Abstract
Interaction with host cells is essential in meningococcal pathogenesis especially at the blood-brain barrier. This step is likely to involve a common regulatory pathway allowing coordinate regulation of genes necessary for the interaction with endothelial cells. The analysis of the genomic sequence of Neisseria meningitidis Z2491 revealed the presence of many repeats. One of these, designated REP2, contains a −24/−12 type promoter and a ribosome binding site 5 to 13 bp before an ATG. In addition most of these REP2 sequences are located immediately upstream of an ORF. Among these REP2-associated genes are pilC1 and crgA, described as being involved in steps essential for the interaction of N. meningitidis with host cells. Furthermore, the REP2 sequences located upstream of pilC1 and crgA correspond to the previously identified promoters known to be induced during the initial localized adhesion of N. meningitidis with human cells. This characteristic led us to hypothesize that at least some of the REP2-associated genes were upregulated under the same circumstances as pilC1 and crgA. Quantitative PCR in real time demonstrated that the expression of 14 out of 16 REP2-associated genes were upregulated during the initial localized adhesion of N. meningitidis. Taken together, these data suggest that these repeats control a set of genes necessary for the efficient interaction of this pathogen with host cells. Subsequent mutational analysis was performed to address the role of these genes during meningococcus-cell interaction.
A striking characteristic of the genomic sequence of the Z2491 meningococcus genome (20) is the abundance and diversity of repetitive DNA elements. Indeed, a thorough analysis of this chromosome revealed almost 2,000 copies of uptake sequences (10-bp sequences required in natural transformation) (6, 7), about 700 neisserial intergenic mosaic elements, approximately 300 full or partial correia elements (2, 14), 43 complete or partial insertion elements, and 26 copies of a 59- to 154-bp repeat initially designated REP2. The role of these sequences remains unknown; however, their frequency suggests a crucial function in meningococcal biology. This hypothesis is reinforced by the fact that REP2 repeats contain a ribosome binding site located 5 to 13 bp upstream of an ATG codon, which is usually localized at the 3′ end of these elements and which is the predicted start codon of an open reading frame (ORF) (20).
Neisseria meningitidis and Neisseria gonorrhoeae are two closely related pathogens. N. meningitidis, a normal inhabitant of the human nasopharynx (R. J. Sim, M. M. Harrison, E. R. Moxon, and C. M. Tang, Letter, Lancet 356:1653-1654, 2000), has the ability, in a small proportion of those colonized, to invade the epithelium, to disseminate within the bloodstream, and to cross the blood-brain barrier. On the other hand, N. gonorrhoeae colonizes and invades the urogenital tract to cause a localized inflammation, gonorrhoea. In both cases the ability to interact with host cells plays a major role in the ability of these bacteria to establish a productive infection. Numerous bacterial attributes have been identified as playing a role in these interactions. Among these, the type IV pili (TFP) play an essential role by allowing the initial adhesion of bacteria with host cells. TFP are believed to mediate adhesion via the adhesin PilC1. We have previously shown that the expression of PilC1 is upregulated during the initial interaction of the bacteria with the cells and that this upregulation is required for a complete adhesiveness mediated by TFP (30). This regulation of the expression of PilC1 is controlled by a 130-bp sequence located upstream of pilC1. Further analysis demonstrated that a 130-bp sequence identical to that found upstream of pilC1 was also present upstream of another ORF designated crgA (4). This 130-bp sequence is also responsible for the upregulation of crgA during the initial contact of the bacterium with the host cell. This 130-bp element was subsequently designated CREN, for contact regulatory element of Neisseria (4).
In this work we demonstrate that the CREN located upstream of pilC1 and crgA correspond to REP2 and that 14 out of the 16 ORFs located downstream of REP2 are upregulated during the initial contact of the bacteria with the cells in a manner similar to that of pilC1 and crgA. This suggests that these 14 REP2-associated genes are coordinately upregulated during the initial interaction of N. meningitidis with the host cells. In an attempt to identify the function of these genes during the bacterial host-cell interaction, we subsequently engineered mutations in all these genes.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study were N. meningitidis Z5463, a serogroup A strain belonging to the same sequence type as the sequenced strain Z2491 (25), and N. meningitidis 2C43, a piliated Opa− Opc− PilC1+ PilC2+ derivative of 8013 (19) and a serogroup C strain. NM98-3 was obtained from M.-K. Taha and is a previously described crgA mutant of 8013 (4). Neisseria strains were grown at 37°C in a 5% CO2 atmosphere on GCB medium (Difco) containing Kellogg's supplement (11) or in GC liquid medium. Escherichia coli strains TOP10 (Invitrogen) were used for DNA cloning and plasmid propagation. E. coli cells were grown on Luria-Bertani agar or in liquid medium. For antibiotic selection of E. coli strains, kanamycin and spectinomycin were used at 60 μg/ml. To select strains derived from meningococcus strain Z5463, the kanamycin concentration was 200 μg/ml and spectinomycin was used at 75 μg/ml.
The sequence of N. meningitidis Z2491 (20) was obtained from the Sanger Center (http://www.sanger.ac.uk/Projects/N_meningitidis/). The sequence of N. meningitidis MC58 (31), a serogroup B isolate, was obtained from The Institute of Genomic Research (http://tigrblast.tigr.org/pub/data/n_meningitidis/). The sequence of N. gonorrhoeae FA1090, an N. gonorrhoeae isolate, was from the University of Oklahoma (http://www.genome.ou.edu/gono.html) and corresponds to the 25 February 2001 release. BLAST searches were performed using BLASTN and the default parameter of each web site.
Antibodies.
Phalloidin-Alexa-488 (Molecular Probes, Eugene, Oreg.) was used to stain the actin cytoskeleton (F-actin). Ezrin was detected using selective rabbit polyclonal antisera kindly provided by P. Mangeat (CNRS, Montpellier, France). In infected monolayers, bacteria were stained using a rabbit polyclonal antibody directed against the meningococcus strain Rou (5). The 20D9 monoclonal antibody recognizes the SB pilin variant produced by 2C43 (22). The antibody used to detect PilC1 protein in Western blot analysis was 18P4 (17).
Cell culture and infection.
Human umbilical vein endothelial cells (HUVECs) (promoCell) were used between passages 1 and 8 and grown in Endo-SFM supplemented with 10% heat-inactivated fetal calf serum (FCS) and 2 mM l-glutamine (Life Technologies), basic fibroblast growth factor (1 ng/ml; Boehringer-Mannheim), heparin (0.5 IU/ml), and endothelial cell growth supplement (1.25 μg/ml) (Sigma). HEC1-B is a human endometrial adenocarcinoma cell line obtained from the American Type Culture Collection and was grown in Dulbecco' modified Eagle medium-Glutamax (Life Technologies) supplemented with 10% heat-inactivated FCS. Cells were seeded at 5 × 104 cells/cm2 in a 24-well culture plate and grown at 37°C in a humidified incubator under 5% CO2 for 2 to 3 days.
Bacteria for RNA preparations were obtained from monolayers infected with 106 bacteria in exponential growth phase. Bacteria were left in contact with the cells for 30 min, the supernatant was then removed, and the wells were gently washed with infection media (RPMI 1640 medium with Glutamax [Life Technologies] supplemented with 10% heat-inactivated FCS). Monolayers were then washed every hour and fresh medium was added. The monolayers were harvested at different time points.
Unless otherwise stated, when the aim of the experiment was to quantify adhesiveness, cells were infected with 105 bacteria in exponential growth phase and left without replacement of the supernatant. The percentage of cell-associated CFU was then determined after 2 and 4 h of infection.
Immunofluorecence and subsequent analysis using confocal microscopy were performed as previously described (5).
RNA extraction.
Bacteria were added to tubes at 65°C containing 700 μl of Trizol reagent (Lifetech) and ∼50 μl of acid-washed glass beads. The mix was vortexed vigorously and freeze-thawed using dry ice. Chloroform (200 μl) was added, and the contents were mixed by vortexing. The tubes were centrifuged at 15,000 × g for 15 min at 4°C, and the aqueous phase was taken and reextracted once before precipitation by addition of 1 volume of isopropanol. The RNA pellet was washed with 75% ethanol, dried, and redissolved in RNase-free TE buffer (10 mM Tris-HCl, 1 mM EDTA-Na [pH 8.0]). DNA was removed by addition of 1 μl of DNase I (Takara) and incubation for 30 min at 37°C, followed by inactivation at 70°C for 10 min. The complete removal of DNA was checked by the absence of signal in a PCR run on the RNA sample.
Real-time PCR assays.
Oligonucleotides were designed using the Primer Express software (PE Applied biosystems) to obtain amplicons from the same size. Reverse transcription was performed using the 3′-oligonucleotides of the genes and the superscript TM II reverse transcriptase (Lifetech): 50 min at 45°C and 10 min at 70°C. Reverse transcription products were diluted to 1/10 and 1/100.
Real-time PCR was run on an ABI Prism 7700 sequence detection system (Perkin-Elmer Biosystems) using SYBR Green PCR Master Mix (PE Biosystems), according to the manufacturer's instructions.
Data analyses for a relative quantification of gene expression were performed by the comparative Ct (threshold cycle) method according to the manufacturer's instructions (user bulletin 2 for the ABI PRISM sequence detection system) and published data (27). The parameter Ct is defined as the cycle number at which fluorescence (which is proportional to the quantity of DNA in the tube during the exponential phase of the PCR) passes the fixed threshold. The relative amount of target, normalized to an endogenous reference (kanamycin resistance gene aphA3 inserted in a non coding region) is given by 2(Ctgene − CtaphA3). The induction factor is then the ratio of the amount of target in different conditions: 2(Ctgene − CtaphA3) infection media/2(Ctgene − CtaphA3).
Experiments were carried out four times. Freshly extracted RNA was used each time. Dilutions of the reverse transcription product were loaded in duplicate for each oligonucleotide couple in every experiments.
The statistical method used to test whether differences observed were significant was the Student test. Differences were considered as significant when P was <0.05.
Construction of mutants.
Mutants were engineered either by in vitro transposition mutagenesis as previously described (21) or by insertion of a spectinomycin resistance cassette into an appropriate restriction site (12). Confirmation of the mutation was performed using PCR and Southern blotting. The precise insertion site of the mutant engineered by in vitro transposition was determined by sequencing.
Transformation of N. meningitidis.
Bacteria grown overnight were resuspended in GC liquid medium containing 5 mM MgCl2 (transformation buffer) so that a 1/10 dilution of this suspension reached an optical density of 0.5 at 550 nm. Aliquots (200 μl) of this bacterial suspension were transferred into the wells of a 24-well tissue culture plate, and approximately 1 μg of DNA was added. These transformation mixtures were incubated for 30 min at 37°C. Then, 1.8 ml of prewarmed transformation buffer was added, and incubation was continued for 2 h at 37°C. Bacteria were then plated onto medium containing the appropriate antibiotic.
RESULTS
Fourteen out of 16 REP2-associated genes are upregulated during meningococcus-cell interaction.
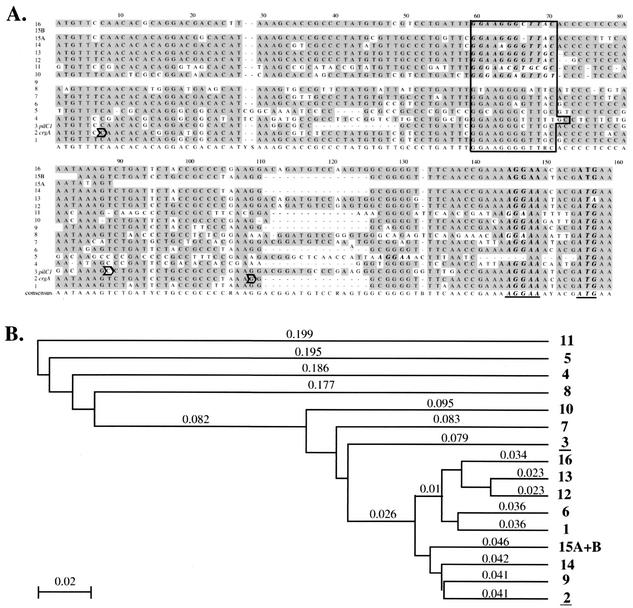
Among the various repeats described in Z2491, we concentrated on REP2. Considering that these sequences contain a ribosome binding site located 5 to 13 bp upstream of an ATG codon, we focused on the 16 out of 26 elements for which this ATG is the predicted start codon of an ORF. These latter frames were designated REP2-associated genes. As shown in Table 1, these ORFs have various predicted functions. Interestingly two of these genes were pilC1, a pilus-associated protein, and crgA, a LysR family transcriptional regulator. Both genes have been reported to be under the control of an element designated CREN. CREN elements are short DNA sequences (130 bp) located upstream of pilC1 and crgA which are involved in the expression of these genes during the initial bacterium-cell interaction (4, 30). A striking observation was that the sequence corresponding to the CREN upstream of these ORFs is identical to that designated as REP2. This prompted us to determine whether upregulation during meningococcus-cell interaction was a feature conserved among REP2-associated genes. This hypothesis was reinforced by the fact that the REP2 consensus sequence is highly homologous to the two identified CREN (Fig. 1A). Indeed, this consensus sequence contains the GG-N8-(A/G)C motif which is very close to the −24/−12 consensus characteristic of a σ54 dependent promoter (1, 32). Though the rpoN gene is inactive in both N. meningitidis and N. gonorrhoeae (13), this promoter has been shown to play a major role in the cell contact regulation of both pilC1 and crgA (4, 29).
TABLE 1.
REP2 located upstream of an ORF in the Neisseria genome Z2491
| REP2 no. | REP2 location in Z2491 genome | Corresponding REP2-associated gene | Proposed function | Presence of REP2 in the same locus of:
|
|
|---|---|---|---|---|---|
| NmMC58 | NgFA1090 | ||||
| 1 | 293648..786cb | NMA0301 (NMB2130) | Unknown | No | Yes |
| 2 | 573248..386 | NMA0601 (NMB1856) | Probable lysR family transcriptional regulator (crgA) | Yes | Yes |
| 3 | 587025..132c | NMA0609 (NMB1847) | Pilus associated protein (pilC1) | Yes | Yes |
| 4 | 792986..3127 | NMA0812 (NMB0607) | Protein export membrane protein (secD) | Yes | Yes |
| 5 | 802752..886c | NMA0820 (NMB0615) | Probable ammonium transporter (amtB) | Yes | Yes |
| 6 | 871802..940c | NMA0902 (NMB0697) | Probable dimethyl adenosine transferase (ksgA) | Yes | Yes |
| 7 | 950026..179 | NMA0987 (NMB0776) | Probable secreted protein | Yes | Yes |
| 8 | 1014984..5136 | NMA1054 (NMB0844) | Possible periplasmic protein | Yes | Yes |
| 9 | 1145604..664c | NMA1204 (NMB0996) | Unknown | Yes | Yes |
| 10 | 1247863..998c | NMA1345 (compcNMB1136 and NMB1174) | Unknown | Yes | Yes |
| 11 | 1447042..180 | NMA1549 (NMB1335) | Unknown | Yes | Yes |
| 12 | 1639218..369 | NMA1708 (NMB1508) | Unknown | Yes | Yes |
| 13 | 1746226..378 | NMA1802 (NMB1605) | DNA topoisomerase IV subunit A (parC) | No | No |
| 14 | 1853556..694 | NMA1923 (NMB1664) | Probable protease | Yes | Yes |
| 15a | 2163243..486 | NMA2210 (NMB0277) | Possible inner membrane virulence factor protein (mviN) | No | Yes |
| 16 | 2177893..8046 | NMA2225 (NMB0262) | Exodeoxyribonuclease small subunit (xseB) | Yes | No |
FIG. 1.
(A) ClustalW alignment of the nucleotidic sequences of REP2. Each REP2 has been assigned a number. Identities are shaded. REP2-2 and REP2-3 correspond to the CREN upstream of crgA and pilC1, respectively. The ribosome binding site (AAGGA) and the predicted start codon are in boldface italic type and are underlined in the consensus sequence. The GG-N8-(A/G)C motif which is very close to the −24/−12 consensus characteristic of a σ54-dependent promoter is boxed. The transcription starting point of crgA (P2) and pilC1 (PC1.3 and PC1.2) are outlined with an arrow (4, 29). (B) Phylogenetic analyses of REP2 sequences. The method used is the unweighted pair group method with arithmetic mean. The method for estimating DNA distances is p-distance. Numbers in boldface type are those assigned to each REP2. The numbers corresponding to pilC1 and crgA are underlined. Other numbers are distances between sequences.
In order to address the above hypothesis, we compared the expression of the REP2-associated ORFs during the course of meningococcal adhesion to that of bacteria grown under identical conditions but in the absence of cells. Pilus-mediated adhesion is essential in the meningococcal life cycle. Adhesion of piliated meningococci has been shown to be a two-step process. The first one leads to the formation of clumps of bacteria associated with microvillus-like structures on the surface of the host cells. The second step corresponds to diffuse adhesion, when bacteria loose their pili, spread on the apical surface of the cell, and become involved in an intimate attachment (23). To test the above hypothesis, we used real-time PCR to quantify the level of expression of the genes located downstream of each REP2 during the two steps of the adhesion process, i.e., at 1 and 4 h (localized adhesion) and 8 h (diffuse adhesion). Results are shown Table 2. As expected the levels of induction of pilC1 and crgA were similar to those previously described: upregulation during the initial localized adhesion and returned to basal levels at late adhesion (4, 30). The expression of 12 out of the 14 remaining ORFs was significantly induced during localized adhesion (P < 0.05) and then decreased at later time points during diffuse adhesion. Induction factors were between 2-and 6-fold, except for three genes NMA1204, NMA1923, and NMA2225, which were induced over 15-fold suggesting a modulation of the regulation through small sequence variations. One REP2-associated gene (NMA1549) did not show any induction at any time point. Another gene (NMA0820 amtB) was induced only at late time points, thus suggesting that it was controlled by a different regulatory pathway. This result is consistent with the observation, shown in Fig. 1B, that the REP2 located in front of amtB and NMA1549 had the lowest homology with the other elements.
TABLE 2.
Cell contact induction of the transcription of REP2-associated genes
| REP2 no. | ORF | Cell contact induction at:
|
||
|---|---|---|---|---|
| 1 h | 4 h | 8 h | ||
| 1 | NMA0301 | 2.5 ± 0.6b | 3.5 ± 0.8b | 2 ± 0.8 |
| 2 | NMA0601 (crgA) | 4.0 ± 0.6b (2.8 ± 1.6)a | 4.2 ± 1.3b (6.2 ± 3.7) | 1.8 ± 0.3 (4.3 ± 2.9) |
| 3 | NMA0609 (pilC1) | 2.7 ± 0.7b (3.9 ± 0.7) | 3.3 ± 0.9b (2.5 ± 0.7) | 1.5 ± 0.5 (1.1 ± 0.2) |
| 4 | NMA0812 (secD) | 4.0 ± 1.2b | 5.5 ± 1.6b | 1.1 ± 0.4 |
| 5 | NMA0820 (amtB) | 1.0 ± 0.4 | 1.0 ± 0.3 | 8.1 ± 2.3b |
| 6 | NMA0902 (ksgA) | 1.1 ± 0.3 | 3.0 ± 0.8b | 2.3 ± 1 |
| 7 | NMA0987 | 3.9 ± 0.8b | 3.7 ± 1.1b | 2.6 ± 0.6b |
| 8 | NMA1054 | 1.6 ± 0.5 | 6.2 ± 2.8b | 0.7 ± 0.2 |
| 9 | NMA1204 | >15b | 7.5 ± 2.6b | 4.7 ± 1.1b |
| 10 | NMA1345 | 6.2 ± 1.2b | 4.5 ± 2b | 3.4 ± 0.9b |
| 11 | NMA1549 | 1.8 ± 1.3 | 1.6 ± 0.5 | 2.0 ± 0.8 |
| 12 | NMA1708 | 2.6 ± 0.5b | 2.9 ± 0.8b | 3.1 ± 1b |
| 13 | NMA1802 (parC) | 2.0 ± 0.5b | 3.5 ± 0.7b | 2.0 ± 0.6 |
| 14 | NMA1923 | >15b | 5.7 ± 2.2b | 0.8 ± 0.3 |
| 15 | NMA2210 (mviN) | 4.7 ± 1.7b | 4.1 ± 1.2b | 2.8 ± 0.9b |
| 16 | NMA2225 (xseB) | >15b | 1.5 ± 0.4 | 2.7 ± 1.5 |
All together these data demonstrate that 14 of the 16 REP2-associated genes, including pilC1 and crgA, are induced during the initial contact with the cells. Considering the high degree of homology between all these sequences and the previously reported data on the promoter of pilC1 and crgA, it is tempting to speculate that the 12 REP2 sequences whose downstream gene is upregulated during the initial localized adhesion correspond to CREN. In addition, this observation suggest that these genes are likely to be coordinately regulated and may be part of a regulon.
Role of the cell contact regulated REP2-associated genes in meningococcus-cell interaction.
Considering the above results, we first hypothesized that some of the REP2-associated ORFs could be directly involved in the initial localized adhesion step. We therefore engineered mutations in all these genes as reported in Table 3. Mutations were obtained for all genes excepting three (NMA0812 secD, NMA1802 parC, and NMA2210 mviN), though numerous attempts were performed, thus suggesting that these ORFs may be encoding proteins essential for bacterial survival. In order to avoid possible phenotypic modifications due to phase or antigenic variation each mutation was reintroduced after amplification of the mutated sequence into the same genetic background. The correct replacement was then confirmed both by amplification of the mutated gene and Southern blotting.
TABLE 3.
Construction of mutants in REP2-associated genes induced during initial cell contact
| ORF | Location on Z2491 genomea | Type of mutation or referenceb |
|---|---|---|
| NMA0301 | 293437-293652 | Insertion of a Spr cassette in a HincII deletion (bases Δ292350-581) |
| NMA0601 (crgA) | 573382-574281 | Insertion of a Spr cassette in Ssp1 site (base 573578) |
| NMA0609 (pilC1) | 583916-587029 c | 17 |
| NMA0812 (secD) | 793120-794976 | None |
| NMA0902 (ksgA) | 871027-871806 c | Insertion of a Spr cassette in a HincII deletion (bases Δ870714-870732) |
| NMA0987 | 950175-950597 | Insertion of a mariner transposon Knr (base 950279) |
| NMA1054 | 1015129-1015455 | Insertion of a mariner transposon Knr (base 1015303) |
| NMA1204 | 1145237-1145608 c | Insertion of a mariner transposon Knr (base 1145344) |
| NMA1345 | 1247280-1247867 c | Insertion of a Spr cassette in SspI site (base 1247743) |
| NMA1708 | 1639365-1640783 | Insertion of a mariner transposon Knr (base 1639896) |
| NMA1802 (parC) | 1746380-1748677 | None |
| NMA1923 | 1853690-1855045 | Insertion of mariner transposon Knr (base 1854352) |
| NMA2210 (mviN) | 2163482-2165020 | None |
| NMA2225 (xseB) | 2178042-2178266 | Insertion of a mariner transposition Knr (base 2178174) |
Numbers in bold-face type correspond to that of the first base of the gene. “c” indicates that the REP2 is on the minus strand according to the sequence of Z2491 (17).
Spr, spectinomycin resistance; Knr, kanamycin resistance.
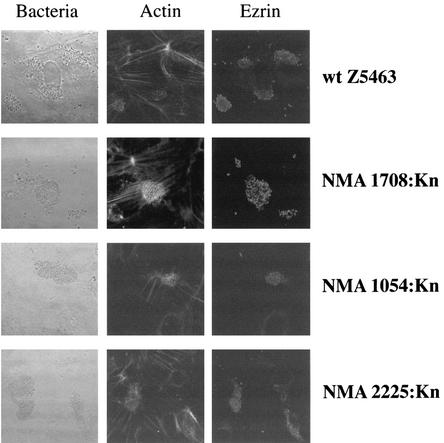
The adhesion of each mutant was tested on both endothelial and epithelial cells during localized adhesion. With the exception of pilC1 whose role in pilus-mediated adhesion is well documented (18), we found that the levels of adhesiveness of the mutants were identical to that of the parental wild-type strain (Table 4). We subsequently assessed the possibility that these genes play a role in the signaling that takes place during meningococcus-cell interaction. As previously shown, following pilus-mediated adhesion, cortical plaques appear at the site of attachment of meningococci, with recruitment of ezrin and cortical actin (15, 16). However, each of the mutants, as for the wild-type strain, induced the formation of cortical plaques, with clumps of adhering bacteria being observed on the apical surface of the cell, and recruitment of actin and ezrin beneath the bacterial colony (Fig. 2 and data not shown). Together these data suggest that, with the exception of pilC1, the REP2-associated genes are not directly involved in the bacterial-cell interaction.
TABLE 4.
Adhesion of mutants in REP2-associated genesa
| Mutated ORFb | Mean adhesions ± SD in:
|
||
|---|---|---|---|
| HUVEC cells
|
HEC1-B cells (4 h) | ||
| 2 h | 4 h | ||
| Wild-type Z2491 | 2.36 ± 0.36 | 6.10 ± 0.42 | 5.56 ± 0.07 |
| NMA0301:Spa | 2.98 ± 0.06 | 5.91 ± 0.05 | 5.96 ± 0.04 |
| NMA0601:Sp (crgA) | 2.83 ± 0.23 | 6.61 ± 0.29 | 4.78 ± 0.1 |
| NMA0609:Sp (pilC1) | 0.015 ± 0.005 | 0.02 ± 0.00 | 0.03 ± 0.005 |
| NMA0902:Sp (ksgA) | 2.59 ± 0.39 | 6.49 ± 0.10 | 5.91 ± 0.75 |
| NMA0987:Knb | 2.35 ± 0.15 | 7.26 ± 0.44 | 7.04 ± 0.15 |
| NMA1054:Kn | 2.44 ± 0.34 | 6.55 ± 0.75 | 7.62 ± 0.18 |
| NMA1204:Kn | 3.22 ± 0.22 | 7.39 ± 0.09 | 7.23 ± 0.14 |
| NMA1345:Kn | 3.03 ± 0.20 | 6.69 ± 0.41 | 7.23 ± 0.14 |
| NMA1708:Kn | 2.70 ± 0.01 | 5.8 ± 0.32 | 6.22 ± 0.47 |
| NMA1923:Kn | 3.06 ± 0.26 | 6.25 ± 0.52 | 6.11 ± 0.71 |
| NMA2225:Kn (xseB) | 2.3 ± 0.56 | 6.03 ± 0.13 | 5.87 ± 0.08 |
Adhesion corresponds to the following ratio: number of cell-associated bacteria/(number of bacteria in the supernatant + number of cell-associated bacteria) × 100).
Mutants were not obtained for ORFs NMA0812 (secD), NMA1802 (parC), and NMA2210 (mviN).
c Sp, insertion of a spectinomycin resistance cassette.
d Kn, insertion of a transposon containing a kanamycin resistance cassette.
FIG. 2.
Actin and ezrin recruitment visualized by confocal examination of infected monolayers of HUVECs. HUVECs were infected with either wild-type bacteria (Z5463) or mutants. Experiments were carried out after 4 h of infection (localized adhesion). The localization of bacterial colonies is shown in the first column by phase contrast. F-actin was labeled with a phalloidin probe (second column) and with ezrin by using a polyclonal antibody (third column). Results obtained with the wild-type strain and three of the mutants are shown. Similar results were obtained with the remaining seven mutants.
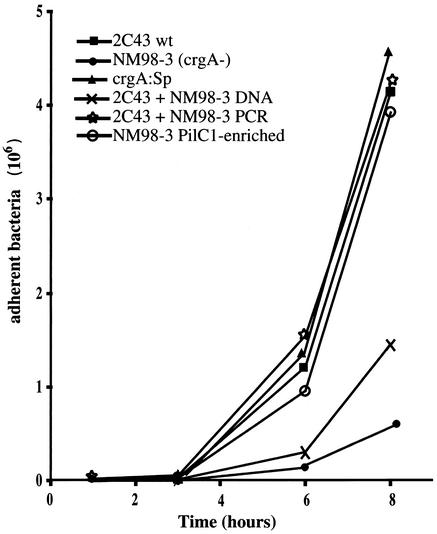
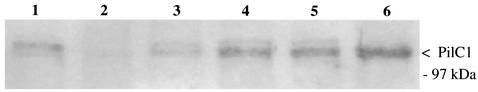
A surprising result was that the crgA mutant was capable of adhering to the same extent as the wild-type strain (Table 4 and Fig. 3) in contrast to what has been previously reported for this gene (4). Mutation in this gene has been described to be responsible for a reduction of meningococcal adhesion by 20 to 50 times after 4 h, and by the lack of switch to diffuse adhesion at later time points. Since our crgA mutant adhered to cells as efficiently as the wild-type strain, we obtained the previously described crgA mutant (NM98-3) of 2C43 (4). As shown Fig. 4, NM98-3 indeed showed a defect in adhesion similar to that previously reported. To investigate this difference, we transformed the crgA mutation from strain NM98-3 into our 2C43 strain, using either a PCR product corresponding to the mutagenized gene or total chromosomal DNA. In an adhesion assay using the different crgA mutants, our strain 2C43 transformed by the PCR product was as adhesive as the wild-type strain, while 2C43 transformed by NM98-3 DNA had an intermediate phenotype (Fig. 4). This led us to hypothesize that, in strain NM98-3, the crgA mutation had been cotransformed with a nonexpressed pilC1 gene, from which it is separated by only 9.5 kb in strains Z2491 and MC58, the two sequenced meningococcal chromosomes. Expression of this gene is controlled by phase variation that places the protein coding sequence in or out of frame with respect to the signal sequence. A Western blot analysis of PilC1 of NM98-3 from bacteria grown on agar plates and after different times of adhesion confirmed our hypothesis, revealing low levels of PilC1 in NM98-3, which increase after 1 and 3 h of contact with cells (Fig. 5). This suggests that pilC1 is out of phase in the majority of the bacteria in the inoculum and that, since PilC1 is required for pilus-mediated adhesion, bacteria with an in-frame gene are selected for during adhesion. This last hypothesis was confirmed by the fact that cell-associated CFU of NM98-3 obtained after 3 h of infection behaved as the wild-type strain and were PilC1+ (Fig. 4). Furthermore sequencing of the poly(G) tract of pilC1 of the different strains showed that NM98-3 harbors 9 Gs and is consequently out of frame, whereas all other strains contain 10 Gs.
FIG. 3.
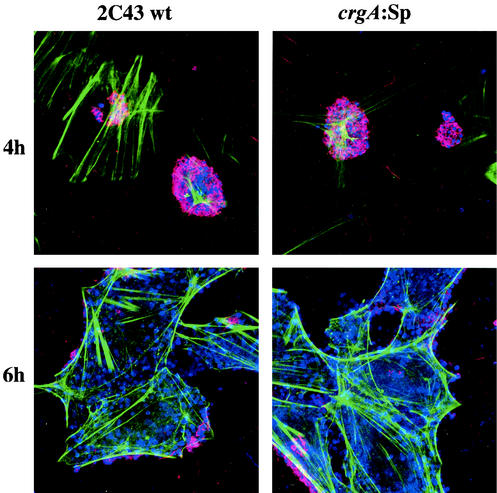
Confocal examination of infected HUVEC monolayers. HUVECs were infected with either wild-type bacteria (2C43) or the crgA:Sp mutant. Confocal microscopy was performed after 4 h (initial adhesion) and 6 h (beginning of diffuse adhesion when pili are already lost) of infection. A polyclonal antibody directed against N. meningitidis ROU was used to reveal bacteria (blue). F-actin was labeled with a phalloidin probe (green), and pilin was labeled by the monoclonal antibody 20D9 (red). Cell-associated CFU are piliated at 4 h and mostly nonpiliated at 6 h.
FIG. 4.
Adhesion of 2C43 and various crgA mutants on HUVECs. Cells were infected for 30 min with 105 bacteria in exponential growth phase and then washed. The monolayer was washed every hour. At different time points, monolayers were harvested and the number of adhesive bacteria was determined. 2C43 is the wild-type bacterium, NM98-3 is the previously described crgA mutant of 2C43 (4), crgA:Sp is the crgA mutant that we engineered in 2C43, 2C43+NM98-3 DNA corresponds to the strain 2C43 transformed with the total chromosomal DNA of NM98-3, 2C43+NM98-3 PCR corresponds to a transformant of strain 2C43 with the PCR product of the crgA mutation of NM98-3, and NM98-3 PilC1-enriched corresponds to cell-associated CFU of NM98-3 after 3 h of infection.
FIG. 5.
Western blot analysis of PilC1. Lane 1, wild-type strain 2C43; lane 2, NM98-3; lane 3, cell-associated CFU of NM98-3 obtained after 1 h of infection; lane 4, cell-associated CFU of NM98-3 obtained after 3 h of infection; lane 5, cell-associated CFU of NM98-3 obtained after 8 h of infection; lane 6, 2C43 crgA:Sp. The antibody used to detect PilC proteins is 18P4 (17). Each well was loaded with similar amounts of protein.
Pili are known to disappear during the transition from localized to diffuse adhesion. It has recently been hypothesized that crgA may control this transition by downregulating piliation (3). Figure 3 shows that the crgA mutant looses its pili during late adhesion stages at a rate similar to that observed with the wild-type strain, thus demonstrating that downregulation of pili induced by crgA is not by itself sufficient to abolish piliation.
DISCUSSION
The interaction between cells and bacteria is an active phenomenon that may implicate a common regulation shared by many genes. In the search of such a regulatory system, the analysis of the genomic sequence of Z2491 (20) revealed a set of repeated sequences, named REP2, which include a ribosome binding site and are found several times located upstream of ORFs. The study of the downstream genes revealed that pilC1 and crgA are among these REP2-associated ORF. The promoters of pilC1 and crgA, named CREN, play a regulatory role during the cell contact (4, 30) and correspond to REP2. The sequence of these repeats is highly conserved and contains the motif GG-N8-(A/G)C, which is a −24/−12 type promoter. The general features of this type of promoter are the conserved −24(GG)/−12(GC) consensus sequence, its recognition by a specific RNA polymerase sigma factor, σ54 (rpoN), and the absolute requirement for a transcriptional regulatory protein to activate the expression of the associated genes. The gene rpoN has been shown to be inactive in the pathogenic neisseriae (13) but the conserved −24(GG)/−12(GC) consensus sequence is involved in the regulation of pilE by PilA in N. gonorrhoeae (28). Moreover, PilA has been shown to be implicated in the induction of pilC1 upon cell contact by interacting directly with the CREN (29, 30). REP2 also share a ribosome-binding site: AAGGA localized 5 to 13 bases before a potential translational start codon ATG. A BLASTN analysis of the other two available neisserial genomic sequences reveals that REP2 sequences are present 22 and 23 times in N. meningitidis MC58 and N. gonorrhoeae FA1090, respectively, and are often localized upstream of ORFs, suggesting a potential role in transcription.
The 130 bp upstream of pilC1 and crgA have been named CREN because they are implicated in the regulation of these genes upon cell contact. The 12 REP2-associated genes which are upregulated during the localized adhesion may correspond to other CRENs. Nine REP2-asociated genes were induced 2- to 6-fold, and three were induced over 15-fold. This difference may be due to the length or to the small differences in repeated sequences (addition, deletion, or modification of bases) that could modify the fixation of regulatory elements. Differences may also be attributable to other factors, such as regulatory motifs upstream of the repeats. For example, upstream of the CREN of pilC1 and crgA are found a variable number of T-N11-A motifs (4) which are binding sites for LysR family transcriptional regulators (26). Another explanation for the different induction levels observed is the mRNA stability. Indeed, mRNA processing and degradation by nucleases depends on their secondary structure, including the presence of stem loops at their 5′ ends (9), the existence of terminators and of repetitive extragenic palindromic sequences at their 3′ ends (10). Our data suggest that the REP2-associated genes which are regulated during the localized adhesion of N. meningitidis are part of a regulon.
Many other genes are certainly modified by contact with cells. A recent report investigating expression changes in N. meningitidis MC58 during interaction with human epithelial cells (16HBE14) revealed that 139 genes are upregulated more than twofold in one of the four time points tested and 55 genes exhibited a difference of more than fourfold (8). Our data are essentially in agreement with these results since some REP2-associated genes were among those 139 genes. However, the large number of genes found to be induced during the interaction with the cells indicate that other regulatory pathways, independent from the one which control the REP2-associated genes, exist. The REP2-associated genes upregulated during the cell contact are one of the regulons which control the meningococcal cell interaction.
The absence of modification of the bacterium-cell interaction observed in strains carrying mutations in one of the REP2-associated genes, apart from PilC1, does not exclude the possibility that these genes are involved in the interaction of the bacteria with host cells. It is possible that subtle changes in behavior of the bacteria may be required for optimal interaction with cell. Indeed, some of the REP2-associated genes could play a role in adaptation of the bacteria to grow on cells, such as the exodeoxyribonuclease xseB, or the topoisomerase parC, or the export protein secD. Equally, it will be important to investigate trans-acting regulatory elements that might coordinate the response of this regulon.
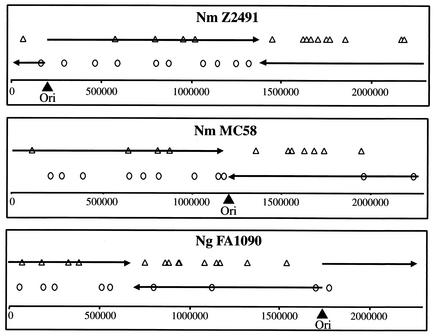
It is interesting to speculate on the evolutionary mechanisms leading to the association of REP2 with ORFs, in view of its function as a regulatory element. Since REP2 sequences are very homologous along their entire length, it seems unlikely that they have evolved separately as promoter sequences before a subset of genes. Rather, they have probably been inserted several times in the neisserial genomes. In support of this hypothesis REP2 precede many genes common to the N. meningitidis and N. gonorrhoeae genomes, but in some cases the same gene is preceded by a REP2 in one strain but not others. The mechanisms of insertion or duplication of many of the repeat sequences in the neisserial genome remain unclear, but the distribution of REP2 sequences in each of the genomes may provide a preliminary indication. The REP2 elements show a distinct preference for the lagging strand (Fig. 6) in contrast to the modest preference of gene polarity for the leading strand, hence suggesting a possible link between chromosomal replication and duplication/insertion of the REP2. Because of the high rates of genetic recombination in the pathogenic Neisseria species, to maintain such a strand preference the process of association of genes with REP2 elements should be an ongoing process. It is then possible that these insertions have been and are being subjected to evolutionary selection. Those conferring a benefit to the bacterium would be maintained (e.g., PilC1), while neutral changes will be seen in only some strains (for example mviN) and unfavorable insertions will be lost due to their host bacteria being nonviable or less competitive.
FIG. 6.
Genomic distribution of REP2 in the three pathogenic Neisseria strains sequenced: Z2491, MC58, and FA1090. REP2 sequences on the plus strand are represented by open triangles, and REP2 sequences on the minus strand are shown by open circles. The origins of replication of neisserial strains (filled triangles) were as given elsewhere (20, 31) and verified (determined for N. gonorrhoeae) by a BLASTN search for clusters of DnaA boxes (24). The circular genome of Neisseria is replicated bidirectionally from this origin to a meeting point. Hence, the continuously synthesized leading strand (black arrows) corresponds to the plus strand (5′ to 3′) in the figures, to the right of the origin of replication, and to the minus strand (3′ to 5′) to the left. The diagrams are linear representations of the circular chromosome.
Acknowledgments
We thank C. R. Tinsley for stimulating discussions and for careful reading of the manuscript. We are grateful to E. Eugène for help with confocal microscopy.
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale, Université Paris V-René Descartes, and the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed]
- 2.Correia, F. F., S. Inouye, and M. Inouye. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194-12198. [PubMed] [Google Scholar]
- 3.Deghmane, A. E., D. Giorgini, M. Larribe, J. M. Alonso, and M. K. Taha. 2002. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 43:1555-1564. [DOI] [PubMed] [Google Scholar]
- 4.Deghmane, A. E., S. Petit, A. Topilko, Y. Pereira, D. Giorgini, M. Larribe, and M. K. Taha. 2000. Intimate adhesion of Neisseria meningitidis to human epithelial cells is under the control of the crgA gene, a novel LysR-type transcriptional regulator. EMBO J. 19:1068-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eugene, E., I. Hoffmann, C. Pujol, P. O. Couraud, S. Bourdoulous, and X. Nassif. 2002. Microvilli-like structures are associated with the internalization of virulent capsulated Neisseria meningitidis into vascular endothelial cells. J. Cell Sci. 115:1231-1241. [DOI] [PubMed] [Google Scholar]
- 6.Goodman, S. D., and J. J. Scocca. 1991. Factors influencing the specific interaction of Neisseria gonorrhoeae with transforming DNA. J. Bacteriol. 173:5921-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grifantini, R., E. Bartolini, A. Muzzi, M. Draghi, E. Frigimelica, J. Berger, G. Ratti, R. Petracca, G. Galli, M. Agnusdei, M. Monica Giuliani, L. Santini, B. Brunelli, H. Tettelin, R. Rappuoli, F. Randazzo, and G. Grandi. 2002. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 20:914-921. [DOI] [PubMed] [Google Scholar]
- 9.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 10.Higgins, C. F., R. S. McLaren, and S. F. Newbury. 1988. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene 72:3-14. [DOI] [PubMed] [Google Scholar]
- 11.Kellogg, D. S., W. L. Peacock, W. E. Deacon, L. Brown, and C. I. Pirkle. 1963. Neisseria gnonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klee, S. R., X. Nassif, B. Kusecek, P. Merker, J. L. Beretti, M. Achtman, and C. R. Tinsley. 2000. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immun. 68:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laskos, L., J. P. Dillard, H. S. Seifert, J. A. Fyfe, and J. K. Davies. 1998. The pathogenic neisseriae contain an inactive rpoN gene and do not utilize the pilE sigma54 promoter. Gene 208:95-102. [DOI] [PubMed] [Google Scholar]
- 14.Liu, S. V., N. J. Saunders, A. Jeffries, and R. F. Rest. 2002. Genome analysis and strain comparison of correia repeats and correia repeat-enclosed elements in pathogenic Neisseria. J. Bacteriol. 184:6163-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merz, A. J., C. A. Enns, and M. So. 1999. Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 32:1316-1332. [DOI] [PubMed] [Google Scholar]
- 16.Merz, A. J., and M. So. 1997. Attachment of piliated, Opa− and Opc− gonococci and meningococci to epithelial cells elicits cortical actin rearrangements and clustering of tyrosine-phosphorylated proteins. Infect. Immun. 65:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morand, P. C., P. Tattevin, E. Eugene, J. L. Beretti, and X. Nassif. 2001. The adhesive property of the type IV pilus-associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N-terminal part of the molecule. Mol. Microbiol. 40:846-856. [DOI] [PubMed] [Google Scholar]
- 18.Nassif, X., J. L. Beretti, J. Lowy, P. Stenberg, P. O'Gaora, J. Pfeifer, S. Normark, and M. So. 1994. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc. Natl. Acad. Sci. USA 91:3769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassif, X., J. Lowy, P. Stenberg, P. O'Gaora, A. Ganji, and M. So. 1993. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol. Microbiol. 8:719-725. [DOI] [PubMed] [Google Scholar]
- 20.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 21.Pelicic, V., S. Morelle, D. Lampe, and X. Nassif. 2000. Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pujol, C., E. Eugene, L. de Saint Martin, and X. Nassif. 1997. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 65:4836-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pujol, C., E. Eugene, M. Marceau, and X. Nassif. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. USA 96:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salzberg, S. L., A. J. Salzberg, A. R. Kerlavage, and J. F. Tomb. 1998. Skewed oligomers and origins of replication. Gene 217:57-67. [DOI] [PubMed] [Google Scholar]
- 25.Sarkari, J., N. Pandit, E. R. Moxon, and M. Achtman. 1994. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol. Microbiol 13:207-217. [DOI] [PubMed] [Google Scholar]
- 26.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 27.Schmittgen, T. D., B. A. Zakrajsek, A. G. Mills, V. Gorn, M. J. Singer, and M. W. Reed. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285:194-204. [DOI] [PubMed] [Google Scholar]
- 28.Taha, M. K., and D. Giorgini. 1995. Phosphorylation and functional analysis of PilA, a protein involved in the transcriptional regulation of the pilin gene in Neisseria gonorrhoeae. Mol. Microbiol. 15:667-677. [DOI] [PubMed] [Google Scholar]
- 29.Taha, M. K., D. Giorgini, and X. Nassif. 1996. The pilA regulatory gene modulates the pilus-mediated adhesion of Neisseria meningitidis by controlling the transcription of pilC1. Mol. Microbiol. 19:1073-1084. [DOI] [PubMed] [Google Scholar]
- 30.Taha, M. K., P. C. Morand, Y. Pereira, E. Eugene, D. Giorgini, M. Larribe, and X. Nassif. 1998. Pilus-mediated adhesion of Neisseria meningitidis: the essential role of cell contact-dependent transcriptional upregulation of the PilC1 protein. Mol. Microbiol. 28:1153-1163. [DOI] [PubMed] [Google Scholar]
- 31.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 32.Thony, B., and H. Hennecke. 1989. The −24/−12 promoter comes of age. FEMS Microbiol. Rev. 5:341-357. [DOI] [PubMed] [Google Scholar]