Abstract
Many DNA viruses concatemerize their genomes as a prerequisite to packaging into capsids. Concatemerization arises from either replication or homologous recombination. Replication is already the target of many antiviral drugs, and viral recombinases are an attractive target for drug design, particularly for combination therapy with replication inhibitors, due to their important supporting role in viral growth. To dissect the molecular mechanisms of viral recombination, we and others previously identified a family of viral nucleases that comprise one component of a conserved, two-component viral recombination system. The nuclease component is related to the exonuclease of phage λ and is common to viruses with linear double-stranded DNA genomes. To test the idea that these viruses have a common strategy for recombination and genome concatemerization, we isolated the previously uncharacterized 34.1 gene from Bacillus subtilis phage SPP1, expressed it in Escherichia coli, purified the protein, and determined its enzymatic properties. Like λ exonuclease, Chu (the product of 34.1) forms an oligomer, is a processive alkaline exonuclease that digests linear double-stranded DNA in a Mg2+-dependent reaction, and shows a preference for 5′-phosphorylated DNA ends. A model for viral recombination, based on the phage λ Red recombination system, is proposed.
Many DNA viruses require DNA replication not only for synthesizing genomes, but also for generating substrates with the correct topology—genomic concatemers—for packaging into infectious viral particles. Under conditions in which replication is inhibited but multiple copies of a viral genome are present, recombination can take over the conversion of monomeric genomic molecules into the concatemeric arrays required for packaging.
In double-stranded DNA (dsDNA) viruses with linear genomes, exemplified by the well-studied phage λ (16), a typical infection begins with injection of linear viral DNA into the host (Escherichia coli), where it takes on a circular conformation. These unit-length molecules replicate in the theta mode but cannot be packaged (45), since they possess a single packaging site (cos). Later in infection, replication switches to the sigma (rolling circle) mode, producing genomic concatemers with multiple cos sites. The packaging endonuclease (Ter) cleaves cos (see reference 7 for a review) and positions the empty viral capsid at one of the nascent DNA ends (cosL). DNA is pumped into the capsid until one genome length of λ DNA is encapsidated and a second cos site is encountered. Packaging is completed when Ter bound to the filled capsid cleaves the DNA at the second cos site and the filled capsid dissociates from Ter.
The general strategy of viral genome packaging by encapsidation of a “headful” of DNA cleaved from a concatemeric array of viral genomes is conserved among linear dsDNA viruses, including the herpesviruses (48), baculovirus (50), and phage SPP1 (42), and is absolutely dependent upon the production of genomic concatemers. As genome concatemerization is essential for viral growth, an alternate pathway to concatemerization is provided by homologous genetic recombination (35). Recombination is promoted by dsDNA ends created by cleavage of the packaging site by the packaging endonuclease (reference 36 and references therein) and at the tips of rolling circle replication intermediates (38). The resulting recombinant product can contain more than one genome equivalent of DNA and at least two packaging sites and can therefore be packaged (11).
In phage λ, two enzymes, λ exonuclease (λ Exo) and beta protein, catalyze homologous recombination. λ Exo is an alkaline exonuclease, and beta protein is a synaptase (a protein that promotes annealing of single-stranded DNA [ssDNA] chains and the pairing of ssDNA with homologous dsDNA in concert with the E. coli RecA protein). λ Exo and beta protein form a specific 1:1 complex (6) and work together as a two-component viral recombinase (29). A search of the nonredundant GenBank database using the PSI-BLAST server (2) with λ Exo as the query sequence revealed over 80 other λ Exo-like proteins from linear dsDNA viruses, with the similarity being limited to five conserved amino acid motifs (3, 30; R. S. Myers and K. E. Rudd, unpublished data). The group of proteins has been variously named the Redα superfamily (30) and the LE family (3). For many of the known or putative exonuclease genes, a nearby gene for a known or putative synaptase (such as λ beta protein, P22 Erf, Rac RecT, or HSV1 UL29) has been identified (19; Myers and Rudd, unpublished). Unlike viral nucleases, which all resemble λ Exo (except RecE, which more closely resembles E. coli RecB [8]), the synaptases fall into several families (19; Myers and Rudd, unpublished). Whereas synaptase sequences are diverse, their structures are very similar, and they assemble into nucleoprotein filaments with ssDNA (e.g., see reference 32).
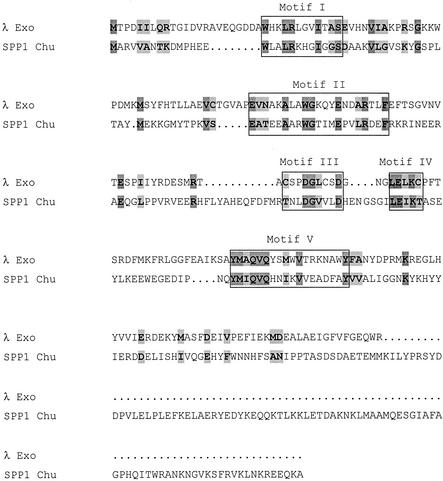
Two-component recombinase pairs include Exo/beta protein from coliphage λ, RecE/T from the Rac prophage of E. coli, UL12/UL29 from human herpesvirus type 1, YqaJ/K from the skin prophage of Bacillus subtilis, open reading frame (ORF) 7(DUF)/BBR29 from a putative prophage in Borrelia burgdorferi, and G34.1P/G35P from phage SPP1. An alignment between the amino acid sequences of SPP1 G34.1P and λ Exo is shown in Fig. 1, with the five Redα superfamily motifs highlighted. SPP1 G35P is very similar to the RecT synaptase from the Rac cryptic prophage of E. coli (1 [and references therein]; Myers and Rudd, unpublished). Moreover, G35P is able to catalyze strand exchange (4). From the sequence alignments, its appears that a putative SPP1 recombinase bridges the λ and Rac recombination systems.
FIG. 1.
Alignment of λ Exo and SPP1 Chu. Motifs shared with the Redα superfamily are boxed (3, 30). Identical and similar amino acids have dark and light shading, respectively. The first aspartate (D) in motif III and the glutamate (E) and lysine (K) in motif IV are found in type II restriction endonucleases and in E. coli RecB (3, 13, 21).
Genes encoding interacting proteins often colocalize in viral genomes. The recT gene is adjacent to and overlaps the recE gene, which specifies a recombination exonuclease. RecE and RecT form a complex and catalyze recombination by a mechanism quite similar to that of the Red system of phage λ. RecE/RecT proteins can substitute for λ Exo and beta protein even though the sequences are completely dissimilar (20). Many of the mechanistic details of the Red system and the RecET system are conserved, including the capacity to support some types of recombination in the absence of the host RecA synaptase (12, 22, 23, 31, 36, 45, 51). The herpesvirus UL12 protein is a known alkaline exonuclease that forms a complex with a synaptase, UL29 (44), and catalyzes recombination in vitro (N. Reuven, A. Staire, R. S. Myers, and S. Weller, submitted for publication). Taken together, the data support our proposal that viruses commonly employ a two-component system for recombination.
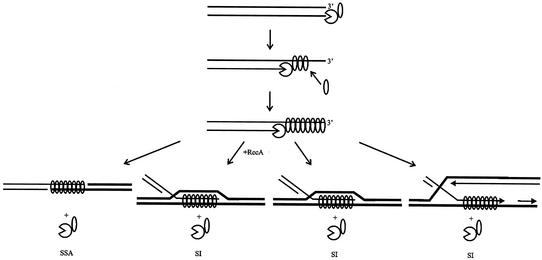
The general model for the mechanism of recombination by viral two-component recombination modules consisting of an exonuclease and a synaptase (which we term “SynExo”) is shown in Fig. 2 and is described below. Recombination is initiated by dsDNA breaks (41, 43), and we propose that this is so in all viruses within the family. The alkaline exonuclease component of the viral recombinase prepares broken dsDNA ends for recombination by 5′ → 3′ resection of one DNA strand. The resulting ssDNA has a 3′ tip. We propose that the synaptase component forms a complex with the alkaline exonuclease and is coordinately loaded onto the nascent ssDNA, thereby coupling DNA digestion to production of a recombinogenic nucleoprotein filament, the synaptasome. The synaptasome structure is conserved even when little sequence homology is evident in comparisons of synaptases (32). We propose that the synaptasome mediates the search for homology that initiates recombination and regulates the processivity of the alkaline exonuclease component (evidence for λ Exo in reference 17 and for herpesvirus UL12 in Reuven et al., submitted). Physical coupling between the nuclease and synaptase components appears to be essential for recombination, even though each component retains its enzymatic activity in the absence of the other (5, 17). For example, homospecific two-component viral recombinase complexes support homologous recombination in E. coli (e.g., RecE/T and λ Exo/beta protein), but heterospecific combinations of the exonuclease and the synaptase (e.g., λ Exo/RecT and RecE/beta protein) do not (29). Since the synaptase component is required in stoichiometric amounts that change in proportion with the number of ssDNA nucleotides exposed and the nuclease that exposes the ssDNA acts catalytically (and processively), the complex of the nuclease to synaptase (1:1 in λ Exo/beta protein [28]) is proposed to represent an intermediate complex in a dynamic process of synaptase assembly on the nascent ssDNA as it is produced by the nuclease. This coordinated assembly and subsequent homologous pairing with a target DNA is proposed to provide synaptic feedback to regulate the exonuclease function. Synaptic feedback could occur via any one of four possible pathways (Fig. 2): single-strand annealing mediated by the viral SynExo, as is proposed to occur in λ (39); strand invasion mediated by the viral SynExo aided by RecA, as is proposed to occur in λ (39); strand invasion mediated by the viral SynExo independent of RecA, as is proposed to occur in the RecET system (31); and strand exchange mediated by the viral synaptase, as is proposed to occur through ssDNA annealing to the lagging-strand template in the replication fork by λ beta protein (10). We are currently dissecting the feedback mechanism, which appears to be a central feature of recombination in general and not specific to the viral recombination mechanism.
FIG. 2.
Model for recombination mediated by the viral SynExo. Linear dsDNA, such as that produced during rolling circle replication by the virus or by the action of the packaging endonuclease on the packaging site, serves as the substrate. The exonuclease is shown resecting the 5′ end while the synaptase binds the resulting 3′-terminated ssDNA. The synaptasome mediates the search for homology promoting ssDNA annealing (SSA) with complementary ssDNA and strand invasion (SI) with homologous dsDNA. SI occurs with and without assistance from the host RecA protein (12, 39, 43). SI is proposed to occur by the synaptase alone in regions of ssDNA (produced, for example, during replication) (10).
We have proposed that the SynExo recombination module is widely distributed throughout linear dsDNA viruses with host ranges in all kingdoms (Myers and Rudd, unpublished). Accordingly, the Redα superfamily would define a group of recombination exonucleases. To test this hypothesis, we compared the properties of the phage SPP1 34.1 gene product to λ Exo. λ Exo is an oligomeric, highly processive, 5′ → 3′ exonuclease with limited endonuclease activity (24, 25, 33). It has an alkaline pH optimum and requires a divalent cation (24, 33). It prefers linear dsDNA with phosphorylated termini (27). We predicted that the SPP1 34.1 gene product shares these properties and tested this prediction by biochemical characterization of the protein.
As predicted, the 34.1 gene product, which we named Chu (after its activity; see below), is oligomeric and digests linear dsDNA in a processive manner. Chu has limited activity on circular ssDNA and no activity on either circular dsDNA or linear ssDNA. It has a pH optimum of 9.0 and requires a divalent cation, which can be provided only by Mg2+. And, like λ Exo, Chu prefers dsDNA with phosphorylated 5′ termini to dsDNA with hydroxyl 5′ termini.
MATERIALS AND METHODS
Strains and media.
All cloning and gene expression used E. coli K-12 derivatives. Bacteria were grown in Luria-Bertani medium (34) for most purposes and SOB/SOC media (14) for transformation. When necessary, growth media were supplemented with antibiotics (50 μg of kanamycin per ml, 100 μg of ampicillin per ml, and 20 μg of chloramphenicol per ml) or isopropyl-1-thio-β-d-galactopyranoside (IPTG) (1 mM).
Chemicals.
All chemicals were reagent grade or better and were purchased from Sigma, Kodak, J. T. Baker, Aldrich, and Mallinckrodt. Divalent-cation concentrations were verified by osmometry using the freezing point depression method (Advanced Instruments). Gel purifications were performed with an UltraClean 15 DNA purification kit (MoBio Laboratories, Inc.). Enzymes used in the cloning were from New England Biolabs.
Cloning.
The 34.1 gene was amplified from bacteriophage SPP1 genomic DNA (generously provided by J. Alonso) with the primers 34.1-Nde (5′-GGAATTCCATATGG*CGAGGGTAGTGGCAAAC-3′) and 34.1-Bam (5′-CGGGATCCT#CATGCCTTTTGCTCCGCCTTTTGCTCCTCCCTCT-3′) corresponding to positions 30532 (*) and 31464 (#) in the SPP1 genome (accession number X97918). The NdeI and BamHI sites are underlined. The 953-bp 34.1 PCR product was resistant to cleavage by NdeI and BamHI (data not shown), perhaps because the engineered restriction sites were not in the correct context for efficient restriction. Therefore, it was blunt-end cloned into pTZ18u (accession number L37352) with SmaI to produce plasmid pTZ18::34.1. Epicurian coli XL10-Gold ultracompetent cells (Stratagene) were transformed to ampicillin resistance, and transformants were identified by colony PCR. The sequence of 34.1 in pTZ18::34.1 is identical to that of 34.1 in the SPP1 complete genome sequence X97918 (CDS 30529-31464) but differs from that of X67865 (CDS CAA48050.1) at the codon corresponding to amino acid 137, which is a serine instead of a tyrosine.
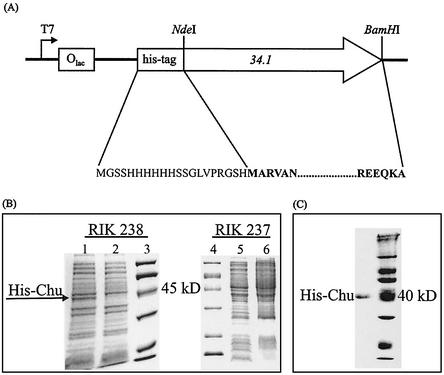
The 34.1 gene was subcloned using standard methods from pTZ18::34.1 into the NdeI and BamHI restriction sites of a derivative of pET28a(+) (Novagen), to produce plasmid pET::34.1 (Fig. 3A). pET::34.1 was used to transform BL21(DE3) (pLysS) cells (40). Transformants were identified by colony PCR.
FIG. 3.
(A) Partial map of plasmid pET::34.1 containing the 34.1 gene, showing the direction of transcription and the locations of the T7 promoter, the His tag, and the NdeI and BamHI sites used for cloning. The amino acid sequence of His-tagged Chu is shown with the Chu sequence in bold. (B) Induction of SPP1 Chu expression. SDS-PAGE of whole-cell lysates of BL21-CodonPlus (DE3)-RP [pET::34.1] cells (RIK 238) 3 and 0 h after induction with IPTG (lanes 1 and 2, respectively) shows the appearance of the expected 38-kDa band. Expression of Chu in BL21(DE3) pLysS [pET::34.1] cells (RIK 237) was not detected. Lanes 5 and 6 show whole-cell lysates 0 and 3 h, respectively, after induction with IPTG. (C) SDS-polyacrylamide gel of pure His-tagged Chu.
Purification of insoluble Chu.
BL21(DE3)(pLysS)(pET::34.1) cells (RIK 237) were grown at 37°C to an optical density at 600 nm (OD600) of 0.55 and induced with 1 mM IPTG. Cells were harvested after an additional 3 h of growth and stored at −80°C. A cell pellet (7.5 g) was sonicated in 10 ml of 20 mM Tris-HCl-1 mM EDTA. Inclusion bodies were collected from the cell lysate by centrifugation at 17,000 × g for 20 min and washed by resuspension in 20 ml of 20 mM Tris-HCl (pH 8.0)-100 mM NaCl-0.5% Triton X-100. Inclusion bodies were again collected by centrifugation at 17,000 × g for 20 min and dissolved for 2 h at 37°C in 20 ml of 8 M urea-20 mM Tris base. Debris was pelleted by centrifugation at 17,000 × g for 10 min. Solubilized inclusion bodies were put over a Macro-Prep High Q strong anion-exchange support column (Bio-Rad) (6 ml) that had been treated with 15 volumes of 100 mM Tris-HCl followed by 40 volumes of 20 mM Tris-HCl and then by 2 volumes of 8 M urea-20 mM Tris-HCl. The column was washed with 13 volumes of 8 M urea-20 mM Tris-HCl and then eluted with a salt gradient from 20 mM NaCl in 8 M urea-20 mM Tris-HCl to 0.5 M NaCl in 8 M urea-20 mM Tris-HCl. Fractions containing protein, as indicated by the OD280, were pooled and then concentrated via ultrafiltration with a Centricon (Amicon) concentrator with a molecular weight cutoff of 10,000. The presence of protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was applied to nickel-nitrilotriacetic acid (Ni-NTA) spin columns (Qiagen) equilibrated with 0.5 M NaCl-8 M urea-20 mM Tris-HCl for further purification. The column was washed three times with 0.6 ml of 20 mM potassium phosphate (pH 6.0)-8 M urea-0.5 M NaCl and twice with 0.6 ml of 20 mM potassium phosphate (pH 5.3)-8 M urea-0.5 M NaCl. His-tagged protein was eluted with 20 mM potassium phosphate (pH 4.0)-8 M urea-0.5 M NaCl.
Pure protein was refolded by gradual removal of the urea by dialysis. The protein was diluted in day 1 refolding buffer (300 mM K2HPO4 [pH 7.0], 0.1 mM K2EDTA, 100 mM KCl, 20% glycerol, 6 M urea) to less than 1 mg/ml. It was then dialyzed at 4°C overnight, against 250 volumes of day 1 refolding buffer. It was dialyzed over five nights in the same buffer with less urea each night as follows: 4 M urea on day 2, 2 M urea on day 3, 1 M urea on day 4, and no urea on day 5. The preparation was stored at −80°C in 300 mM K2HPO4 (pH 7.0)-0.1 mM K2EDTA-100 mM KCl-20% glycerol at a protein concentration of 9.7 μM monomers, as estimated by OD280.
Purification of soluble Chu.
The level of Chu protein expression in BL21(DE3) pLysS [pET::34.1] (RIK 237) was not detectable by SDS-PAGE of whole-cell lysates (Fig. 3B). To increase the yield, we transformed BL21-CodonPlus-RP (Stratagene) which encodes tRNAs for rare arginine codons (six AGG codons and three AGA codons) found in the 34.1 gene. This strain produced detectable levels of soluble protein. BL21-CodonPlus(DE3)-RP(pET::34.1) cells (RIK 238) were grown at 30°C to an OD600 of 0.5 to 1.35 and induced with 1 mM IPTG (Fig. 3B). Cells were harvested after an additional 3 h of growth at 30°C and stored at −80°C. The cell pellet was lysed in 5 volumes of a solution containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, and 1 mg of lysozyme (Amersham)/ml and incubated on ice until the solution appeared viscous. The lysate was centrifuged for 30 min at 17,000 × g or 60 min at 20, 000 × g, and the supernatant fluid was centrifuged again for 10 min at 17,000 × g or 30 min at 20, 000 × g. Approximately the same volume as the cell pellet of a 50% slurry of Ni-NTA resin (Qiagen) was added to the supernatant fluid. His-tagged protein was allowed to bind to Ni-NTA resin at 4°C by rotating the sample for 1 h or inverting several times every 15 min. The resin was then either washed four times with an equal volume of 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-20 mM imidazole, once with 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-40 mM imidazole, once with 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-60 mM imidazole, and once with 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-80 mM imidazole or washed twice with 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-20 mM imidazole and twice with 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-40 mM imidazole. His-tagged protein was eluted with 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-250 mM imidazole. The preparation was stored on ice at 4°C at a protein concentration of 1.0 μM monomers, as estimated by SDS-PAGE.
Gel filtration.
The molecular mass of native Chu enzyme was determined by fast-performance liquid chromatography (FPLC) gel filtration on a Superose 6 HR 10/30 column (Pharmacia) calibrated with catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), and RNase A (13.7 kDa) (Pharmacia). Samples were passed through the column in 50 mM K2HPO4 buffer, pH 7.0, with 0.1 mM K2EDTA, 100 mM NaCl, and 5% glycerol at a constant flow rate of 0.2 ml/min. Fractionation was monitored at 280, 230, and 214 nm.
Enzyme assays.
Standard reaction conditions for nuclease assays were as follows: 50 mM CHES (2-[N-cyclohexylamino]ethanesulfonic acid; pH 9.0) (except when pH was tested), 50 mM KCl, 5 mM magnesium acetate (except when Mg2+ concentration was tested), 5% glycerol, and 1 mM dithiothreitol at 37°C. The reaction volume was 50 μl. The reaction time was 30 min except in the titrations used to determine the fractional activity of Chu, in which it was 3 or 10 min, and in time courses. Buffers were MES (pH 6.0), HEPES (pH 7.0, 7.5, and 8.0), Tris-HCl (pH 8.5), and CHES (pH 9.0, 9.2, 9.4, 9.6, 9.8, and 10.0).
Linear dsDNA was prepared by alkaline lysis miniprep (26) of pUC19 and complete digestion with PstI, followed by purification with either phenol-chloroform, ether, and two ethanol precipitations or an UltraClean PCR clean-up spin column (MoBio Tech). DNA was resuspended in Tris-HCl, pH 8.0, with or without 0.1 mM EDTA. It was quantified by measuring the absorbance at 260 nm. Circular dsDNA was prepared by alkaline lysis miniprep of pUC19. Preparations were treated with RecBCD (generously provided by M. Jockovich) to digest any linear DNA species, gel purified, and quantified by measuring absorbance at 260 nm. RecBCD digests were incubated at 37°C for 30 min and contained 10 mM magnesium acetate, 1 mM dithiothreitol, 200 μM ATP, 0.2 mg of bovine serum albumin per ml, and 3.5 nM RecBCD. Circular ssDNA (M13) was generously provided by A. Nimonkar. Dephosphorylated pUC19 DNA was prepared by treatment of PstI-cut pUC19 with calf intestinal phosphatase (New England Biolabs) per the manufacturer's instructions. Dephosphorylated DNA was purified by UltraClean PCR clean-up spin column, checked for 100% dephosphorylation by ligation with Quick T4 DNA ligase (New England Biolabs), and quantified by measuring the absorbance at 260 nm. Control DNA for the experiment to determine the preference of Chu for particular DNA ends was also passed through an UltraClean PCR clean-up spin column.
Linear dsDNA was labeled with the fluorescent dye PicoGreen (Molecular Probes) at a concentration of 1.6× or 229× (product is supplied at a 2000× strength). The dye fluoresces with a higher intensity when bound to dsDNA than ssDNA or nucleotides. Reaction rates were measured by a decrease in fluorescence upon DNA digestion. A linear relationship exists between fluorescence and moles of dsDNA (Molecular Probes). Reactions were stopped with EDTA (13 to 20 mM), and assay mixtures were incubated with PicoGreen in the dark for no less than 3 min. DNA was quantified by excitation at 484 nm and scanning the emission between 510 and 545 nm in a spectrofluorometer (QuantumMaster; Photon Technology International). The final reaction conditions were estimated by measuring the difference in fluorescence between 1.15 nmol of dsDNA (starting conditions) and 0.575 nmol of ssDNA (boiled and quickly chilled on ice) as an approximation of final reaction conditions. Fluorescence contribution from nucleotides is negligible (K. Subramanian, personal communication). Light output was measured by integrating the area under the fluorescence output curve from 510 to 545 nm, converted to moles of nucleotides digested.
The substrate specificity tests were evaluated by agarose gel electrophoresis. Reactions were performed with standard conditions. Samples were mixed with loading buffer and electrophoresed through 1 or 2% agarose in Tris-acetate EDTA buffer at 2 to 3.5 V/cm. Gels were stained with ethidium bromide, and images were recorded with a charge-coupled device camera and a UV transilluminator by using the AlphaImager 2000 (Alpha Innotech Corp.).
Sequence alignments.
Primary sequence alignments were performed using ClustalW (46) with minor adjustments made by hand.
RESULTS
Cloning and expression of 34.1 of SPP1.
Based on the high degree of similarity of G34.1P of phage SPP1 from B. subtilis to the alkaline exonuclease λ Exo of phage λ (Fig. 1) and the proximity of the 34.1 gene to a synaptase gene, we predicted that G34.1P is a processive alkaline exonuclease that requires a divalent cation and prefers phosphorylated 5′ termini. To test this hypothesis, we determined the biochemical characteristics of the recombinant G34.1P protein purified as a polyhistidine fusion protein from E. coli. The gene was amplified from genomic DNA by PCR and cloned into a derivative of expression vector pET28a(+) to form plasmid pET::34.1 (Fig. 3A). The nucleotide sequence of the cloned gene corresponds to ORF 34.1 in the complete genome sequence of SPP1 in GenBank (X97918). Induction of expression of 34.1 in transformed E. coli produced a band at ∼40 kDa corresponding to the expected mass of 38,107 Da, which was not present in uninduced cells (Fig. 3B). We have named this protein Chu after its activity, as it “chews” DNA (see below).
Purification of Chu.
We initially expressed Chu in a standard lab strain, and it displayed so little induction as to be undetectable by inspection of Coomassie brilliant blue (CBB)-stained gels. Furthermore, the Chu protein so produced was found entirely as insoluble protein aggregates. We developed two purification schemes to isolate apparently homogeneous (>95%, as determined by SDS-PAGE and CBB staining) Chu enzyme (Fig. 3C). The first approach provided most of the material used in the characterization of Chu activity. Washed protein aggregates were dissolved in urea and then subjected to anion exchange and immobilized nickel affinity column chromatography to purify denatured Chu protein. This preparation was refolded by gradual dilution of denaturant, resulting in recovery of exonuclease activity.
To determine if refolded Chu retained the activity of the native enzyme, we purified Chu under nondenaturing conditions. When overexpressed in E. coli, Chu represents 5 to 6% of the total cell protein. We managed to obtain a small amount of native and soluble Chu by centrifugation to remove the insoluble protein, followed by a single step of immobilized nickel chromatography. The purification procedure resulted in 19% recovery of the total Chu protein (by mass) as determined by scanning the image of a CBB-stained SDS-polyacrylamide gel. The protein obtained was more than 95% pure, as assessed by CBB staining of an SDS-polyacrylamide gel.
The oligomeric state of Chu was determined by FPLC gel filtration. The native enzyme is an oligomer that eluted with an apparent molecular mass of 146 to 212 kDa. This observation is consistent with that made for λ Exo, which is known to be a trimer (21, 47) but elutes as a pentamer or hexamer under our conditions (K. Subramanian and R. S. Myers, unpublished data), possibly due to its toroidal structure (21). For the purposes of calculating percent active enzyme, we assumed Chu to be a trimer, since the structure of λ Exo determined by X-ray crystallography is trimeric (21). For all enzyme assays, Chu was titrated to determine the concentration of active enzyme.
Activity of the Chu preparation.
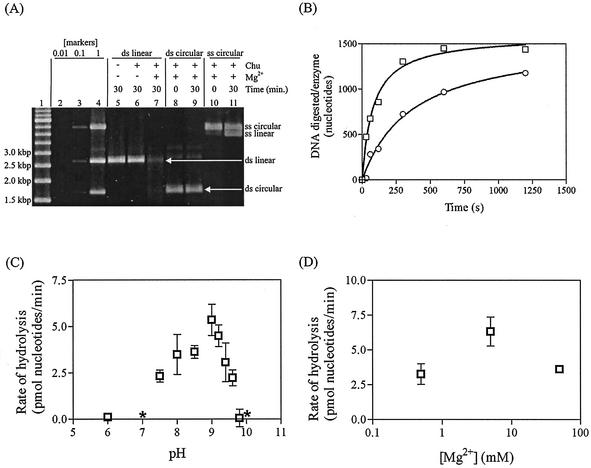
We established that Chu is a dsDNA exonuclease using a quantitative fluorescence-based assay that we developed. Figure 4A shows raw data obtained from a typical nuclease assay in which the progression of the reaction is monitored as dsDNA is converted to ssDNA. The fluorescence contribution from nucleotides is negligible (Subramanian, personal communication). A linear relationship exists between fluorescence and moles of dsDNA (Molecular Probes), so for each enzyme reaction in this study, the fluorescence contribution from ssDNA was subtracted from the signal, and remaining fluorescence was converted to moles of dsDNA. Percent digestion was then calculated, using full-length dsDNA as a point of reference.
FIG. 4.
SPP1 Chu digests linear dsDNA. PstI-cut pUC19 was digested under standard reaction conditions, fluorescently labeled, and quantified by excitation at 484 nm and scanning of the emission between 510 and 545 nm in a spectrofluorometer (QuantumMaster; see Materials and Methods). (A) Raw data obtained from the spectrofluorometer. Because the dye fluoresces with a higher intensity when bound to dsDNA than ssDNA or nucleotides, a decrease in fluorescence reflects digestion of DNA. Titrations were performed with various concentrations of refolded Chu against 2.3 nM DNA ends. Reaction time was 30 min. The dsDNA curve represents the fluorescence output from a reaction in which a quantity of EDTA sufficient to quench the reaction was added prior to the addition of enzyme. Curves 1 through 8 contained 20, 40, 60, 80, 120, 240, 160, and 200 nM Chu, respectively. The ssDNA curve represents data from a reaction mixture that contained 0.5 equivalent of denatured input DNA to imitate limit digest conditions. DNA products are illustrated to the right of the chart, with Chu represented as a circle with a slice missing. (B and C) Time course of DNA digestion by native Chu under saturation conditions initiated with enzyme and Mg2+, respectively. Samples were withdrawn and quenched with EDTA at various times. DNA and Chu concentrations were 2.3 nM DNA ends and at least 2.3 nM Chu (as determined empirically by titration). (D and E) Titrations of native and refolded Chu, respectively, against 2.3 nM DNA ends. Reaction time was 10 min.
A time course experiment in which reactions were initiated with native enzyme (Fig. 4B) revealed that the rate has two linear components, a low initial rate followed by a higher persistent rate. The high rate (3 nucleotides per s) was similar to the rate obtained (2 nucleotides per s) when enzyme and DNA were premixed and reactions were initiated with Mg2+ (Fig. 4C).
The percent active enzyme was calculated from titrations of enzyme against constant linear dsDNA concentration, in which we presumed that saturation of DNA digestion rate was achieved at a 1:1 stoichiometry of active Chu to DNA ends (Fig. 4D and E). The concentration of Chu at which saturation was reached suggested that the native preparation was 100% active and the refolded preparation was 10% active, assuming that the quaternary structure of the enzyme is trimeric. Assuming a trimeric structure seems reasonable in light of the data from FPLC suggesting that Chu is an oligomer (see above) and the high degree of similarity in sequence (3, 30; Myers and Rudd, unpublished) and biochemical properties (this study) to λ Exo, which appears to be trimeric by gel filtration (47) and X-ray crystallography (21). One unit of activity is defined as the amount of enzyme required to release 1 μmol of nucleotides in 1 min at 37°C under the standard reaction conditions described in Materials and Methods. Native and refolded Chu had exonuclease activities of 5.4 and 0.39 U/mg, respectively.
Substrate specificity of Chu.
We observed dsDNA exonuclease activity of Chu by using a gel-based assay (Fig. 5A, lanes 5 and 7). Linear dsDNA was digested in the presence but not in the absence of Chu. The nuclease activity requires a divalent cation: DNA was digested in the presence but not in the absence of Mg2+ (Fig. 5A, lanes 6 and 7). Chu has no dsDNA endonuclease activity, as circular dsDNA was not cut by Chu in 30 min (Fig. 5A, lanes 8 and 9). Chu has weak ssDNA endonuclease activity, since circular ssDNA was converted to linear ssDNA of a single size (Fig. 5A, lanes 10 and 11). One unit of activity is defined as the amount of enzyme required to convert 1 μmol of DNA molecules from circular to linear form in 1 min at 37°C under the standard reaction conditions described in Materials and Methods. Refolded Chu had an endonuclease activity of 4.3 × 10−4 U/mg. Chu has no ssDNA exonuclease activity detectable in 30 min under standard conditions—the linear ssDNA in Fig. 5A, lane 11, was not digested further, as determined by densitometry.
FIG. 5.
SPP1 Chu enzyme characterization. (A) DNA (2.3 nM ends or 1.15 nM molecules) was digested with refolded Chu (at least 2.3 nM, as determined by titration) for 30 min at 37°C under standard reaction conditions (see Materials and Methods). The gel was scanned to determine DNA quantities with an AlphaImager 2000. Lanes 1 through 4, molecular weight markers. Lane 1, 500-bp ladder; lanes 2, 3, and 4, combination of the substrates used in the assays, i.e., linear dsDNA, circular dsDNA, and circular ssDNA, at initial concentrations of 0.01×, 0.1×, and 1×, respectively. Lane 5, control without Mg2+ or Chu. (B) Phosphorylated (squares) or dephosphorylated (circles) PstI-cut pUC19 (0.151 nM ends) was incubated with Chu (at least 0.151 nM, as determined by titration) for 2 min at 37°C, and reactions were then initiated with 5 mM Mg2+. DNA was digested under standard reaction conditions for 0, 0.5, 1, 2, 5, or 10 min. The 0-min sample was quenched with 20 mM EDTA prior to the addition of Mg2+. (C) PstI-cut pUC19 (2.3 nM ends) was digested with refolded Chu (at least 2.3 nM, as determined by titration) for 30 min at 37°C under standard conditions, except that pH was varied. (D) PstI-cut pUC19 (2.3 nM ends) was digested with refolded Chu (at least 2.3 nM, as determined by titration) for 30 min at 37°C under standard conditions, except that Mg2+ concentration was varied. *, the mean value was below zero, but the rate fell within the range of 0.
Chu prefers dsDNA with phosphorylated 5′ termini to dsDNA with hydroxyl 5′ termini (Fig. 5B). The difference in rates is 26-fold in the first 30 s and 2.4-fold in the first minute (Fig. 5B). We propose that the rates of digestion are equivalent once the first turnover has occurred and that the rate on dephosphorylated DNA appears to be lower because of the asynchronous release of stalled enzyme-substrate complexes into the active form.
Optimum pH.
We determined that the exonuclease activity of Chu, like that of λ Exo and UL12 of HSV1, has an alkaline pH optimum. Activity was optimum at pH 9.0 (Fig. 5C). λ Exo has a pH optimum of about 9.2 to 9.5 (24, 33). At the estimated cytoplasmic pH of B. subtilis, approximately 7.5 (18), the activity in our assay system is 43% of the maximum.
Effect of divalent cations.
The exonuclease activity of Chu was absolutely dependent on the presence of Mg2+ (Fig. 5A, lanes 6 and 7) with an optimum concentration in the range of 5 mM (Fig. 5D). A number of other divalent cations were tested (Ca2+, Co2+, Cu2+, Mn2+, Ni2+, and Zn2+), but no enzyme-dependent DNA degradation was observed above the enzyme-independent background at concentrations between 0.5 and 50 mM (data not shown). λ Exo also requires a divalent cation for activity (24).
Processivity of Chu.
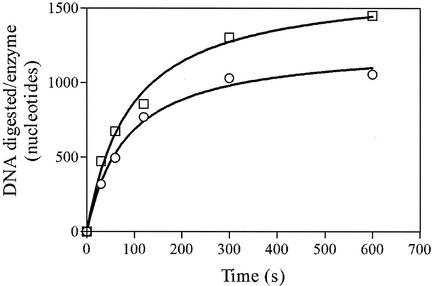
The processivity of Chu was determined (Fig. 6). The average amount of digestion by each enzyme-substrate complex before dissociation was measured using heparin to trap excess enzyme. Assays were conducted under conditions in which DNA ends are saturated with enzyme (as determined by titration). Controls (not shown) were included in which the enzyme was added after the heparin to ensure that the concentration of heparin added was adequate to trap all of the enzyme. If the enzyme acts distributively, DNA digestion will quickly reach a limit. If the enzyme acts processively, an amount of DNA equivalent to the average processivity of the enzyme will be digested before the limit is reached. As seen in Fig. 6, an average of 1,000 nucleotides were digested before the limit was reached, indicating that Chu is a processive enzyme with an average processivity of 1,000 bp.
FIG. 6.
Chu is a processive enzyme. Refolded Chu (0.151 nM, as determined by titration) was incubated with PstI-cut pUC19 DNA (0.151 nM ends) for 2 min at 37°C, and reactions were then initiated with 5 mM Mg2+. Sufficient heparin to trap all of the enzyme was added after 10 s (open circles). DNA was digested under standard reaction conditions (see Materials and Methods) for 0, 0.5, 1, 2, 5, or 10 min. Reactions were quenched with 20 mM EDTA. The 0-min sample was quenched with EDTA prior to the addition of Mg2+. Heparin was added after quenching in the controls (squares). The digestion of 1,000 bp by a single turnover of Chu indicates that the enzyme is processive.
The enzymatic properties of Chu can be summarized as follows. It is a dsDNA exonuclease and a weak ssDNA endonuclease. It has no ssDNA exonuclease or dsDNA endonuclease activities. It prefers a pH of 9.0, phosphorylated 5′ DNA ends, and Mg2+ as a divalent cation.
DISCUSSION
Viruses with linear dsDNA genomes infect organisms from every major branch of the tree of life. Based on the ubiquity of two-component viral recombinases (which we term the SynExo family of two-component recombinases) apparent from bioinformatic analysis of these viral genomes, we propose that homologous recombination plays an important role in viral growth and development. Interestingly, these recombinases are restricted to viral sequences; the cellular genomes that encode family members appear to have integrated them along with other viral sequences (e.g., the Rac prophage-encoded RecE/T system in E. coli, the YqaJ/K system encoded by the B. subtilis skin element, and ORF 7 (DUF)/BBR29 from a putative prophage comprising a small plasmid of B. burgdorferi [19]). Since these recombinases are specific to viruses (including pathogens) and since diverse viruses require recombination nuclease function for optimal growth (including λ [37], human herpesvirus type 1 [13], and Paramecium bursaria chlorella virus 1 [J. Van Etten, personal communication]), viral recombination nucleases may be a significant target for new antiviral drugs to be used in concert with more traditional DNA synthesis inhibitors.
One way to determine structural features of enzymes that contribute to a conserved mechanism is to let nature do the mutagenesis experiment. By identifying highly conserved features of homologous but distantly related protein sequences, it may be possible to identify potent targets for antiviral drugs and generate insight into mechanistic details. Therefore, in order to challenge the Redα superfamily model and to extend our understanding of the conserved features of the viral recombination mechanism, we identified a family member with a homologous ORF of unknown function, isolated the gene, expressed it in a heterologous system (E. coli), and showed that the gene encodes a bona fide alkaline exonuclease with properties similar to those of viral nucleases from distantly related taxa.
We report here the characterization of Chu, an enzyme from phage SPP1 of B. subtilis belonging to the Redα superfamily of recombination exonucleases. Based on similarity to λ Exo and proximity of the gene to the synaptase gene, 35 (1; Myers and Rudd, unpublished), we predicted that Chu is a processive oligomeric alkaline exonuclease requiring a divalent cation and preferring phosphorylated 5′ termini. The prediction was borne out. Chu digests linear dsDNA processively, has limited activity on circular ssDNA, and demonstrates a preference for 5′-phosphoryl over 5′-hydroxyl DNA termini. Chu exists as an oligomer in solution and has an alkaline pH optimum and an absolute requirement for a divalent cation, which can be supplied only by Mg2+. Based on these properties, the ssDNA binding activity of G35P (4), and the reduction of linear genomic concatemer formation in temperature-sensitive 35 mutants (49), we predict that Bacillus phage SPP1 employs a recombination mechanism similar to that of coliphage λ and herpesvirus (Reuven et al., submitted).
The SynExo functional unit may represent a recurrent theme in linear dsDNA viruses. It may have evolved as a portable module (15) that can function in diverse hosts without requiring extensive interaction with host-specific functions. This general strategy of encoding a simple portable recombination module may confer a great adaptive advantage on viruses exploring new niches. This same property may be exploited by genetic technologists to promote in vivo genetic engineering (“recombineering” [9]) in genetically recalcitrant organisms such as humans, certain model organisms, and organisms of agricultural importance, plants and animals alike. If some connectivity remains between viral recombinase modules and host proteins, one may wish to employ recombinase modules from viruses that coevolved with the hosts. Examples of this might include using herpesvirus UL12/29 recombinase to engineer mammals, avian species, fish, and reptiles, phage recombinases to engineer bacteria (SPP1 Chu/G35P for gram-positive bacteria, λ Exo/beta protein for gram negative-bacteria), and algal virus (PBCV1) or Arabidopsis (unknown viral species) recombinases to engineer plants.
Acknowledgments
This work was supported by grants from the American Cancer Society and the Florida Biomedical Research Program to R.S.M.
We are grateful to Juan Alonso for SPP1 genomic DNA, Amitabh Nimonkar and Paul Boehmer for M13 DNA, Maria-Elena Jockovich for RecBCD enzyme, Arun Malhotra for the use of his Phastgel and FPLC systems, T. K. Harris for the use of his FPLC system, Jim Van Etten for access to his unpublished observations, Sandy Weller for helping spread the work into areas outside our ken, Keith Brew for the refolding protocol, T. J. Ragan for technical assistance with FPLC, and Krithika Subramanian and all the members of the Myers lab for engaging discourse and critical review of the manuscript.
REFERENCES
- 1.Alonso, J. C., G. Lüder, A. C. Stiege, S. Chai, F. Weise, and T. A. Trautner. 1997. The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204:201-212. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayora, S., R. Missich, P. Mesa, R. Lurz, S. Yang, E. H. Egelman, and J. C. Alonso. 2002. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 277:35969-35979. [DOI] [PubMed] [Google Scholar]
- 5.Berger, I., and A. Cohen. 1989. Suppression of recA deficiency in plasmid recombination by bacteriophage lambda beta protein in RecBCD− ExoI− Escherichia coli cells. J. Bacteriol. 171:3523-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, D. M., and C. M. Radding. 1971. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J. Biol. Chem. 246:2502-2512. [PubMed] [Google Scholar]
- 7.Catalano, C. E., D. Cue, and M. Feiss. 1995. Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol. 16:1075-1086. [DOI] [PubMed] [Google Scholar]
- 8.Chang, H. W., and D. A. Julin. 2001. Structure and function of the Escherichia coli RecE protein, a member of the RecB nuclease domain family. J. Biol. Chem. 276:46004-46010. [DOI] [PubMed] [Google Scholar]
- 9.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769-779. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, H. M., D. Yu, T. DiTizio, and D. L. Court. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98:6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feiss, M., and D. A. Siegele. 1979. Packaging of the bacteriophage lambda chromosome: dependence of cos cleavage on chromosome length. Virology 92:190-200. [DOI] [PubMed] [Google Scholar]
- 12.Gillen, J. R., D. K. Willis, and A. J. Clark. 1981. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J. Bacteriol. 145:521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein, J. N., and S. K. Weller. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442-457. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 15.Hartwell, L. H., J. J. Hopfield, S. Leibler, and A. W. Murray. 1999. From molecular to modular cell biology. Nature 402(Suppl.):C47-C52. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix, R. W., J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.). 1983. Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Hill, S. A., M. M. Stahl, and F. W. Stahl. 1997. Single-strand DNA intermediates in phage lambda's Red recombination pathway. Proc. Natl. Acad. Sci. USA 94:2951-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer, L. M., E. V. Koonin, and L. Aravind. 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser, K., and N. E. Murray. 1979. Physical characterisation of the “Rac prophage” in E. coli K12. Mol. Gen. Genet. 175:159-174. [DOI] [PubMed] [Google Scholar]
- 21.Kovall, R., and B. W. Mathews. 1997. Toroidal structure of lambda-exonuclease. Science 277:1824-1827. [DOI] [PubMed] [Google Scholar]
- 22.Kusano, K., N. K. Takahashi, H. Yoshikura, and I. Kobayashi. 1994. Involvement of RecE exonuclease and RecT annealing protein in DNA double-strand break repair by homologous recombination. Gene 138:17-25. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., G. Karakousis, S. K. Chiu, G. Reddy, and C. M. Radding. 1998. The beta protein of phage lambda promotes strand exchange. J. Mol. Biol. 276:733-744. [DOI] [PubMed] [Google Scholar]
- 24.Little, J. W. 1967. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J. Biol. Chem. 242:679-686. [PubMed] [Google Scholar]
- 25.Little, J. W., I. R. Lehman, and A. D. Kaiser. 1967. An exonuclease induced by bacteriophage lambda. I. Preparation of the crystalline enzyme. J. Biol. Chem. 242:672-678. [PubMed] [Google Scholar]
- 26.Maloy, S. R. 1990. Experimental techniques in bacterial genetics. Jones and Bartlett, Boston, Mass.
- 27.Mitsis, P. G., and J. G. Kwagh. 1999. Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Res. 27:3057-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muniyappa, K., and C. M. Radding. 1986. The homologous recombination system of phage lambda. Pairing activities of beta protein. J. Biol. Chem. 261:7472-7478. [PubMed] [Google Scholar]
- 29.Muyrers, J. P., Y. Zhang, F. Buchholz, and A. F. Stewart. 2000. RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 14:1971-1982. [PMC free article] [PubMed] [Google Scholar]
- 30.Myers, R. S., and K. E. Rudd. 1998. Mining DNA sequences for molecular enzymology: the Redα superfamily defines a set of recombination nucleases, p. 49-50. In Proceedings of the 1998 Miami Nature Biotechnology Winter Symposium. Oxford University Press, Oxford, United Kingdom.
- 31.Noirot, P., and R. D. Kolodner. 1998. DNA strand invasion promoted by Escherichia coli RecT protein. J. Biol. Chem. 273:12274-12280. [DOI] [PubMed] [Google Scholar]
- 32.Passy, S. I., X. Yu, Z. Li, C. M. Radding, and E. H. Egelman. 1999. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc. Natl. Acad. Sci. USA 96:4279-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radding, C. M. 1966. Regulation of lambda exonuclease. I. Properties of lambda exonuclease purified from lysogens of lambda T11 and wild type. J. Mol. Biol. 18:235-250. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Skalka, A. 1971. Origin of DNA concatemers during growth, p. 535-547. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Smith, G. R. 1983. General recombination, p. 175-209. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Stahl, F. W., M. M. Stahl, and R. E. Malone. 1978. Red-mediated recombination of phage lambda in a recA− recB− host. Mol. Gen. Genet. 159:207-211. [Google Scholar]
- 38.Stahl, F. W., K. D. McMilin, M. M. Stahl, and Y. Nozu. 1972. An enhancing role for DNA synthesis in formation of bacteriophage lambda recombinants. Proc. Natl. Acad. Sci. USA 69:3598-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl, M. M., L. Thomason, A. R. Poteete, T. Tarkowski, A. Kuzminov, and F. W. Stahl. 1997. Annealing vs. invasion in phage lambda recombination. Genetics 147:961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi, N., and I. Kobayashi. 1990. Evidence for the double-strand break repair model of bacteriophage lambda recombination. Proc. Natl. Acad. Sci. USA 87:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavares, P., R. Lurz, A. Stiege, B. Rückert, and T. A. Trautner. 1996. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol. 264:954-967. [DOI] [PubMed] [Google Scholar]
- 43.Thaler, D. S., M. M. Stahl, and F. W. Stahl. 1987. Double-chain-cut sites are recombination hotspots in the Red pathway of phage lambda. J. Mol. Biol. 195:75-87. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, M. S., M. Gao, D. M. Knipe, and K. L. Powell. 1992. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 66:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomason, L. C., D. S. Thaler, M. M. Stahl, and F. W. Stahl. 1997. In vivo packaging of bacteriophage lambda monomeric chromosomes. J. Mol. Biol. 267:75-87. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Oostrum, J., J. L. White, and R. M. Burnett. 1985. Isolation and crystallization of lambda exonuclease. Arch. Biochem. Biophys. 243:332-337. [DOI] [PubMed] [Google Scholar]
- 48.Vlazny, D. A., A. Kwong, and N. Frenkel. 1982. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc. Natl. Acad. Sci. USA 79:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiese, F., S. Chai, G. Lüder, and J. C. Alonso. 1994. Nucleotide sequence and complementation studies of the gene 35 region of the Bacillus subtilis bacteriophage SPP1. Virology 202:1046-1049. [DOI] [PubMed] [Google Scholar]
- 50.Wu, Y., G. Liu, and E. B. Carstens. 1999. Replication, integration, and packaging of plasmid DNA following cotransfection with baculovirus viral DNA. J. Virol. 73:5473-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokochi, T., K. Kusano, and I. Kobayashi. 1995. Evidence for conservative (two-progeny) DNA double-strand break repair. Genetics 139:5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]