate pathway, it could also be induced in a similar manner. Accordingly, P. putida ATCC 12633 was grown on medium containing, individually, glucose, R,S-mandelic acid, or R,S-mandelamide. Following cell lysis, the cell extracts were assayed for the ability to hydrolyze R,S-mandelamide. The results shown in Table 2 clearly indicate that this strain is able to utilize both mandelate and mandelamide as the sole carbon source and that the strain possesses MAH activity. Furthermore, a sevenfold-higher level of activity was found in the cells grown on R,S-mandelamide, while growth on mandelate also provided a modest (twofold) boost in MAH activity. We also examined extracts of P. putida ATCC 17453 (a strain lacking the mandelate pathway), which was unable to grow on mandelamide-containing minimal media. However, when the same strain was conjugated with pSCR4, the plasmid carrying the mandelate pathway genes, it was able to grow with mandelamide as the sole carbon source. We also demonstrated that, unlike ATCC 12633, which has a basal level of MAH activity even when grown on glucose, P. putida ATCC 17453 grown on glucose possesses no MAH activity (Table 2). Conversely, when conjugated with pSCR4, the latter not only grows on mandelamide, but also possesses a level of MAH activity similar to that of ATCC 12633.
TABLE 2.
MAH activity in crude cell extractsa
| P. putida strain | Carbon source | MAH activity (nmol/min/mg)b |
|---|---|---|
| ATCC 12633 | d-Glucose | 15.6 ± 1.2 |
| ATCC 12633 | R,S-Mandelate | 31.0 ± 1.1 |
| ATCC 12633 | R,S-Mandelamide | 103 ± 3.0 |
| ATCC 12633 | R,S-Mandelonitrile | —c |
| ATCC 17453 | d-Glucose | NDd |
| ATCC 17453 | R,S-Mandelamide | —c |
| ATCC 17453 (pSCR4) | R,S-Mandelamide | 113 ± 5.2 |
Cells were grown on M9 minimal medium containing either d-glucose, R,S-mandelate, or R,S-mandelamide (20 mM).
Assays were carried out at 30°C and pH 7.8 as described in Materials and Methods. The data are the mean ± standard deviation of at least three individual experiments.
—, strain unable to grow.
ND, no activity detected.
Taken together, these data suggest that the mdlY gene product, MAH, can be used to support growth on mandelamide. However, it is still uncertain as to whether MAH is part of the mandelate pathway. The other enzymes in the pathway are equally induced by their substrates and R,S-mandelate, with levels of induction ranging between 500- and 2,000-fold (10). The results in Table 2 show that the level of induction by R,S-mandelate is lower than that brought about by R,S-mandelamide. Furthermore, the twofold increase in MAH activity is significantly lower than the increases R,S-mandelate produces in the other mandelate pathway enzymes. Laverack and Clarke (18) suggested that their “mandelamidase” was constitutively expressed, an observation more in keeping with the results in Table 2. Finally, P. putida ATCC 12633 is unable to grow on mandelonitrile, the most obvious precursor to mandelamide. Overall, the results show that the mdlY gene product can certainly support growth on R,S-mandelamide but do not provide definitive proof that it is a member of the mandelate pathway. Certainly, if it is a member, it is not induced in the same manner as the other enzymes in the pathway.
Sequencing of mdlD and identification of its gene product as a BADH.
The final open reading frame on the restriction fragment encoding the enzymes of the mandelate pathway was located 71 bp downstream of mdlY, was transcribed in the same direction, and comprised 1,308 bp. A BLAST search, again using the full-length sequence of 436 residues, indicated that the mdlD gene product was likely to be an aldehyde dehydrogenase, probably of the class 3 variety (29). The suggestion that the mdlD gene product would be an aldehyde dehydrogenase was not particularly surprising, because both NAD+- and NADP+-dependent BADHs have always been associated with the mandelate pathway in P. putida (7, 10). These enzymes are induced by mandelate and benzoylformate, whereas another P. putida BADH is induced only in the presence of benzaldehyde or benzyl alcohol (33).
There are at least 13 distinct families of aldehyde dehydrogenases (29). Most are specific for NAD+, and a few are specific for NADP+. The class 3 aldehyde dehydrogenases, for which the enzyme from rat liver is the structural paradigm (19), are characterized by an ability to use NAD+ or NADP+ (29). Alignment of the sequences of P. putida BADH and the rat liver enzyme (not shown) shows that the two share about 40% sequence identity and 50% sequence similarity, and as expected, the catalytic residues are identical. These include the catalytic thiol and the general base, Cys249 and Glu 337, respectively, as well as the aspartic acid residue Asp253, which seems to link two conserved motifs in class 3 enzymes (11). Overall, BADH shares about 50% sequence similarity with several class 3 aldehyde dehydrogenases, including the aldehyde dehydrogenase from Pseudomonas oleovorans (14). The majority of aromatic aldehyde dehydrogenases from Pseudomonas sp. are not class 3 enzymes, although they are closely related phylogenetically (29). In keeping with that, BADH shares approximately 40% similarity (33% identity) with the aldehyde dehydrogenases from other pseudomonads.
Cloning, expression, purification, and characterization of BADH.
To confirm that the mdlD gene product was an NAD(P)+-dependent benzaldehyde dehydrogenase, it was necessary to express and purify it. The basic methodology employed to amplify the mdlX gene was also used to amplify mdlD. Again, good expression of soluble protein was observed, and, as for MAH, expression was induced at 28°C.
A three-step purification protocol provided protein that was judged to be better than 98% homogenous (Fig. 4). The molecular weight of the protein, determined by electrospray mass spectrometry, was 47,425 ± 5, which is consistent with the 47,431 deduced from the amino acid sequence. As predicted, BADH was able oxidize benzaldehyde by using either NAD+ or NADP+ as the cofactor. BADH was only marginally active with acetaldehyde, while longer-chain aliphatic aldehydes were poor substrates (data not shown). The apparent Km values for benzaldehyde were in the low micromolar range (Table 3), suggesting that, in all likelihood, this was the natural substrate. The enzyme exhibits greater affinity for NAD+ than NADP+ (data not shown), similar to the rat class 3 aldehyde dehydrogenase (28), but unlike the P. aeruginosa enzymes betaine aldehyde dehydrogenase (37) and glucose-6-phosphate dehydrogenase (20), both of which have greater affinity for NADP+. The data in Table 3 indicate that the enzyme has marginally greater affinity for benzaldehyde when NADP+ is the cofactor, but both the kcat and the overall efficiency of the enzyme are greater in the presence of NAD+. Similar results are observed for rat aldehyde dehydrogenase (28) and glucose-6-phosphate dehydrogenase (20), yet betaine aldehyde dehydrogenase shows greater efficiency in the presence of NADP+ (37). Clearly there is considerable variation in the nucleotide specificity in this class of enzymes, but, overall, our data confirm that the mdlD gene product is, indeed, an NAD(P)+-dependent BADH. Finally, an NAD(P)+-dependent BADH is induced when P. putida ATCC 17453, conjugated with pSCR4, is grown on R-mandelate. This enzyme was purified and subjected to N-terminal sequencing. The sequence of the first 15 amino acids of this enzyme, MNYLSPAKIDSLFSA, is identical to those deduced for the mdlD gene product (P. Guidinger, unpublished results).
TABLE 3.
Kinetic parameters for BADHa
| Cofactorb | Km for benzalde- hyde (μM) | kcat (min−1) | kcat/Km (min−1 M−1) |
|---|---|---|---|
| NAD+ | 63.4 ± 1.5 | (8.24 ± 0.32) × 103 | 1.3 × 108 |
| NADP+ | 39.9 ± 4.4 | (2.55 ± 0.10) × 103 | 6.39 × 107 |
Experiments were carried out at 25°C and pH 8.5 as described in Materials and Methods. The data are the mean ± standard deviation of at least three individual experiments.
Cofactor concentrations were maintained at 1 mM.
Acknowledgments
This work was supported by NIH GM40570 (to J.A.G. and G.L.K.).
Thanks are due to Amy Tsou, Susan Schafer, Stephen Ransom, and Peter Guidinger for assistance during the early stages of this project. We also thank Pan-Fen Wang for carrying out the mass spectrometry.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babbitt, P. C., M. S. Hasson, J. E. Wedekind, D. R. Palmer, W. C. Barrett, G. H. Reed, I. Rayment, D. Ringe, G. L. Kenyon, and J. A. Gerlt. 1996. The enolase superfamily: a general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry 35:16489-16501. [DOI] [PubMed] [Google Scholar]
- 3.Babbitt, P. C., G. T. Mrachko, M. S. Hasson, G. W. Huisman, R. Kolter, D. Ringe, G. A. Petsko, G. L. Kenyon, and J. A. Gerlt. 1995. A functionally diverse enzyme superfamily that abstracts the alpha protons of carboxylic acids. Science 267:1159-1161. [DOI] [PubMed] [Google Scholar]
- 4.Brammar, W. J., and P. H. Clarke. 1964. Induction and repression of Pseudomonas aeruginosa amidase. J. Gen. Microbiol. 37:307-319. [DOI] [PubMed] [Google Scholar]
- 5.Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528-530. [DOI] [PubMed] [Google Scholar]
- 6.Chebrou, H., F. Bigey, A. Arnaud, and P. Galzy. 1996. Study of the amidase signature group. Biochim. Biophys. Acta 1298:285-293. [DOI] [PubMed] [Google Scholar]
- 7.Fewson, C. A. 1988. Microbial metabolism of mandelate: a microcosm of diversity. FEMS Microbiol. Rev. 4:85-110. [DOI] [PubMed] [Google Scholar]
- 8.Gunsalus, C. F., R. Y. Stanier, and I. C. Gunsalus. 1953. The enzymatic conversion of mandelic acid to benzoic acid. III. Fractionation and properties of the soluble enzymes. J. Bacteriol. 66:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasson, M. S., A. Muscate, M. J. McLeish, L. S. Polovnikova, J. A. Gerlt, G. L. Kenyon, G. A. Petsko, and D. Ringe. 1998. The crystal structure of benzoylformate decarboxylase at 1.6 Å resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes. Biochemistry 37:9918-9930. [DOI] [PubMed] [Google Scholar]
- 10.Hegeman, G. D. 1966. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J. Bacteriol. 91:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hempel, J., I. Kuo, J. Perozich, B. C. Wang, R. Lindahl, and H. Nicholas. 2001. Aldehyde dehydrogenase. Maintaining critical active site geometry at motif 8 in the class 3 enzyme. Eur. J. Biochem. 268:722-726. [DOI] [PubMed] [Google Scholar]
- 12.Hirrlinger, B., A. Stolz, and H.-J. Knackmuss. 1996. Purification and properties of an amidase from Rhodococcus erythropolis MP50 which enantioselectively hydrolyzes 2-arylpropionamides. J. Bacteriol. 178:3501-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan, A. 1969. The determination of urea, ammonia, and urease. Methods Biochem. Anal. 17:311-324. [DOI] [PubMed] [Google Scholar]
- 14.Kok, M., R. Oldenhuis, M. P. van der Linden, C. H. Meulenberg, J. Kingma, and B. Witholt. 1989. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J. Biol. Chem. 264:5442-5451. [PubMed] [Google Scholar]
- 15.Koo, C. W. 1997. Mandelamide hydrolase from Pseudomonas putida. Ph.D. thesis. University of California, San Francisco.
- 16.Koo, H. M., S. O. Choi, H. M. Kim, and Y. S. Kim. 2000. Identification of active-site residues in Bradyrhizobium japonicum malonamidase E2. Biochem. J. 349:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krallmann-Wenzel, U. 1985. An improved method of ammonia determination, applicable to amidases and other ammonia-producing enzyme systems of mycobacteria. Am. Rev. Respir. Dis. 131:432-434. [DOI] [PubMed] [Google Scholar]
- 18.Laverack, P. D., and P. H. Clarke. 1979. Selection of mandelamidase constitutive mutants in continuous culture of Pseudomonas putida. Biotechnol. Lett. 1:353-358. [Google Scholar]
- 19.Liu, Z. J., Y. J. Sun, J. Rose, Y. J. Chung, C. D. Hsiao, W. R. Chang, I. Kuo, J. Perozich, R. Lindahl, J. Hempel, and B. C. Wang. 1997. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4:317-326. [DOI] [PubMed] [Google Scholar]
- 20.Ma, J.-F., P. W. Hager, M. L. Howell, P. V. Phibbs, and D. J. Hassett. 1998. Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat). J. Bacteriol. 180:1741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayaux, J. F., E. Cerebelaud, F. Soubrier, D. Faucher, and D. Pétré. 1990. Purification, cloning, and primary structure of an enantiomer-selective amidase from Brevibacterium sp. strain R312: structural evidence for genetic coupling with nitrile hydratase. J. Bacteriol. 172:6764-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra, B., J. A. Gerlt, P. C. Babbitt, C. W. Koo, G. L. Kenyon, D. Joseph, and G. A. Petsko. 1993. A novel structural basis for membrane association of a protein: construction of a chimeric soluble mutant of (S)-mandelate dehydrogenase from Pseudomonas putida. Biochemistry 32:12959-12967. [DOI] [PubMed] [Google Scholar]
- 23.Neidhart, D. J., G. L. Kenyon, J. A. Gerlt, and G. A. Petsko. 1990. Mandelate racemase and muconate lactonizing enzyme are mechanistically distinct and structurally homologous. Nature 347:692-694. [DOI] [PubMed] [Google Scholar]
- 24.Omeir, R. L., G. Arreaza, and D. G. Deutsch. 1999. Identification of two serine residues involved in catalysis by fatty acid amide hydrolase. Biochem. Biophys. Res. Commun. 264:316-320. [DOI] [PubMed] [Google Scholar]
- 25.Patricelli, M. P., and B. F. Cravatt. 2000. Clarifying the catalytic roles of conserved residues in the amidase signature family. J. Biol. Chem. 275:19177-19184. [DOI] [PubMed] [Google Scholar]
- 26.Patricelli, M. P., and B. F. Cravatt. 1999. Fatty acid amide hydrolase competitively degrades bioactive amides and esters through a nonconventional catalytic mechanism. Biochemistry 38:14125-14130. [DOI] [PubMed] [Google Scholar]
- 27.Patricelli, M. P., M. A. Lovato, and B. F. Cravatt. 1999. Chemical and mutagenic investigations of fatty acid amide hydrolase: evidence for a family of serine hydrolases with distinct catalytic properties. Biochemistry 38:9804-9812. [DOI] [PubMed] [Google Scholar]
- 28.Perozich, J., I. Kuo, B. C. Wang, J. S. Boesch, R. Lindahl, and J. Hempel. 2000. Shifting the NAD/NADP preference in class 3 aldehyde dehydrogenase. Eur. J. Biochem. 267:6197-6203. [DOI] [PubMed] [Google Scholar]
- 29.Perozich, J., H. Nicholas, B. C. Wang, R. Lindahl, and J. Hempel. 1999. Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 8:137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petsko, G. A., G. L. Kenyon, J. A. Gerlt, D. Ringe, and J. W. Kozarich. 1993. On the origin of enzymatic species. Trends Biochem. Sci. 18:372-376. [DOI] [PubMed] [Google Scholar]
- 31.Ransom, S. C., J. A. Gerlt, V. M. Powers, and G. L. Kenyon. 1988. Cloning, DNA sequence analysis, and expression in Escherichia coli of the gene for mandelate racemase from Pseudomonas putida. Biochemistry 27:540-545. [DOI] [PubMed] [Google Scholar]
- 32.Shin, S., T. H. Lee, N. C. Ha, H. M. Koo, S. Y. Kim, H. S. Lee, Y. S. Kim, and B. H. Oh. 2002. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. EMBO J. 21:2509-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevenson, I. L., and J. Mandelstam. 1965. Induction and multi-sensitive end-product repression in two converging pathways degrading aromatic substances in Pseudomonas fluorescens. Biochem. J. 96:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsou, A. Y. 1990. A study of the genes encoding the enzymes in the mandelate pathway in Pseudomonas putida. Ph.D. thesis. University of Maryland, College Park.
- 35.Tsou, A. Y., S. C. Ransom, J. A. Gerlt, D. D. Buechter, P. C. Babbitt, and G. L. Kenyon. 1990. Mandelate pathway of Pseudomonas putida: sequence relationship involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry 29:9856-9862. [DOI] [PubMed] [Google Scholar]
- 36.Tsou, A. Y., S. C. Ransom, J. A. Gerlt, V. M. Powers, and G. L. Kenyon. 1989. Selection and characterization of a mutant of the cloned gene for mandelate racemase that confers resistance to an affinity label by greatly enhanced production of enzyme. Biochemistry 28:969-975. [DOI] [PubMed] [Google Scholar]
- 37.Velasco-Garcia, R., L. Gonzalez-Segura, and R. A. Munoz-Clares. 2000. Steady-state kinetic mechanism of the NADP+- and NAD+-dependent reactions catalysed by betaine aldehyde dehydrogenase from Pseudomonas aeruginosa. Biochem. J. 352:675-683. [PMC free article] [PubMed] [Google Scholar]
- 38.Wheelis, M. L., and R. Y. Stanier. 1970. The genetic control of dissimilatory pathways in Pseudomonas putida. Genetics 66:245-266. [DOI] [PMC free article] [PubMed] [Google Scholar]