Abstract
Enterobacteria have developed numerous constitutive and inducible strategies to sense and adapt to an external acidity. These molecular responses require dozens of specific acid shock proteins (ASPs), as shown by genomic and proteomic analysis. Most of the ASPs remain poorly characterized, and their role in the acid response and survival is unknown. We recently identified an Escherichia coli gene, asr (acid shock RNA), encoding a protein of unknown function, which is strongly induced by high environmental acidity (pH < 5.0). We show here that Asr is required for growth at moderate acidity (pH 4.5) as well as for the induction of acid tolerance at moderate acidity, as shown by its ability to survive subsequent transfer to extreme acidity (pH 2.0). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western analysis of acid-shocked E. coli cells harboring a plasmid-borne asr gene demonstrated that the Asr protein is synthesized as a precursor with an apparent molecular mass of 18 kDa. Mutational studies of the asr gene also demonstrated the Asr preprotein contains 102 amino acids. This protein is subjected to an N-terminal cleavage of the signal peptide and a second processing event, yielding 15- and 8-kDa products, respectively. Only the 8-kDa polypeptide was detected in acid-shocked cells containing only the chromosomal copy of the asr gene. N-terminal sequencing and site-directed mutagenesis revealed the two processing sites in the Asr protein precursor. Deletion of amino acids encompassing the processing site required for release of the 8-kDa protein resulted in an acid-sensitive phenotype similar to that observed for the asr null mutant, suggesting that the 8-kDa product plays an important role in the adaptation to acid shock. Analysis of Asr:PhoA fusions demonstrated a periplasmic location for the Asr protein after removal of the signal peptide. Homologues of the asr gene from other Enterobacteriaceae were cloned and shown to be induced in E. coli under acid shock conditions.
Enteric bacteria often encounter acid stress conditions in a variety of pathogenic and natural situations. Proteome analysis of bacterial cell cultures grown in laboratory liquid medium and subjected to acid stress revealed that over 30 Escherichia coli proteins and 48 to 60 Salmonella enterica serovar Typhimurium proteins are acid inducible, depending on the growth conditions (3, 9, 37, 38). These proteins are proposed to be components of acid survival and resistance systems, functioning in biochemical reactions such as neutralizing of external acid, adjusting cellular catabolism to a new environment, performing DNA repair and membrane biogenesis, acting as chaperones, and contributing to the microbial pathogenesis (1, 3, 21, 36, 37, 38). Only a limited number of acid-inducible proteins, however, have been characterized and had their individual functions dissected (8). The best-investigated examples are bacterial acid-inducible amino acid decarboxylase-based systems, which function to neutralize acidic pH by consuming intracellular protons and producing less-acidic products via decarboxylation reactions. Several acid-inducible amino acid decarboxylase systems, such as cadAB (encoding lysine decarboxylase CadA and its antiporter CadB) (30, 33), adiA (encoding arginine decarboxylase) (39), gad (encoding glutamate decarboxylases GadA and GadB, antiporter GadC, and regulator protein GadX) (11, 15, 24, 36, 43) of E. coli, cadAB of S. enterica serovar Typhimurium and Vibrio vulnificus, and cadA of Vibrio cholerae (17, 31, 33), have been reported. Some of the inducible decarboxylase systems, such as cadAB of E. coli, function at relatively mild acidity, while others, such as adiA and gad of E. coli, significantly contribute to resistance at extreme acidity (11, 15, 28). The glutamate decarboxylase system (gad) has been extensively studied and demonstrated to be an important player in the acid resistance of enteric pathogens such as E. coli, Listeria monocytogenes, and Shigella flexneri (14, 36, 45). A variety of inducible amino acid decarboxylases reflect the overall complexity of acid response and survival systems in bacteria. Searches for bacterial regulatory factors controlling such systems in S. enterica serovar Typhimurium have revealed the RpoS, Fur, PhoP, and OmpR proteins, which are responsible for the regulation of different sets of acid shock proteins (ASPs) (5, 6, 7, 18, 19). These ASPs are induced when Salmonella growing in minimal medium is exposed to moderate acidity (pH 4.5), and this induction is required for survival during subsequent exposures to extreme acidity (pH 3.0), a phenomenon known as acid tolerance response (ATR) (19). Similar acid adaptation systems appear to exist in E. coli, although their components are much less defined (22, 25, 27, 34). Besides the ATR, E. coli is equipped with additional acid resistance systems which help the stationary-phase cells to survive at an extremely low pH (pH 2.0) (11, 27).
We recently identified the E. coli asr (acid shock RNA) gene, as it was strongly induced by acid shock (pH < 5.0). To further characterize the asr gene and its product and elucidate the role in stress response, we decided to compare an asr mutant strain and a wild-type strain in different conditions. To search for possible functional links for asr, we also decided to clone the homologous genes from other enteric pathogens in E. coli and test whether they were induced by acid treatment.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise indicated, E. coli cells were grown in Luria-Bertani (LB) medium (35). When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1, and kanamycin, 60 μg ml−1. Acid induction was performed as follows: cells from overnight cultures were diluted 1:1,000 with a low-phosphate-glucose-salts medium, LPM (4) [50 mM Tris base, 20 mM KCl, 7.5 mM (NH4)2SO4, 240 μM MgCl2, 18.5 μM CaCl2, 6.5 μM FeCl3, 0.2% glucose, 0.6% Bacto peptone], which was buffered with MOPS (4-morpholinepropanesulfonic acid) (pH 7.0) to a final concentration of 0.1 M. Cells were grown at 37°C with rotary aeration to an A600 of 0.5 and then resuspended in the same volume of LPM medium, adjusted by HCl to pH 4.5.
TABLE 1.
E. coli strains and plasmids used in this study
| Strains and plasmids | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| JC7623 | recB21 recC22 sbcB15 thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rfbD1 rpsL21 kdgK51 xyl-5 mtl-1 argE3 thi-1 | 13 |
| MC4100 | F−araD139 Δ(argF-lac)205 flbB5301 ptsF25 relA1 rpsL150 deoC1 | 10 |
| JE14 | MC4100 asr::Kan | This study |
| JE15 | MC4100 with deletion of amino acids 29-31 of Asr and Kanr fragment, cloned into the MluI site upstream of the asr sequence | This study |
| JE16 | MC4100 with deletion of amino acids 67-68 of Asr and Kanr fragment, cloned into the MluI site upstream of the asr sequence | This study |
| JE17 | MC4100 containing Kanr fragment, cloned into the MluI site upstream of asr sequence | This study |
| BL21 | F−ompT(rB− mB−) | 41 |
| Plasmids | ||
| pET28b | Expression vector | Novagen |
| pUC19 | Cloning vector | 46 |
| pGB2 | Low-copy-number cloning vector | 12 |
| pUC4K | Cloning vector | Pharmacia |
| pAS1 | 2.13-kb BglI fragment from λ 311 cloned into pUC19 | 42 |
| pAS2 | 1.3-kb BglI-PvuII fragment from lambda phage 311 cloned into pUC19 | 42 |
| pAS11 | 706-bp Csp61 fragment from pAS2 cloned into SmaI site of pTZ19 | This study |
| pLAS2 | 1.3-kb EcoRI-HindIII fragment from pAS2 cloned into EcoRI-HindIII site of pGB2 | This study |
| pAP1 | 0.35-kb DraI-XhoI asr DNA fragment obtained by PCR, cloned between the NcoI and XhoI sites of pET28b | This study |
| pDM1 | Derivative of pAP1 containing a 1.36-kb phoA DNA fragment without 26 N-terminal amino acids cloned in frame into the PstI site of pAP1 | This study |
| pDM2 | Derivative of pAP1 containing a 1.36-kb phoA DNA fragment without 26 N-terminal amino acids cloned in frame into the XhoI site of pAP1 | This study |
| pDM3 | 1.36-kb phoA DNA fragment without 26 N-terminal amino acids cloned into Xho1 site of pET28b | This study |
| pSS1 | Derivative of pAS11 with deletion of amino acids 29-31 in Asr | This study |
| pSS3 | Derivative of pAS11 with deletion of amino acids 67-68 in Asr | This study |
| pSS4 | Derivative of pAS1 containing Kanr determinant cloned into the MluI restriction site upstream asr DNA sequence | This study |
| pSS5 | Derivative of pAS1 containing Kanr determinant cloned into the MluI restriction site upstream of the asr DNA sequence and deletion of amino acids 29-31 in Asr | This study |
| pSS6 | Derivative of pAS1 containing Kanr determinant cloned into the MluI restriction site upstream of the asr DNA sequence and deletion of amino acids 67-68 in Asr | This study |
| pST5 | 570-bp Eco241-GsuI S. enterica serovar Typhi asr DNA fragment cloned into the SmaI site of pUC19 | This study |
| pKP5 | 728-bp Bsp143I K. pneumoniae asr DNA fragment cloned into the SmaI site of pUC19 | This study |
| pEC5 | 900-bp Bsp143I E. cloacae asr DNA fragment cloned into the SmaI site of pUC19 | This study |
| pLST5 | 570-bp S. enterica serovar Typhi asr DNA fragment cloned into EcoRI-HindIII site of pGB2 | This study |
| pLKP5 | 728-bp K. pneumoniae asr DNA fragment cloned into EcoRI-HindIII site of pGB2 | This study |
| pLEC5 | 470-bp E. cloacae asr DNA fragment cloned into EcoRI-HindIII site of pGB2 | This study |
For growth kinetics at low pH, cultures were grown in LPM medium buffered at different pH values either by sodium citrate (0.05 M) or homopiperazine-N,N′-bis-2(ethanesulfonic acid) (HOMOPIPES) (0.1 M). Growth was monitored by measuring turbidity with a C075 colorimeter (Werner-Glas AB, Stockholm, Sweden). For ATR analysis, cultures were grown at 37°C in LPM medium (pH 7.0) to an A600 of 0.3 for the log-phase ATR assay and to an A600 of 1.5 for the stationary-phase ATR assay. To induce acid adaptation (adapted cells), cells were resuspended in fresh LPM medium, adjusted by HCl to pH 4.45, and incubated for 2 h. Control (unadapted) and adapted cell cultures were inoculated at approximately 2 × 108 to 3 × 108 cells ml−1 into LPM medium acidified by HCl to a pH of 2.0. Cultures were incubated for 2 h. Viable cells were calculated by plating aliquots of serially diluted cultures onto LB (pH 7.0) plates. The number of surviving microorganisms was expressed as a percentage of the cell number at time zero (immediately after challenge to pH 2.0).
DNA and RNA manipulation and plasmid construction.
Plasmid DNA preparation, restriction endonuclease digestion, DNA ligation, and DNA and RNA gel electrophoresis were performed according to standard protocols (35). Restriction enzymes and other DNA- and RNA-modifying enzymes were from AB Fermentas (Vilnius, Lithuania). PCR was performed using proofreading DNA polymerase Pfu (AB Fermentas). All enzymes were used as recommended by the suppliers.
Using primers H1 (5′-GGGTAATTTAAAAAACAAATTG-3′) and H2 (5′-TTACTGTTGAACTCGAGCGCTGC-3′), which contain DraI and XhoI restriction sites (underlined), respectively, to construct the plasmid pAP1, the E. coli asr gene was amplified from plasmid pAS2. The resulting fragment was cut with DraI and XhoI and inserted between the NcoI and XhoI sites of vector pET28b (Novagen, Madison, Wis.). In this construct, a six-His tag is attached to the C terminus of the Asr protein. Induction and purification of Asr-His6 protein with Nickel-NTA agarose (Qiagen, Hilden, Germany) was performed according to the manufacturer's recommendations.
To obtain plasmids pDM1 and pDM2, the E. coli phoA gene was amplified from genomic DNA by PCR using the primer pair PH1 (5′-CCAGAAACTGCAGTTCTGGAA-3′) and PH2A (5′-GGTTTTCTGCAGGCCCCAGA-3′) and the primer pair PP2 (5′-ACACCACTCGAGCCTGTTCTGGAA-3′) and PP4 (5′-GGGCGCGCTCGAGTTTCAGCCC-3′), respectively. The amplified DNA contained the phoA gene lacking the sequence coding for 26 amino acids of signal peptide. Primers PH1-PH2A and PP2-PP4 introduced, respectively, PstI and XhoI restriction sites (underlined) at both ends of the amplified phoA DNA; the resulting PCR fragments were cut with PstI and XhoI, respectively, and inserted into pAP1 digested with PstI or XhoI. The resulting plasmids contained the phoA gene sequence placed in frame after residue 32 (pDM1) and at the carboxy terminus (after residue 111) of the Asr protein (pDM2), respectively. In both constructs, the reading frame was ended by a six-residue histidine tag. Control plasmid pDM3 was constructed similarly by PCR with primers PP2-PP4 (see above). The amplified DNA was digested with XhoI and inserted into an XhoI site of expression vector pET28b.
To construct plasmids pSS1 and pSS3 (from which amino acids 29 to 31 [Phe-Ala-Ala] and 67 to 68 [Gln-Ala], respectively, were deleted at the proposed cleavage sites of the Asr protein precursor), plasmid pAS11 was PCR amplified using primer pair SP2 (5′-GGCGGCAGAAGACAGACC-3′) and SP3 (5′-GAGACTACGACCACACCTG-3′) and primer pair Sig1 (5′-GCTAAAAAGCATCATAAAAAT-3′) and Sig2 (5′-CGCTTTCTGGGCAGGTGCT-3′). The PCR products were purified, ligated, and transformed into E. coli JE14 cells. The mutations were verified by DNA sequencing.
Plasmid pSS4 was constructed by cloning the Kanr cassette isolated from pUC4K (Pharmacia, North Peapack, N.J.) into the MluI site of pAS1 plasmid, which is located 404 nucleotides upstream from the asr transcription start. Plasmids pSS5 and pSS6 were obtained by recloning Bsp1407I-Bpu1102I and PstI-Bpu1102I DNA fragments (from which amino acids 29 to 31 and 67 to 68 of asr from pSS1 and pSS3 had been deleted) into similarly digested pSS4. To construct both an E. coli strain containing the same mutations on the chromosome and a control strain harboring a Kanr cassette, plasmids pSS5, pSS6, and pSS4 (control) were linearized with ScaI and introduced into E. coli strain JC7623 by electrotransformation. Plasmid-free Kanr recombinants were analyzed for the presence of the mutations and the Kanr cassette by PCR and subsequent restriction analysis of the reaction product. The two mutations and Kanr marker were further transferred into strain MC4100 by using phage P1vir, yielding deletion mutant strains JE15 and JE16 and control strain JE17, respectively.
To construct plasmid pLAS2, a 1.3-kb DNA fragment containing the asr gene from pAS2 was cut with EcoRI and HindIII and inserted directly into the EcoRI-HindIII site of low-copy-number cloning vector pGB2.
Plasmids pST5, pKP5, and pEC5, which contain homologues of the asr gene of S. enterica serovar Typhi, K. pneumoniae, and E. cloacae, respectively, were obtained as follows: genomic DNA of S. enterica serovar Typhi was digested with restriction endonucleases Eco24I-GsuI, and genomic DNAs of K. pneumoniae and E. cloacae were digested with restriction endonuclease Bsp143I and separated on a 1% agarose gel. Southern analysis of restriction digests identified a ∼570-bp S. enterica serovar Typhi DNA fragment, a ∼730-bp K. pneumoniae DNA fragment, and a ∼900-bp E. cloacae DNA fragment hybridizing with a 32P-labeled E. coli 213-bp asr DNA fragment, which was obtained by PCR with primers VA5 (5′-TTCCCAGGGATATAGTTATTT-3′) and VA12 (5′-TGCTTTATGCTGTTTTTTATG-3′). The DNA fragments were eluted from the gel and ligated into the SmaI site of pUC19. After transformation into E. coli JE14, clones were identified (using a 32P-labeled 213-bp E. coli asr DNA fragment) by colony hybridization. Plasmids pST5, pKP5, and pEC5, with chromosomal inserts of S. enterica serovar Typhi, K. pneumoniae, and E. cloacae asr, were subjected to DNA sequencing in an ALFwin Sequence Analyser 2.00 (Pharmacia).
Plasmids pLST5, pLKP5, and pLEC5 were obtained by recloning EcoRI-HindIII DNA fragments from plasmids pST5, pKP5, and pEC5 into the low-copy-number vector pGB2 digested with EcoRI-HindIII.
Protein analysis.
E. coli proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS)-20% polyacrylamide gels (2). After electrophoresis, proteins were transferred (using the semidry blotting system [Millipore, Bedford, Mass.]) onto Protran-X membranes (Schleicher & Schuell, Dassel, Germany). Rabbit polyclonal antibodies against Asr-His6 protein were prepared and used as described earlier (42). Western analysis was performed using goat horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Amersham, Buckinghamshire, United Kingdom). An ECL detection kit (NEN Life Science Prod., Boston, Mass.) was used for visualization.
Alkaline phosphatase activity was determined as described previously with para-nitrophenyl-phosphate as a substrate (42).
Determination of N-terminal sequence.
Using a Procise sequencer (Applied Biosystems, Foster City, Calif.), N-terminal sequence determination was performed by Edman degradation after the transfer of the proteins onto polyvinylidene difluoride membranes (Schleicher & Schuell).
Nucleotide sequence accession numbers.
The newly determined gene sequences used in this work were submitted to the EMBL database with accession numbers AF405542 to AF405544.
RESULTS
The E. coli asr mutant is affected in its ability to grow at acidic pH.
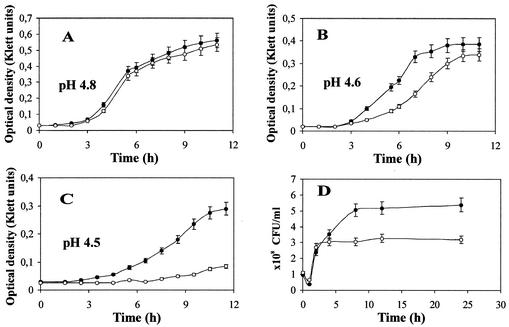
To identify a phenotype for an asr mutant, we analyzed the growth and survival of an asr knockout mutant at different acid shock conditions. In previous work, Sužiedėlienė et al. observed no difference between the asr mutant and the wild type when cells were grown in different media at pH 5.0 (42). However, we have also observed that transcription of the asr gene increases gradually with the decrease of pH and is maximal at pH 4.5 to 4.0 (results not shown), suggesting that asr is required for growth at more acidic conditions. The cultures of the asr strain (JE14) and the wild-type strain (MC4100) were grown in LPM medium buffered with sodium citrate (0.05 M) at pH 4.8, 4.6, and 4.5 for a period of 12 h. The pH of the cultures was maintained throughout the experiment within 0.01 units (grown to an A600 of 0.2) or within 0.02 units (grown to an A600 of 0.5) of the starting pH. As expected, growth of the asr mutant strain and growth of the wild-type strain were nearly the same at pH 4.8 (Fig. 1A). However, when both strains were examined at pH 4.6, a growth reduction of the asr mutant strain was observed after approximately 4 to 5 h of acid exposure (Fig. 1B). The growth defect of the asr mutant was even more pronounced at pH 4.5, exhibiting an approximately fourfold difference at A600 after several hours of growth compared to that of the parent strain (Fig. 1C). The asr mutant strain and the wild-type strain showed the same difference in growth whether the LPM medium was buffered to pH 4.5 with citrate or with HOMOPIPES, indicating that citrate was not causing the observed growth defect. Similarly, the addition of 1 mM phosphate to the LPM medium had no effect on the growth difference observed between asr and its parent strain, suggesting that phosphate limitation is not needed for the observed asr phenotype. We also compared the abilities of both strains to grow in LB medium and in glucose minimal medium buffered with HOMOPIPES (0.1 M) or citrate (0.05 M) at pH 4.9, 4.7, and 4.5. Growth of the asr mutant and growth of the parent strain in acidic LB medium were identical at all pH values tested, whereas in glucose minimal medium acidified at pH 4.7 and 4.5, both strains grew very poorly and did not give reproducible results (data not shown).
FIG. 1.
Growth kinetics and survival of asr mutant JE14 (open symbols) and parental MC4100 strain (filled symbols) at low pH. Cultures were grown in LPM medium buffered at pH 4.8 (A), pH 4.6 (B), and pH 4.5 (C), and growth was monitored as described in Materials and Methods. For the survival assay (D), cultures were grown in LPM medium (pH 7.0) to an A600 of 0.3 (1 × 108 CFU ml−1) and resuspended in LPM medium (pH 4.5); survival at different time points was defined as the CFU ml−1. The values shown are the means of at least three independent experiments. Bars indicate standard deviations (SD).
To verify that the defect in growth of the asr strain (JE14) was caused by the loss of this gene, we reintroduced the asr gene on a plasmid into this strain. We observed that overexpression of asr on a high-copy-number plasmid (pAS2) led to lysis of the cells after several hours of exposure to low pH. Complementation of the JE14 strain with a low-copy-number asr plasmid (pLAS2) rescued the growth at pH 4.6 and 4.5 to the level of the wild-type parental strain (data not shown), indicating that the phenotype was directly connected with the asr gene product.
We next examined the viability of mutant and parent strains during 24 h of acid stress at pH 4.5. MC4100 and JE14 strains were grown in LPM medium (pH 7.0) to an A600 of 0.3 (∼1 × 108 CFU/ml) and then resuspended in fresh LPM medium (pH 4.5), and viable cells of both strains were monitored over the next 24 h. Whereas during the first 5 h of acid exposure both strains were equally viable, the asr mutant culture reached a twofold lower bacterial count (compared to the parent strain) after longer incubation time (Fig. 1D). The observed acid sensitivity of the asr mutant suggests that the asr gene, while able to respond to a relatively mild pH (pH 5.0) as shown by transcription analysis (42), is required for growth and/or survival, in particular at lower pH values (pH 4.5) during a longer exposure to acidity.
The asr mutant exhibits an impaired log-phase ATR.
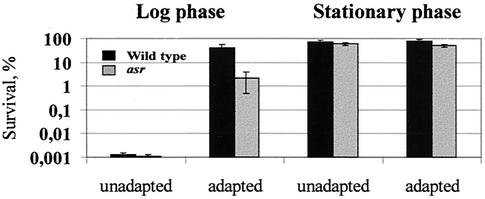
The high expression of asr at low pH values (pH 4.5), and the ability of its product to support bacterial growth at this pH, suggested that Asr protein also represents an ASP required for cell protection during extreme acidity. We tested the acid tolerance of the asr mutant strain and its parent strain at pH 2.0 after an initial adaptation of both strains at pH 4.45. MC4100 and JE14 cells grown into log phase or into stationary phase (in LPM medium at pH 7.0) were incubated in LPM medium at pH 4.45 (adapted) for 2 h or shifted immediately (unadapted) to pH 2.0 for 2 h. Whereas a direct acid challenge of the log-phase cultures at pH 2.0 gave ∼0.001% survival of either strain, a prior adaptation to mild acid at pH 4.45 resulted in ∼20-fold more survivors of the wild-type strain (40% survival) compared to the asr mutant strain (2% survival) (Fig. 2). Statistical treatment revealed that the difference in acid survival observed between the acid-adapted asr mutant and wild-type strains is significant (P < 0.05). Sensitivity to acid for cells grown in stationary phase was nearly the same in adapted and unadapted cultures, presumably due to acid resistance, a phenomenon reported for stationary-phase E. coli (11). In summary, it seems as if the Asr protein supports growth of the cells at moderate acidity (pH 4.5) and is also required for the tolerance response, which leads to protection of the cells during extreme acid shock (pH 2.0).
FIG. 2.
Log- and stationary-phase ATR of asr null mutant (JE14) and its parent MC4100. Cultures were grown in LPM medium (pH 7.0) to an optical density at A600 of 0.3 (log-phase culture) or 1.5 (stationary-phase culture). Unadapted cultures were immediately shifted to pH 2.0. Adapted cultures were shifted to pH 4.45 for 2 h prior to the challenge to pH 2.0 for 2 h as described in Materials and Methods. The percent of survival after 2 h of severe acid shock was calculated using CFU. A 100% viability was determined according to the viable counts obtained immediately after the challenge to pH 2.0. The values shown are the means of at least three independent experiments. Bars indicate SD.
Synthesis of Asr protein in acid-shocked E. coli cells.
The open reading frame (ORF) deduced from the asr DNA sequence is capable of coding for a protein composed of 111 amino acids with a theoretical mass of 11.6 kDa and a pI of ∼10. The most likely candidate for a translation start site is the third AUG codon in E. coli asr gene (Fig. 3). It is preceded by a good Shine-Dalgarno sequence, which is conserved in the asr gene homologues (see below) of all of our examined organisms (GAGGG; data not shown). The importance of this AUG was confirmed by making individual codon mutations of the three potential initiation codons in the E. coli asr gene. The translation of the Asr protein was blocked only in the case of the mutation of the third AUG (data not shown). Therefore, the E. coli Asr should be considered not as a 111-residue but as a 102-residue protein with a calculated molecular mass of 10.6 kDa.
FIG. 3.
Sequence alignment of the E. coli Asr protein with its homologues from E. cloacae, K. pneumoniae, S. enterica serovars Typhi, Paratyphi, Typhimurium, and Enteritidis, and Y. pestis. Identical amino acids are indicated by stars, conservative substitutions are indicated by stacked double dots, and semiconservative substitutions are indicated by single dots. The signal peptide is boxed. Numbers that follow the aligned regions refer to the numbers of amino acid positions within each ORF deduced from DNA sequence. The cleavage site of the E. coli Asr signal peptide and the cleavage site of the 8-kDa polypeptide, as determined by N-terminal protein sequencing, are indicated by thin and thick arrows, respectively. The black triangle indicates the probable translation start codon of E. coli Asr, as determined by site-directed mutagenesis.
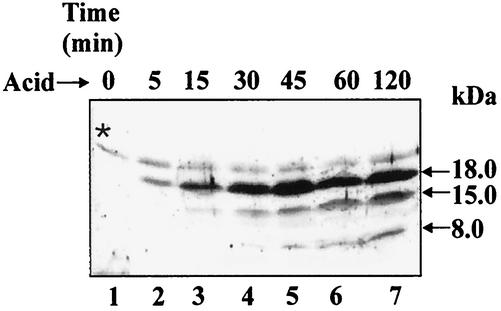
Using the T7 RNA polymerase system and subsequent SDS-polyacrylamide gel electrophoresis (PAGE) analysis of 35S-labeled proteins, expression of the asr gene readily identified a band corresponding to a protein of 18 kDa (42). This discrepancy in theoretical and observed sizes of the Asr is most probably due to the high number of basic amino acids in the protein. To further characterize the Asr protein, polyclonal antibodies were prepared against recombinant Asr-His6 (see Materials and Methods) and the Asr protein in acid-induced E. coli cells was examined by Western analysis. asr null mutant strain JE14, harboring plasmid pAS2 with a cloned asr gene containing its native promoter and terminator, and the parent strain (MC4100) were grown and acid induced as described earlier (42). The synthesis of a polypeptide with an apparent molecular mass of 18 kDa occurred within 5 min after the shift from pH 7.0 to 4.5 (Fig. 4, lane 2). The Western analysis also revealed two additional smaller polypeptides in acid-induced bacterial cells with apparent molecular masses of ∼15 kDa and ∼8 kDa (Fig. 4, lanes 3 to 7).
FIG. 4.
Synthesis of the E. coli Asr protein under acid shock conditions. JE14/pAS2 cells were grown in LPM medium (pH 7.0) to an optical density at A600 of 0.5, and then the pH of the medium was shifted from pH 7.0 to 4.5. At different times after acidification of the medium (as indicated on the top of the panel), 0.5-ml samples were taken (lanes 1 to 7) and total bacterial lysates (30 μg protein) were separated on an SDS-20% polyacrylamide gel and subjected to Western analysis using antiserum raised against Asr-His6. The approximate band sizes in kilodaltons calculated from molecular mass markers (data not shown) are indicated on the right. The star indicates a nonspecific background.
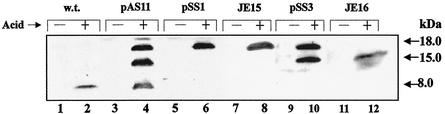
Computer analysis of the ORF (111 amino acids [aa]) deduced from the asr DNA sequence identified a potential 30-amino-acid signal peptide (42). According to the experimental determination of the initiation codon for Asr protein, its signal sequence consists of 21 aa (Fig. 3). Thus, the polypeptide of 18 kDa most likely represents the Asr precursor protein while the faster-migrating polypeptide of 15 kDa corresponds to the processed protein without the signal sequence. Interestingly, we did not detect polypeptides of 18 and 15 kDa at any time point during an acid shock of E. coli wild-type cells carrying only a chromosomal copy of the asr gene. Western analysis revealed only a small amount of 8-kDa polypeptide in these cells (Fig. 5, lane 2). Most likely, the precursor Asr protein is rapidly converted into the 8-kDa polypeptide.
FIG. 5.
Analysis of cleavage of the Asr protein precursor. E. coli strain MC4100 (w.t.), strain JE14 (asr null mutant) containing plasmids pAS11, pSS1, and pSS3, and strains JE15 and JE16 (asr deletion mutants) were grown in LPM medium to an optical density at A600 of 0.5, and then the pH of the medium was shifted from pH 7.0 to 4.5, whereas that of the control cultures remained unchanged (pH 7.0), and incubation was continued for 1 h. Total bacterial lysates from acid-induced (lanes 2, 4, 6, 8, 10, and 12) and control (lanes 1, 3, 5, 7, 9, and 11) cultures were separated on a SDS-20% polyacrylamide gel, and Western analysis was performed using anti-Asr-His6 antiserum. The approximate band sizes in kilodaltons calculated from molecular mass markers (data not shown) are indicated on the right.
To determine the N termini of the 15- and 8-kDa polypeptides, Asr-His6 protein was expressed in BL21 cells (41) and subsequently purified by affinity chromatography with Ni-NTA agarose. SDS-PAGE analysis of the purified Asr-His6 fraction revealed three polypeptides with sizes about the same (accounting for the presence of the C-terminal histidine tags) as described above. All polypeptides carried an intact C-His6 terminus, as determined by assays using commercial (Invitrogen, San Diego, Calif.) anti-His6 C-terminal antibodies (data not shown). The N-terminal amino acid sequences of the 15- and 8-kDa polypeptides were determined to be AETTTTPAPT and AAKKHHKNTK, respectively. The first sequence is in perfect agreement with that of the mature Asr protein lacking the signal peptide, indicating a cleavage between residues Ala-30 and Ala-31 (Fig. 3; numbering of amino acids corresponds to the longest ORF deduced from the asr DNA sequence [111 aa]). The second sequence matched the deduced sequence of Asr from residues 68 to 75, indicating a cleavage between residues Gln-67 and Ala-68 (Fig. 3).
Asr is localized in the periplasm.
Computer-based analysis of the amino acid sequence of the Asr polypeptide indicated no apparent membrane-spanning region, suggesting that the protein is exported into the periplasm (32). To examine this, we fused the gene coding for alkaline phosphatase (phoA), missing its signal sequence, to the N terminus (after aa 32) or the C terminus (after aa 111) of the asr protein in the pAP1 plasmid. The resulting plasmids, pDM1 and pDM2, were introduced into E. coli BL21 cells. After induction with IPTG (isopropyl-β-d-thiogalactopyranoside), cells were assayed for alkaline phosphatase activity. Cells with fusion plasmids pDM1 and pDM2 displayed 1,847 ± 220 and 4,807 ± 550 activity units, respectively (those with control plasmid pDM3 displayed 28 ± 6 U), showing that the alkaline phosphatase moiety of the hybrid proteins was exported into the periplasm. The lower alkaline phosphatase activity present in cells harboring the pDM1 fusion plasmid compared to that in cells with the pDM2 plasmid was possibly due to the insertion of phoA sequence in close proximity to the signal peptidase cleavage site of Asr protein, resulting in some reduction of cleavage and export.
Analysis of processing of Asr protein.
To verify the processing sites of the Asr precursor, we deleted amino acids 29 to 31 and 67 to 68 (Fig. 3) from the pAS11 plasmid (see Materials and Methods). The resultant plasmids pSS1 and pSS3 were transformed into the asr mutant strain JE14. Cells were acid shocked (pH 4.5) for 1 h, and proteins were analyzed by SDS-PAGE and Western analysis. In the cells harboring plasmid pSS1 with the deletion of the presumed cleavage site for the signal peptidase, only the 18-kDa (and not the 15- and 8-kDa) polypeptide was detected (Fig. 5, lane 6). In cells with the mutation of the second potential Asr cleavage site (plasmid pSS3), both the 18- and 15-kDa polypeptides accumulated (but not the 8-kDa derivative) (Fig. 5, lane 10).
To confirm that the presence of the observed 8-kDa polypeptide was a result of Asr processing when the gene is also expressed as a single copy from the chromosome, the deletion constructs of amino acids 29 to 31 and 67 to 68 were introduced into the chromosome of strain JC7623 by homologous recombination (see Materials and Methods) and subsequently transduced into strain MC4100, yielding strains JE15 and JE16, respectively. Western analysis of acid-stressed cells showed that removal of amino acids 29 to 31 (JE15) caused an accumulation of the 18-kDa preprotein (Fig. 5, lane 8), whereas deletion of amino acids 67 to 68 resulted in accumulation of the 15-kDa polypeptide (Fig. 5, lane 12), thus providing evidence that the 8-kDa polypeptide was a result of a processing of this longer polypeptide, which had undergone the removal of a signal sequence.
The 8-kDa derivative of Asr is functional in acid-stressed E. coli.
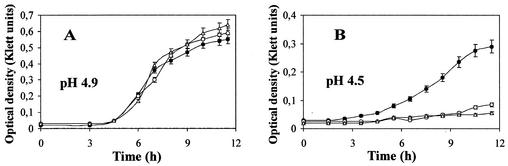
We next asked whether the 8-kDa protein is functional in E. coli under stress conditions. An asr null mutant (strain JE14), the asr mutant with the deletion of amino acids 67 to 68 (strain JE16), and their parent (strain JE17) were tested for growth ability in LPM medium at pH 4.5 and pH 4.9. There was no significant difference in growth rate between the strains at pH 4.9 (Fig. 6A). At pH 4.5, however, both JE16 and JE14 exhibited impaired growth compared with that of the wild type (Fig. 6B). Altogether, the results suggest that the Asr protein must be processed to the 8-kDa form to become functional.
FIG. 6.
Growth kinetics of the asr null mutant JE14 (open circles), the asr deletion mutant JE16 (open triangles), and the parental JE17 strain (filled circles) at low pH. Cultures were grown in LPM medium buffered at pH 4.9 (A) and pH 4.5 (B). Culture growth was monitored by measuring turbidity as described in Materials and Methods. The values shown are the means of at least three independent experiments. Bars indicate SD.
Cloning and expression analysis of asr gene homologues from other Enterobacteriaceae.
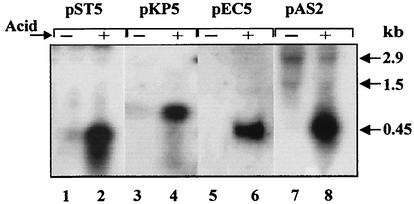
A recent comparative analysis of the E. coli K-12 genome with the genomes of Klebsiella pneumoniae and the Salmonella enterica serovars Typhimurium, Typhi, and Paratyphi led to the conclusion that there were no close homologues of the E. coli asr gene (29). Nevertheless, we found proteins with significant homologies in Enterobacter cloacae (76% amino acid identity), Klebsiella pneumoniae (63% amino acid identity), Salmonella enterica serovars Typhimurium, Typhi, Paratyphi, and Enteritidis (∼58% amino acid identity), and Yersinia pestis (54% amino acid identity). An alignment of these proteins by the CLUSTAL W multiple sequence alignment program (http://clustalw.genome.ad.jp/) is presented in Fig. 3. The promoter region, which has been found to be important for acid regulation of the E. coli asr gene, is also highly conserved among all the examined bacteria (data not shown). This finding prompted us to test whether the asr gene homologues in other Enterobacteriaceae are acid regulated. The putative asr homologues from K. pneumoniae, E. cloacae, and S. enterica serovar Typhi were inserted into pUC19 vector, yielding plasmids pKP5, pEC5, and pST5, respectively (see Materials and Methods). After transformation into E. coli JE14 cells and acid induction, total RNA was examined by Northern analysis. As shown in Fig. 7, lanes 2, 4, and 6, an acid shift from pH 7.0 to 4.5 induces the synthesis of specific transcripts in JE14 cells harboring plasmids pST5 (S. enterica serovar Typhi), pKP5 (K. pneumoniae), and pEC5 (E. cloacae) as well as in control cells with the pAS2 plasmid harboring the E. coli asr gene (Fig. 7, lane 8). These mRNAs are long enough to encode the polypeptides which were deduced from the putative gene sequences. Antibodies developed against the E. coli Asr-His6 protein detected bands of ∼18, ∼22, and 15 kDa in acid-induced E. coli JE14 cells harboring plasmids pEC5 (E. cloacae), pKP5 (K. pneumoniae), and pST5 (S. enterica serovar Typhi), respectively. E. cloacae and S. enterica serovar Typhi asr gene homologues cloned into low-copy-number plasmids (pLEC5 and pLST5; Table 1) rescued the growth of E. coli asr mutant strain JE14 to the level of wild-type strain (MC4100/pGB2) in LPM medium (pH 4.6), while the plasmid with the K. pneumoniae asr gene (pLKP5) did not restore its acid resistance (data not shown).
FIG. 7.
Expression analysis of asr gene homologues from S. enterica serovar Typhi, K. pneumoniae, and E. cloacae. The results of Northern blot analysis of total cellular RNA isolated from E. coli JE14 cells with plasmids pST5, pKP5, and pEC5 are shown. Cells were grown in LPM medium (pH 7.0) (−) to an optical density at A600 of 0.5, and then the pH of the medium was shifted from pH 7.0 to 4.5 (+). A total of 20 μg of RNA was loaded into each lane. The 32P-labeled 570-bp Eco24I-GsuI S. enterica serovar Typhi DNA fragments (lanes 1 and 2), 730-bp Bsp143I K. pneumoniae DNA fragments (lanes 3 and 4), and 900-bp Bsp143I E. cloacae DNA fragments (lanes 5 and 6) were used as hybridization probes. Total RNA from acid-induced (lane 8) and control (lane 7) JE14/pAS2 cells was probed with 32P-labeled 706-bp Csp6I E. coli asr DNA for comparison. Positions of the RNA markers are indicated on the right.
DISCUSSION
We have shown that E. coli induces the asr gene in response to environmental acidity (pH < 5.0) (42). Here we demonstrate that this gene encodes a 102-amino-acid long preprotein with a molecular mass of 10.6 kDa and a pI of ∼10.5, which is secreted into the periplasm and processed a second time to yield an 8-kDa product. We show that this 8-kDa product is needed for bacterial growth at moderate acidity and, furthermore, that asr is needed for the adaptation process at pH 4.5 that enables log-phase-grown E. coli cells to survive at extremely low pH (pH 2.0).
Recent proteomic and biochemical analyses of E. coli cells exposed to acidic pH have revealed a large number of inducible ASPs. These proteins belong to different groups, including those of cytoplasmic enzymes in amino acid catabolism, membrane-bound transporters, periplasmic proteins, and extracellular components (9, 34, 38), although the exact function is not yet known for many of them. The large number of ASPs may reflect the need for different acid-defense systems, depending on the conditions to which cells are exposed, e.g., the level of acidity, the growth phase, and the availability of nutrients or the O2 supply (3, 8). Our present observations, showing the Asr protein is required for growth of E. coli in acidified LPM medium, but not in LB medium, further extends understanding of the complexity of the acid response in this organism. asr also appears to be the most highly inducible of all genes in the bacterial genome when cells grown in LPM medium are shifted from neutral pH (7.0) to acidic pH (4.5), as determined by ongoing microarray analysis (V. Šeputienė, unpublished data). Notably, this was true when cells were grown in acidified minimal medium (44) and LPM medium, but not in LB medium (42), indicating that maximal induction of asr might be required for acid protection in specific environment.
ATR, also known as the acid habituation phenomenon, was first observed in S. enterica serovar Typhimurium and E. coli (19, 22) and was later reported in a broad range of other enteric bacteria (for reviews, see references 18 and 20). When cells are left to adapt at a pH just below 5.0, numerous ASPs are synthesized, and these are needed to efficiently protect the cells from a subsequent treatment with otherwise lethal levels of acid (8). Studies of acid survival in S. enterica serovar Typhimurium, E. coli, and S. flexneri demonstrated that different types of ATR systems exist in log-phase and stationary-phase cells (27). S. enterica serovar Typhimurium, for example, induces approximately 60 ASPs during log phase and 48 ASPs during stationary phase following acid treatment, and only a few of these proteins are common between the groups (3).
The ATR appeared generally less effective in E. coli than in S. enterica serovar Typhimurium (27). The same study also revealed a large strain variation; for example, the frequently used E. coli laboratory strain MC4100 was shown to adapt to acid very poorly. However, our work demonstrates that exponentially grown MC4100 cells adapt well in supplemented minimal medium, such as LPM.
Only a few of the individual proteins induced during the ATR have been shown to be needed for survival in acids. The S. enterica serovar Typhimurium RpoS, PhoP, and Fur proteins have all been shown to be acid induced and to be needed for the log-phase ATR (7, 23, 26). Recently, the OmpR response regulator protein was reported to be required (as a first ASP) for efficient induction of the stationary-phase ATR of S. enterica serovar Typhimurium (5, 6). Which specific ASPs are needed for the ATRs in E. coli is even less defined.
Cells in the exponential phase induce the asr gene at pH 5.0 to 4.5, which leads to protection when cells are subsequently transferred to pH 2.0. Cells growing in stationary phase also induce the asr gene at pH 5.0 to 4.5, as determined by Northern analysis (data not shown), but asr appears not to be required for survival when cells are transferred to pH 2.0. In the latter case, efficient acid resistance systems such as glutamate and arginine decarboxylase systems, which have been shown to protect stationary grown E. coli cells from extreme acid (pH 2.0) even without adaptation, most likely predominate (11). According to recent reports, neither of these low-pH-inducible systems appears to function in protecting exponential-phase E. coli cells grown in minimal medium or LB glucose medium from strong acid, even though some of them, such as glutamate decarboxylases, are induced in these conditions (11). We did not succeed in finding a link between these decarboxylase systems and asr, since neither the presence nor absence of glutamate (1.5 mM) and arginine (0.6 mM) in the challenge medium (glucose minimal medium, pH 2.0) had an effect on the relative log-phase ATR survival of the asr mutant strain compared to that of its parent strain when grown and adapted in LPM medium. In a similar experiment, the asr mutant strain and its parent strain showed nearly equally (80 to 100%) robust glutamate- and arginine-dependent stationary-phase acid resistance (data not shown), indicating that gad and adiA acid resistance systems function independently of asr.
The fact that Asr is also needed for full bacterial growth at the pH values at which adaptation occurs (4.7 to 4.5) suggests that the Asr protein is functional over a wide pH range.
Our database searches did not reveal any significant sequence similarity between Asr and proteins with known functions (42). We showed Asr is present in the periplasm or, alternatively, has domains protruding into the periplasm. The periplasm is the first barrier to external acidification (16), and the high number of basic amino acids, such as lysine, suggests that Asr acts by sequestering protons in this compartment. In this context, it might act as a chaperone to protect essential proteins against acid. The abundant acid-inducible 110-amino-acid periplasmic E. coli protein HdeA, like its counterpart in Shigella, is needed for full acid resistance and is essential for virulence. HdeA displays a chaperone-like activity by preventing the aggregation of acid-denatured proteins (21, 45). This proposed role in the periplasm is also supported by the fact that asr is induced only by external, but not internal, acidification (42).
The Asr precursor protein is rapidly formed in the cytoplasm following acid treatment but undergoes a removal of the signal peptide and processing to an 8-kDa polypeptide. Processing of the Asr precursor is required for its functional activity, since an E. coli mutant defective in the production of the 8-kDa derivative and the asr null mutant are equally acid sensitive. The 15-kDa form is basic all along the protein, but it may be that conversion to the shorter 8-kDa peptide facilitates the association with acid-denatured proteins. The mechanism by which the second cleavage occurs is unclear, since the cleavage site does not resemble any typical consensus motif of known peptidases (the Proteases information database can be found at the following Web address: http://merops.iapc.bbsrc.ac.uk/). The well-studied periplasmic stress protease DegP (40) is not involved in the Asr cleavage, as determined from the analysis of Asr in acid-induced degP mutant cells (data not shown). The fact that most amino acids encompassing this cleavage site (Fig. 3) are conserved in the Asr homologues in other Enterobacteriaceae suggests that this protease can also be found in other bacterial species.
The observation that the Asr protein is converted into a functional product by two-step cleavage could also be an indication of an autocatalytic processing. However, we have not been able to detect any specific degradation of affinity-purified Asr after prolonged incubation in 0.05 M citrate buffer (pH 5.0) or 0.05 M Tris buffer (pH 7.0) at 20°C and 37°C. Testing the ability of Asr to cleave other substrates, such as azocasein, also did not show any such protease activity (data not shown).
The amino acid sequence of E. coli Asr was found to be 76 to 54% identical with its homologues from E. cloacae, K. pneumoniae, S. enterica (serovars Typhimurium, Typhi, Paratyphi, Enteritidis), and Y. pestis. While amino acid homology appears throughout the whole range of proteins, there is substantial variation in the length of the deduced Asr polypeptides, which range from 139 residues in K. pneumoniae to 82 residues in S. enterica serovars Typhimurium and Enteritidis. The asr promoter region is also highly conserved (data not shown) in the examined species, suggesting that all of the genes are acid regulated. This was tested by cloning these asr homologues into E. coli and by confirming they are induced by low pH. E. cloacae and S. enterica serovar Typhi asr cloned into the low-copy-number plasmids pLEC5 and pLST5 rescued the growth defect of E. coli asr mutant in LPM medium at pH 4.6, while K. pneumoniae asr (pLKP5) was not able to restore its acid resistance to the wild-type level.
These bacteria exhibit different sensitivities to acidity, and different types of molecular systems to combat acidity have been proposed (11, 27). The asr homologues might well play the same role in other enteric bacteria as in E. coli. However, our asr null mutant of S. enterica serovar Typhimurium (strain ST 14028) did not exhibit any acid-sensitive phenotype when grown in culture (data not shown) indicating that at least in Salmonella, E. coli, and K. pneumoniae, the relative roles of asr may be distinct.
Acknowledgments
The Lithuanian state research program “Molecular Principles of Biotechnology” (grant 219/2490/-2), The Swedish Institute, and The Royal Swedish Academy of Sciences supported this work. V.S. was a fellow of the Lithuanian State Science and Studies Foundation.
REFERENCES
- 1.Adams, P., R. Fowler, N. Kinsella, G. Howell, M. Farris, P. Coote, and C. D. O'Conor. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, C. W., P. R. Baum, and R. F. Gesteland. 1973. Processing of adenovirus 2-induced proteins. J. Virol. 12:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 4.Baily, S. C., and D. Apirion. 1977. Identification of lipopolysaccharides and phospholipids of Escherichia coli in polyacrylamide gels. J. Bacteriol. 131:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang, I. S., J. P. Audia, Y. K. Park, and J. W. Foster. 2002. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol. 44:1235-1250. [DOI] [PubMed] [Google Scholar]
- 6.Bang, I. S., B. H. Kim, J. W. Foster, and Y. K. Park. 2000. OmpR regulates The stationary-phase acid-tolerance response of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:2245-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 9.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casabadan, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Churchward, G., D. Belin, and Y. A. Nagamine. 1984. pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 221:165-171. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, A., and J. Clark. 1986. Synthesis of linear plasmid multimers in Escherichia coli K-12. J. Bacteriol. 167:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 15.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 16.Dilworth, M. J., and A. R. Glenn. 1999. Problems of adverse pH and bacterial strategies to combat it, p. 4-18. In D. J. Chadwick and G. Cardew (ed.), Bacterial responses to pH (Novartis Foundation Symposium 221). Wiley, Chichester, United Kingdom. [DOI] [PubMed]
- 17.Eun Rhee, J., J. Haeng Rhee, P. Youl Ryu, and S. Ho Choi. 2002. Identification of the cadBA operon from Vibrio vulnificus and its influence on survival to acid stress. FEMS Microbiol Lett. 208:245-251. [DOI] [PubMed] [Google Scholar]
- 18.Foster, J. W. 2001. Acid stress responses of Salmonella and E. coli: survival mechanisms, regulation, and implication for pathogenesis. J. Microbiol. 39:89-94. [Google Scholar]
- 19.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, J. W., and M. Moreno. 1999. Inducible acid tolerance mechanisms in enteric bacteria, p. 55-74. In D. J. Chadwick and G. Cardew (ed.), Bacterial responses to pH (Novartis Foundation Symposium 221). Wiley, Chichester, United Kingdom. [DOI] [PubMed]
- 21.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 22.Goodson, M., and R. J. Rowbury. 1989. Habituation to normal lethal acidity by prior growth of Escherichia coli at a sublethal acid pH value. Lett. App. Microbiol. 8:77-79. [Google Scholar]
- 23.Hall, H. K., and J. W. Foster. 1996. The role of Fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickey, E. W., and I. N. Hirshfield. 1990. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl. Environ. Microbiol. 56:1038-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 27.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClelland, M., L. Florea, K. Sanderson, S. W. Clifton, J. Parkhill, C. Churcher, G. Dougan, K. Wilson, and W. Miller. 2000. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acid Res. 28:4974-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng, S.-Y., and G. N. Bennett. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 32.Nakai, K., and M. Kanehisa. 1991. Expert system for the predicting protein localization sites in gram-negative bacteria. Proteins Struct. Funct. Genet. 11:95-110. [DOI] [PubMed] [Google Scholar]
- 33.Park, Y. K., B. Bearson, S. H. Bang, I. S. Bang, and J. W. Foster. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20:605-611. [DOI] [PubMed] [Google Scholar]
- 34.Rowbury, R. J. 1999. Acid tolerance induced by metabolites and secreted proteins, and how tolerance can be counteracted, p. 93-106. In D. J. Chadwick and G. Cardew (ed.), Bacterial responses to pH (Novartis Foundation Symposium 221). Wiley, Chichester, United Kingdom. [DOI] [PubMed]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 37.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at extreme pH, p. 1539-1549. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 38.Stancik, L., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stim, K. P., and G. N. Bennett. 1993. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J. Bacteriol. 175:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studier, F. W., and B. A. Moffat. 1986. Use of bacteriophage T7 RNA polymerase to direct high-level expression of cloned genes. J. Mol. Biol. 198:113-130. [DOI] [PubMed] [Google Scholar]
- 42.Sužiedėlienė, E., K. Sužiedėlis, V. Garbenčiūtė, and S. Normark. 1999. The acid-inducible asr gene in Escherichia coli: transcriptional control by the phoBR operon. J. Bacteriol. 181:2084-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterman, S. R., and P. L. Small. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect. Immun. 64:2808-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanisch-Perron, C., J. Vierra, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]