Abstract
Most flagellar proteins of Salmonella are exported to their assembly destination via a specialized apparatus. This apparatus is a member of the type III superfamily, which is widely used for secretion of virulence factors by pathogenic bacteria. Extensive studies have been carried out on the export of several of the flagellar proteins, most notably the hook protein (FlgE), the hook-capping protein (FlgD), and the filament protein flagellin (FliC). This has led to the concept of two export specificity classes, the rod/hook type and the filament type. However, little direct experimental evidence has been available on the export properties of the basal-body rod proteins (FlgB, FlgC, FlgF, and FlgG), the putative MS ring-rod junction protein (FliE), or the muramidase and putative rod-capping protein (FlgJ). In this study, we have measured the amounts of these proteins exported before and after hook completion. Their amounts in the culture supernatant from a flgE mutant (which is still at the hook-type specificity stage) were much higher than those from a flgK mutant (which has advanced to the filament-type specificity stage), placing them in the same class as the hook-type proteins. Overproduction of FliE, FlgB, FlgC, FlgF, FlgG, or FlgJ caused inhibition of the motility of wild-type cells and inhibition of the export of the hook-capping protein FlgD. We also examined the question of whether export and translation are linked and found that all substrates tested could be exported after protein synthesis had been blocked by spectinomycin or chloramphenicol. We conclude that the amino acid sequence of these proteins suffices to mediate their recognition and export.
Bacteria swim by rotating their flagella, which consist of at least three substructures: a basal body, a hook, and a thin helical filament (24). The basal body is imbedded in the cell surface, whereas the hook and filament extend outwards from the cell. Flagellar assembly begins with the basal body, followed by the hook and finally the filament.
Almost all of the external components of the flagellum cross the cytoplasmic membrane into the central channel of the nascent flagellar structure by a specialized apparatus, which consists of six integral-membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, and FliR) and three cytoplasmic proteins (FliH, FliI, and FliJ) (28). The membrane components are presumed to be located in a central pore within the basal-body MS ring and function as a protein-conducting device; experimental evidence supports this presumption in the cases of FlhA, FliP, and FliR (8, 18, 28). FliI is an ATPase which provides the energy for the translocation of export substrates across the cytoplasmic membrane (7, 36). FliH is a regulatory protein that is believed to prevent FliI from hydrolyzing ATP until the flagellar export apparatus is competent to link this hydrolysis to the translocation of export substrates (10, 29). FliJ functions as a general chaperone to prevent export substrates from premature aggregation in the cytoplasm (25).
The flagellar export apparatus switches its substrate specificity upon completion of hook assembly. At least two components, the integral-membrane component FlhB and the hook length control protein FliK, are believed to be involved in this process (21, 26, 40). In the first phase, FlhB supports the assembly of the rod and hook structures and presumably, therefore, the export of their component proteins. The export properties of the hook-type proteins (the hook protein, FlgE, the hook-capping protein, FlgD, and the hook length control protein, FliK) have been firmly established (27, 28, 33). However, this is not the case for the rod-type proteins (see below).
Upon completion of the hook structure, the C-terminal domain of FliK communicates with the C-terminal cytoplasmic domain of FlhB (FlhBC), resulting in a conformational change of FlhBC, which is responsible for the specificity switching to filament-type substrates: the anti-σ factor (FlgM), the first hook-filament junction protein (FlgK), the second hook-filament junction protein (FlgL), the filament-capping protein (FliD), and flagellin (FliC) (21).
The presumption that rod-type proteins belong to the same export specificity class as hook-type proteins is based on the fact that during flagellar morphogenesis, rod assembly precedes hook assembly (16, 38, 39) and on the fact that hook-type proteins can still be exported to the periplasm in rod mutants (28). However, because of difficulties in detecting the rod-type proteins, there has been no direct experimental evidence as to whether they are predominantly exported prior to hook-basal body completion (28). In the present study, we have been able to demonstrate that the putative MS ring-rod junction protein (FliE), the rod proteins (FlgB, FlgC, FlgF, and FlgG), and the putative rod-capping protein (FlgJ) are exported efficiently prior to the completion of hook assembly and poorly thereafter. This confirms that they belong to the same export specificity class as the hook-type proteins and therefore that the term “rod/hook type” is an appropriate one overall with respect to the export process.
Another issue is whether, in vivo, flagellar protein export occurs cotranslationally or whether the fully translated protein (dissociated from the ribosome) is competent to associate with the export apparatus and undergo translocation. It is known from in vitro experiments that export substrates bind to soluble components of the flagellar export apparatus, namely, FlhAC, FlhBC, FliH, FliI, and FliJ and the substrate-specific chaperones FlgN, FliS and FliT (2, 3, 9, 30, 36), suggesting that export is not obligatorily linked to translation. By the use of the protein synthesis inhibitor spectinomycin, we show that this is indeed the case.
MATERIALS AND METHODS
Bacterial strains, plasmids and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Luria broth (LB), soft tryptone agar plates, and M9 medium-0.3% Casamino Acids-1% glycerol (M9 complete medium) were prepared as described before (28). Ampicillin and spectinomycin (Wako) were added at final concentrations of 50 μg/ml and 100 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strains and plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| Salmonella | ||
| JR501 | Conversion of E. coli-derived plasmids to compatibility with Salmonella | 35 |
| SJW1103 | Wild type for motility and chemotaxis | 41 |
| SJW1353 | flgE | 33 |
| SJW2177 | flgK | 13 |
| Plasmids | ||
| pET22b | Cloning vector | Novagen |
| pTrc99A | Cloning vector | Pharmacia |
| pTrc99AFF4 | Cloning vector | 32 |
| pTrc99AET | Cloning vector (see Materials and Methods) | This study |
| pMM202 | pTrc99AFF4/FlgG | This study |
| pMM204 | pET19b/His-FlgG | 31 |
| pMM501 | pET19b/His-FlgJ | This study |
| pMM504 | pTrc99AFF4/FlgJ | This study |
| pMM507 | pET22b/FlgJ | 11 |
| pMM711 | pET19b/His-FlgD | 29 |
| pMM1001 | pTrc99AFF4/FliE | 31 |
| pMM1002 | pET19b/His-FliE | 30 |
| pMM1101 | pTrc99AFF4/FlgB | 31 |
| pMM1102 | pET19b/His-FlgB | 30 |
| pMM1201 | pTrc99AFF4/FlgC | This study |
| pMM1202 | pET19b/His-FlgC | 31 |
| pMM1301 | pTrc99AFF4/FlgF | This study |
| pMM1302 | pET19b/His-FlgF | 31 |
| pMM1901 | pET19b/His-FlgL | 30 |
| pMM1903 | pTrc99AFF4/FlgL | This study |
| pRCD1 | pTrc99A/FlgD | This study |
| pTH090 | pTrc99AET/FlgJ | This study |
| pTH130 | pET22b/FliE | This study |
| pTH190 | pTrc99AET/FliE | This study |
| pTH230 | pET22b/FlgB | This study |
| pTH290 | pTrc99AET/FlgB | This study |
| pTH330 | pET22b/FlgC | This study |
| pTH390 | pTrc99AET/FlgC | This study |
| pTH430 | pET22b/FlgG | This study |
| pTH490 | pTrc99AET/FlgG | This study |
| pTH530 | pET22b/FlgF | This study |
| pTH590 | pTrc99AET/FlgF | This study |
| pTH730 | pET22b/FlgD | This study |
| pTH790 | pTrc99AET/FlgD | This study |
| pTH1230 | pET22b/FlgL | This study |
| pTH1290 | pTrc99AET/FlgL | This study |
DNA manipulations.
Procedures for DNA manipulation in vitro were carried out as described previously (28). Primers used in the constructions are listed in Table 2. pET22b-based plasmids were derived from the corresponding pET19b-based plasmids by preparing NdeI-BamHI fragments and subcloning them into pET22b. pTrc99AFF4-based plasmids were constructed by inserting a fragment containing the gene of interest flanked by an NdeI site (containing the start codon) and a BamHI site (downstream of the stop codon). The genes were transcribed from the pTrc promoter and translated using the ribosome-binding sequence deriving from pTrc99AFF4.
TABLE 2.
Primers used in this study
| Primer name | Primer sequencea |
|---|---|
| SFLGG-NdeA | GGC ATA TGA TGA TCA GGT CAT TAT GGA |
| SFLGG-BamB2 | GCC GGT GGC GGA TCC CCC ACC GGC |
| SFLGJ-NdeA1 | GGA AAT CAT CAT ATG ATC GGA GAC |
| SFLGJ-BamB1 | AGC GAC TAC GGA TCC TTG AGC AAT |
| SFLIE-NdeA1 | CAG GGG AAT CAT ATG GCA GCA ATA |
| SFLIE-BamB1 | GTG TTG TCA GGA TCC GCT TAA ACC |
| SFLGB-NdeA1 | TGC GGA GGA CAT ATG CTC GAC AGG |
| SFLGB-BamB2 | TAA CAG CGC GGA TCCTTA GTT TCC |
| SFLGC-NdeA1 | GGA AAC TAA CAT ATG GCG CTG TTA |
| SFLGC-BamB1 | GAC ATA CGC GGA TCC TTT ACT GGC |
| SFLGF-NdeA1 | CGG GAT AGC CAT ATG GAT CGC GCA |
| SFLGF-BamB1 | AAA ATG TGG ATC CTT AAC TCA TCG |
| SFLGD-NdeA | AAG GAG GCG CAT ATG TCT ATT GCC |
| SFLGD-BamB | CTG ACT CCT GGA TCC TGT AAG GGC |
| SFLGL-NdeA | GGA GAA GGA TCA TAT GCG TAT CAG |
| SFLGL-BamB | TTC GTG ATA GGA TCC AAA AAG AGG |
Restriction sites are indicated by underlining; start and stop codons (where present on the primer) are indicated by italics.
To get higher expression levels, modified pTrc99A-based plasmids were constructed as follows. First, an XbaI-HindIII fragment was prepared from a pET22b-based plasmid containing the intended downstream gene. This fragment was then inserted into the XbaI-HindIII sites of pTrc99A. The genes were thus transcribed from the pTrc promoter but were translated using the ribosome-binding sequence derived from pET22b, resulting in very high levels of expression. We describe these as pTrc99AET-based plasmids.
Analysis of multicopy effects of export substrates on swarming.
SJW1103 was transformed with various plasmids, and the resulting transformants were inoculated onto soft tryptone agar plates containing ampicillin and incubated at 30°C for 6 h.
Preparation of culture supernatants.
Cells carrying pTrc99AFF4-based plasmids encoding untagged flagellar proteins were grown in 3 ml of M9 complete medium containing ampicillin at 30°C with shaking, and 2 ml was harvested at an optical density at 600 nm (OD600) of 0.6 and washed twice with M9 complete medium. Then, the cells were resuspended in 2 ml of M9 complete medium containing ampicillin and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), incubated at 30°C for 1 h (FlgF, FlgG, FlgJ, FlgD, and FlgL) or 3 h (FliE, FlgB, and FlgC), and fractionated into cell pellet and culture supernatant by low-speed centrifugation. Trichloroacetic acid was added to the supernatants to 10%, the supernatants were incubated at 4°C overnight and centrifuged for 30 min at 4°C, and the pellets were washed with 1 ml of ice-cold acetone and resuspended in sodium dodecyl sulfate (SDS) sample buffer containing 8 M urea.
Treatment of cells with spectinomycin.
To examine the effect of spectinomycin on flagellar protein export, cells harboring pTrc99A-based plasmids were grown with shaking in 5 ml of LB containing ampicillin at 37°C until the cell density had reached an OD600 of ca. 0.6, at which point IPTG was added to a final concentration of 1 mM and incubation was continued for another 2 h with shaking. Then spectinomycin was added to a final concentration of 0 or 100 μg/ml, and incubation was continued further for 30 min without shaking, by which time the arrest of cell growth of the spectinomycin-containing sample was essentially complete (in growth experiments starting with an initial OD600 of 0.24, the spectinomycin-containing sample had reached a value of 0.33 after 30 min and 0.40 after 2 h; by comparison, the untreated culture continued to grow, reaching an OD600 of 1.12 after 2 h). The spectinomycin-untreated and -treated cells were washed twice with phosphate-buffered saline, suspended in 5 ml of M9 complete medium containing ampicillin and 0 or 100 μg of spectinomycin/ml, respectively, and incubated at 37°C. During this last incubation period, samples were taken at 0, 30, 60, and 90 min and were centrifuged to obtain the cell pellets and culture supernatants. Cell pellets were resuspended in SDS-loading buffer normalized by the cell density to give a constant amount of cells. The proteins in the culture supernatants were precipitated by 10% trichloroacetic acid, suspended in Tris-SDS loading buffer, and heated at 95°C for 5 min, as described previously (28). Ca. 1% of the cell pellets and 50% of the supernatants were used for analysis.
Immunoblotting.
Immunoblotting with anti-FlgG, -FlgD, -FlgL, and -FliC antibodies was carried out as described previously (27). Immunodetection was performed with an ECL immunoblotting detection kit (Amersham International).
RESULTS
Export assay of rod-type proteins.
Komoriya et al. (19) demonstrated that they were able to detect three of the rod proteins (FlgC, FlgF, and FlgG) in the culture supernatants of Salmonella cells. This encouraged us to attempt export assays, before and after hook completion, of all four rod proteins (FlgB, FlgC, FlgF, and FlgG) and also of the muramidase and putative rod-capping protein (FlgJ) and the putative MS ring-rod junction protein (FliE), which was believed to be an export substrate, although this had never been directly demonstrated (30). A known hook-type protein, the hook-capping protein FlgD, and a known filament-type protein, the hook-filament junction protein FlgL, were used as controls. pTrc99AFF4-based plasmids (see Materials and Methods) were constructed encoding these proteins. For hosts, SJW1353 (flgE) was used as an example of a strain blocked in the process of hook assembly which therefore remained in the hook-type specificity state and SJW2177 (flgK) was used as an example of a strain in which hook assembly has been completed and therefore the switch to the filament-type specificity has occurred; this strain also has the advantage that because there is no structure beyond the hook (FlgK being the first-hook-filament junction protein), export into the culture supernatant is especially efficient.
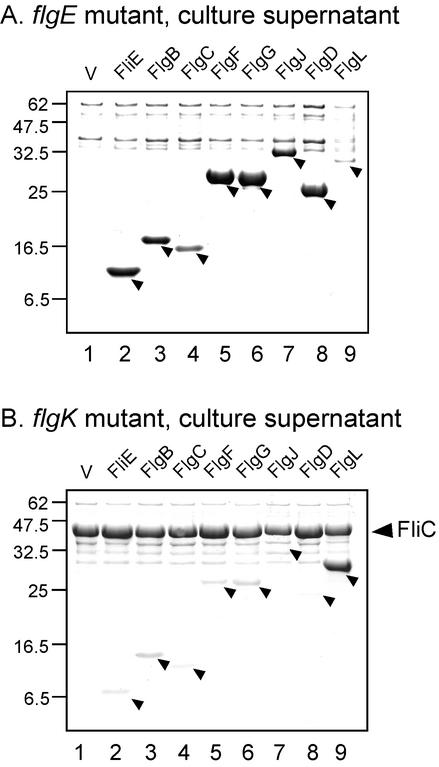
In the culture supernatants, the amounts of FliE, FlgB, FlgC, FlgF, FlgG, and FlgJ and the hook-type protein FlgD were much higher with the flgE mutant (Fig. 1A) than with the flgK mutant (Fig. 1B). The opposite was true for the filament-type protein FlgL, for which amounts in the culture supernatant were much higher with the flgK mutant than with the flgE mutant. Presumably because of its high level of production from the chromosomal fliC gene, flagellin was seen with the flgK mutant, regardless of the plasmid used; this was not true with the flgE mutant, with which expression of fliC is repressed by the anti-sigma factor FlgM. Using His-tagged versions and anti-His immunoblotting, we measured the cellular levels of the various proteins in the two hosts and found no difference (data not shown). Thus, the export specificity properties of FliE, the rod proteins, and FlgJ are the same as those of hook-type proteins and opposite from those of filament-type proteins. We conclude that the rod-type proteins are exported predominantly during rod and hook assembly and that the broad term “rod/hook-type specificity” has now been validated experimentally.
FIG. 1.
Coomassie-stained gels of the culture supernatants of SJW1353 (flgE) (A) and SJW2177 (flgK) (B) mutants carrying pTrc99AFF4-based plasmids producing various flagellar proteins under induction with 1 mM IPTG. The proteins produced are identified above each lane, and their positions are indicated by arrowheads. V, vector alone. Molecular mass markers (in kilodaltons) are shown to the left.
Multicopy effects on swarming of wild-type SJW1103 cells.
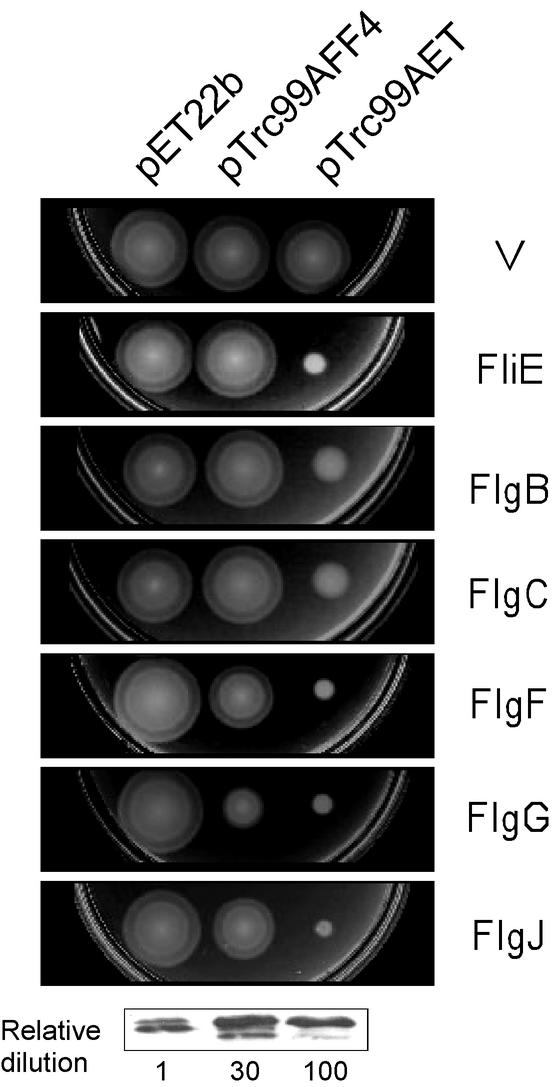
The fact that the putative MS ring junction protein, the four rod proteins, and the putative rod-capping protein belong to the same class as the three hook-type proteins means that there is at a minimum a set of nine different proteins that conceivably are being exported down a narrow channel simultaneously. This raises a logistical issue, especially if one of the export substrates is being overproduced. To investigate whether interference occurs under these conditions, we examined the consequence for the motility of wild-type cells of overproducing FliE, FlgB, FlgC, FlgF, FlgG, and FlgJ. We used three kinds of plasmids: (i) pET22b-based plasmids, (ii) pTrc99AFF4-based plasmids, and (iii) pTrc99AET-based plasmids. Members of the last class of plasmids have an optimized ribosome-binding site deriving from pET22b. Protein production levels differ considerably among these plasmids (pET-based plasmids do not express at high levels with hosts lacking T7 RNA polymerase). Using anti-FlgG antibody, for example, we carried out immunoblots of cellular levels and estimate that the relative FlgG production levels were in the approximate ratio of 1:30:100 (Fig. 2, bottom panel).
FIG. 2.
Multicopy effects of rod-type proteins. The results of assays for determination of the swarming ability of the wild-type strain SJW1103 transformed with pET22b-, pTrc99AFF4-, and pTrc99AET-based plasmids are shown. The individual panels correspond to plasmids encoding the proteins shown at the right. V, vector alone. (In the third column, pTrc99A, not pTrc99AET, was used). Cells were inoculated onto soft tryptone agar plates containing 0.1 mM IPTG and incubated for 6 h at 30°C. Bottom panel: intracellular levels of FlgG produced in the same hosts as in the upper panels. A fixed number of cells was serially diluted at 1/1, 1/3, and 1/10 through to 1/300 and subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting with polyclonal anti-FlgG antibody.
Salmonella wild-type strain SJW1103 was transformed with these plasmids, inoculated onto soft tryptone agar plates containing ampicillin and 0.1 mM IPTG, and incubated at 30°C for 6 h (Fig. 2). pET22b-based plasmids had essentially no effect on the swarming of wild-type cells. With pTrc99AFF4-based plasmids, swarming was inhibited to various degrees: FliE, FlgB, and FlgC had no effect, whereas FlgG caused ca. 70% inhibition. With pTrc99AET-based plasmids, strong inhibition was seen in all cases, ranging from ca. 70% (e.g., with FlgB or FlgC) to ca. 90% with several of the other proteins. Thus, high levels of overproduction of FliE, FlgB, FlgC, FlgF, FlgG, and FlgJ result in strong multicopy effects on the motility of the wild-type cells. We also obtained the same results with the hook-capping protein FlgD, the hook-length control protein FliK, and the hook-filament junction proteins FlgK and FlgL (data not shown). Failure to swarm was in all cases a consequence of failure to assemble flagella, as judged by dark-field light microscopy (23) (data not shown). These results suggest that excess of one export substrate either interferes directly with export of other substrates or sequesters some other essential components of the export system.
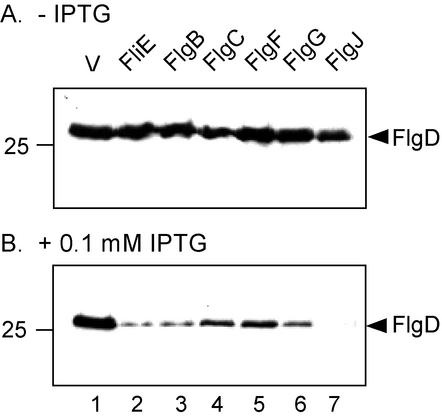
By immunoblotting with anti-FlgD antibody (Fig. 3), we analyzed the amount of the hook-capping protein FlgD in the culture supernatants of SJW1353 (flgE) cells carrying pTrc99AET-based plasmids and grown exponentially at 30°C in M9 complete medium containing ampicillin and 0.1 mM IPTG.
FIG. 3.
Immunoblotting (using anti-FlgD antibody) of the culture supernatants from SJW1353 (flgE) cells transformed with pTrc99AET-based plasmids and grown at 37°C in the presence (+) and absence (−) of 0.1 mM IPTG. The proteins produced by the various plasmids are shown above each lane. V, vector alone (pTrc99A). The positions of FlgD are indicated by arrowheads. Molecular mass markers (in kilodaltons) are shown to the left.
In the absence of IPTG, FlgD was detected in all of the culture supernatants (Fig. 3A). However, in the presence of 0.1 mM IPTG, FlgD was detected strongly in the culture supernatant with the pTrc99A vector control (Fig. 3B, lane 1) but weakly or not at all when the host carried pTrc99AET-based plasmids encoding the putative MS ring-rod junction protein FliE, the rod proteins, or the putative rod-capping protein FlgJ. At this concentration of IPTG, growth and intracellular levels of FlgD were essentially unaffected (data not shown). Thus, overproduction of any one of these proteins abolishes or severely reduces the export of FlgD and, by extension (based on the swarm data presented in Fig. 2), the other rod/hook-type proteins.
Effect of spectinomycin on flagellar protein export.
To test whether flagellar protein export in vivo occurs cotranslationally or posttranslationally, we investigated the export of substrates in the presence of spectinomycin, an antibiotic which prevents protein synthesis by inhibiting the elongation factor G cycle and the peptidyl tRNA translocase reaction (4, 5). For substrates, we chose FlgG, FlgD, and FlgL as examples of rod-type, hook-type, and filament-type substrates, respectively. SJW1353 (flgE) was the host for examining FlgG and FlgD export, and SJW2177 (flgK) was the host for examining FlgL export. To detect these proteins more efficiently, we transformed the flgE and flgK mutants with pTrc99AFF4-based plasmids pMM202 (FlgG), pRCD1 (FlgD), and pMM1903 (FlgL), respectively.
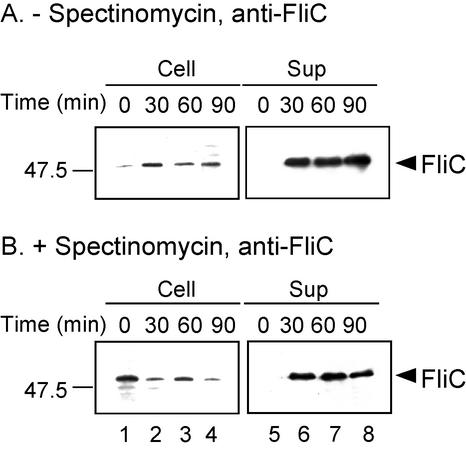
The resulting transformants were grown exponentially in LB and induced with IPTG. Then, one portion was treated with spectinomycin and incubation was continued for 30 min, at which point growth had essentially ceased (see Materials and Methods). The cells were then washed with phosphate-buffered saline, resuspended in M9 complete medium plus 0 or 100 μg of spectinomycin/ml, and incubated at 37°C. Thus, protein detected in the supernatants of the spectinomycin-containing cultures must have been exported posttranslationally. Cellular and culture supernatant fractions were prepared at the zero time point of this last incubation and at 30, 60, and 90 min thereafter. Proteins in these fractions were subjected to SDS-polyacrylamide gel electrophoresis and were analyzed by immunoblotting with anti-FlgG, -FlgD, and -FlgL antibodies (Fig. 4).
FIG. 4.
Immunoblotting (using anti-FlgG [A and B], anti-FlgD [C and D], and anti-FlgL [E and F] antibodies) of the cells and culture supernatants of SJW1353 (flgE) carrying pMM202 (FlgG) (A and B), SJW1353 (flgE) carrying pRCD1 (FlgD) (C and D), and SJW2177 (flgK) carrying pMM1903 (FlgL) (E and F), respectively, in the presence (+) and absence (−) of 100 μg of spectinomycin/ml. The positions of FlgG, FlgD, and FlgL are indicated by arrowheads. The high-molecular-mass bands in the left panels of Fig. 4A, B, and C represent either aggregates or a cross-reacting species. Molecular mass markers (in kilodaltons) are shown to the left.
Intracellular levels of FlgG decreased over the 90-min interval both in the presence and in the absence of spectinomycin (left panels of Fig. 4A and B); whether this was because of instability or export is not clear. Intracellular levels of FlgD and FlgL remained more or less constant in the absence of spectinomycin (Fig. 4C and E, left panels) but decreased somewhat in its presence (Fig. 4D and F, left panels). In the culture supernatants, increasing amounts of FlgG, FlgD, and FlgL were detected both in untreated cells (Fig. 4A, C, and E, right panels) and in spectinomycin-treated cells (Fig. 4B, D, and F, right panels). In the case of FlgG, the amounts did not increase after 30 min in both untreated and treated cells. In the cases of FlgD and FlgL, the amounts continued to increase in the untreated cells but reached a plateau at about 30 min in the spectinomycin-treated cells.
Choosing flagellin (FliC), since it is the most abundant of the exported substrates (Fig. 5), and using strain SJW2177 (flgK), which permits flagellin export but not polymerization, we also tested whether chromosomally encoded flagellar proteins could still be exported in the presence of spectinomycin. Intracellular levels of FliC remained roughly constant in untreated cells (Fig. 5A, left panel) and decreased in treated cells (Fig. 5B, left panel). Substantial amounts of FliC were seen in the culture supernatants of untreated cells, increasing rapidly over the first 30 min and remaining almost constant thereafter (Fig. 5A, right panel); a similar pattern was observed with the treated cells (Fig. 5B, right panel). We are uncertain why they did not continue to rise.
FIG. 5.
Anti-FliC immunoblot of the cells and culture supernatants from SJW2177 (flgK) in the presence (+) and absence (−) of spectinomycin. The positions of flagellin (FliC) are indicated by arrowheads. Molecular mass marker (in kilodaltons) is shown to the left.
Similar results regarding flagellar protein export were obtained when chloramphenicol (an inhibitor of the peptidyl transferase activity of the 50S subunit of the ribosome) in a 100-μg/ml concentration was used (data not shown).
DISCUSSION
Almost all flagellar proteins are exported by the type III flagellar export apparatus, which consists of six integral-membrane components and three cytoplasmic components (28). A switch in the substrate specificity of the export apparatus occurs upon completion of hook assembly. On the basis of this difference, its export substrates have been grouped into two classes, the rod/hook-type class and the filament-type class. However, the inclusion of the rod-type proteins with the hook-type proteins has until now been based on circumstantial evidence and has not had a firm and direct experimental basis.
FliE, FlgB, FlgC, FlgF, FlgG, and FlgJ are most efficiently exported prior to completion of hook-basal body assembly.
they had established previously that hook-type proteins could be exported into the periplasm regardless of whether rod protein export was occurring (28). At that time, they tried unsuccessfully to detect the export of rod-type proteins (the putative MS ring-rod junction protein FliE and the rod proteins FlgB, FlgC, FlgF, and FlgG) into the periplasm. In the present study, we have looked at export into the culture supernatant rather than the periplasm and have improved expression conditions such that we can readily detect export of FliE, FlgB, FlgC, FlgF, and FlgG and the putative rod-capping protein FlgJ in both flgE and flgK mutant backgrounds. We show that their export occurs much more efficiently prior to hook completion (Fig. 1) than thereafter, indicating that their export properties are similar to those of hook-type proteins. This finding strongly supports our prior supposition that the rod-type proteins and the hook-type proteins belong to a single export class; i.e., there is no specificity switch such as occurs between the hook-type proteins and the filament-type proteins.
FliE is probably the first flagellar substrate to be exported.
In a previous study, Minamino et al. showed that FliE is required for export of hook-type substrates (28, 31). This fact, coupled with biochemical evidence that FliE physically interacts with export components (30) and genetic evidence that it interacts with a known export substrate, the proximal rod protein FlgB (31), has led to the strong presumption that FliE is an export substrate. We have now shown directly that it is an export substrate and that it belongs to the rod/hook-type specificity class. Minamino et al. have suggested that it functions as an MS ring-rod junction protein and enables the export of other substrates (31), properties that would make it both an export substrate and a component of the export apparatus. We suggest therefore that FliE is the first export substrate recognized by the flagellar export apparatus. However, since we have no reason to believe that the export apparatus, and specifically FlhB, undergoes a change between export of FliE and export of subsequent rod/hook-type substrates, we prefer at this point to keep it within that class rather than placing it in a class by itself.
Overproduction of some export substrates interferes with the export of the others.
Overproduction of FliE, FlgB, FlgC, FlgF, FlgG, and FlgJ impaired the motility of wild-type cells (Fig. 2). Failure to swarm was in all cases a consequence of failure to export flagellar proteins (Fig. 3), indicating that interference occurs between export substrates. The simplest explanation of these data is that of competition for a pathway with a finite capacity. We have no evidence to suggest that an ordered export for different substrates exists within the rod/hook-type specificity class.
Export of flagellar structural proteins is not obligatorily coupled to protein translation.
In both the Yersinia type III secretion system and the Salmonella flagellar export system, multiple signals (amino acid and mRNA) have been proposed to be used (1, 17, 37), and indeed, Karlinsey et al. (17) have concluded that, at least in the case of FlgM, export is cotranslational.
It has been shown in vitro that flagellar export substrates interact with soluble components or cytoplasmic domains of membrane components of the export apparatus such as FlhAC and FlhBC (30). However, with the exception of FlgM, it has not been established whether, in vivo, substrate recognition by the export apparatus is in any way dependent on translation. We found that translation-inhibited spectinomycin-treated cells carrying pMM202, pRCD1, or pMM1903 retained the ability to export FlgG (a rod-type protein), FlgD (a hook-type protein), or FlgL (a filament-type protein) into the culture supernatants (Fig. 4). This was also true for FliC (flagellin) encoded on the chromosome (Fig. 5). Therefore, we conclude that flagellar protein export can occur posttranslationally.
There is independent evidence to support this conclusion. Loss of FliS, FlgN, and FliT severely impairs the export of FliC, FlgK and FlgL, and FliD, respectively, suggesting that they function as substrate-specific chaperones to facilitate the export of their cognate substrates (2, 3, 9, 42). These chaperones bind to the C-terminal regions of their cognate substrates (2, 3), implying that protein synthesis is complete prior to export.
The importance of amino acid sequence for recognition for export is supported by several lines of evidence. N-terminal polypeptides of flagellar proteins retain the ability to go through the flagellar export pathway (15, 20, 22, 27). Also, single amino acid substitutions within the N-terminal region of the anti-sigma factor FlgM (itself a substrate for export) severely impair its export (6). It appears that the N-terminal amino acid sequences of export substrates function as the signal.
The amino acid sequences of flagellar proteins show some sequence similarities at their termini (12, 14). These reflect their similarities as axial components of the flagellum rather than signals for the export, however, because they are required for flagellar assembly but not for export (20). Therefore, as suggested before (6, 15, 20, 22), a common higher-order motif in the export substrates mediates recognition by the type III flagellar export apparatus. The existence of two export specificities, rod/hook type and filament type, implies that their recognition signals must be different. What that difference is is not obvious from the protein sequences and awaits a better understanding of the interactions between export substrates and components of the export apparatus.
For the flagellar genes encoded on the plasmids used in the present study, the 5′-untranslated region of the mRNA derived from the vector and so an obligatory role of this region in the export recognition process appears to be ruled out.
While we have shown that export of flagellar structural proteins can occur posttranslationally under our conditions, this does not necessarily imply that it cannot occur cotranslationally. In the Shigella flexneri type III secretion system, for example, Page et al. (34) have found that the secretion of what they describe as the stored form of a particular protein, IpaA, is dependent on a chaperone-like molecule, Spa15, and that this secretion is independent of ongoing protein synthesis. Without Spa15, however, secretion is delayed in the absence of chloramphenicol and abolished in its presence. They suggest that Spa15 is necessary for secretion of fully synthesized and stored IpaA but that cotranslational secretion of IpaA can also occur in a Spa15-independent fashion. Whether such complexities exist in the flagellar export system is not known.
Acknowledgments
T. Hirano and T. Minamino contributed equally to this work.
We acknowledge Kouhei Ohnishi and Shin-Ichi Aizawa for providing anti-FlgD and anti-FlgG antibody, respectively, and May Kihara and Ryan Chu for technical assistance.
This work has been supported by USPHS grant AI12202 (to R.M.M.).
REFERENCES
- 1.Anderson, D., and O. Schneewind. 1997. A mRNA signal for type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Auvray, F., J. Thomas, G. M. Fraser, and C. Hughes. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, J. C. Q., J. Thomas, G. M. Fraser, and C. Hughes. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol. Microbiol. 39:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilgin, N., A. A. Richter, M. Ehrenberg, A. E. Dahlberg, and C. G. Kurland. 1990. Ribosomal RNA and protein mutants resistant to spectinomycin. EMBO J. 9:735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature (London) 407:340-348. [DOI] [PubMed] [Google Scholar]
- 6.Chilcott, G. S., and K. T. Hughes. 1998. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from amino-terminus into the anti-σ28 domain. Mol. Microbiol. 30:1029-1040. [DOI] [PubMed] [Google Scholar]
- 7.Fan, F., and R. M. Macnab. 1996. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271:31981-31988. [DOI] [PubMed] [Google Scholar]
- 8.Fan, F., K. Ohnishi, N. R. Francis, and R. M. Macnab. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035-1046. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, G. M., J. C. Q. Bennett, and C. Hughes. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569-580. [DOI] [PubMed] [Google Scholar]
- 10.González-Pedrajo, B., G. M. Fraser, T. Minamino, and R. M. Macnab. 2002. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol. Microbiol. 45:967-982. [DOI] [PubMed] [Google Scholar]
- 11.Hirano, T., T. Minamino, and R. M. Macnab. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312:359-369. [DOI] [PubMed] [Google Scholar]
- 12.Homma, M., D. J. DeRosier, and R. M. Macnab. 1990. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J. Mol. Biol. 213:819-832. [DOI] [PubMed] [Google Scholar]
- 13.Homma, M., H. Fujita, S. Yamaguchi, and T. Iino. 1984. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in the hook-associated proteins. J. Bacteriol. 159:1056-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homma, M., K. Kutsukake, M. Hasebe, T. Iino, and R. M. Macnab. 1990. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211:465-477. [DOI] [PubMed] [Google Scholar]
- 15.Iyoda, S., and K. Kutsukake. 1995. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol. Gen. Genet. 249:417-424. [DOI] [PubMed] [Google Scholar]
- 16.Jones, C. J., and R. M. Macnab. 1990. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J. Bacteriol. 172:1327-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlinsey, J. E., J. Lonner, K. L. Brown, and K. T. Hughes. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487-497. [DOI] [PubMed] [Google Scholar]
- 18.Kihara, M., T. Minamino, S. Yamaguchi, and R. M. Macnab. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komoriya, K., N. Shibano, T. Higano, N. Azuma, S. Yamaguchi, and S.-I. Aizawa. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767-779. [DOI] [PubMed] [Google Scholar]
- 20.Kornacker, M. G., and A. Newton. 1994. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N-terminus. Mol. Microbiol. 14:73-85. [DOI] [PubMed] [Google Scholar]
- 21.Kutsukake, K., T. Minamino, and T. Yokoseki. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176:7625-7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuwajima, G., I. Kawagishi, M. Homma, J.-I. Asaka, E. Kondo, and R. M. Macnab. 1989. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc. Natl. Acad. Sci. USA 86:4953-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macnab, R. M. 1976. Examination of bacterial flagellation by dark-field microscopy. J. Clin. Microbiol. 4:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol. 1. ASM Press, Washington, D.C.
- 25.Minamino, T., R. Chu, S. Yamaguchi, and R. M. Macnab. 2000. Role of FliJ in flagellar protein export in Salmonella. J. Bacteriol. 182:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minamino, T., H. Doi, and K. Kutsukake. 1999. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci. Biotechnol. Biochem. 63:1301-1303. [DOI] [PubMed] [Google Scholar]
- 27.Minamino, T., B. González-Pedrajo, K. Yamaguchi, S.-I. Aizawa, and R. M. Macnab. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295-304. [DOI] [PubMed] [Google Scholar]
- 28.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minamino, T., and R. M. Macnab. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37:1494-1503. [DOI] [PubMed] [Google Scholar]
- 30.Minamino, T., and R. M. Macnab. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052-1064. [DOI] [PubMed] [Google Scholar]
- 31.Minamino, T., S. Yamaguchi, and R. M. Macnab. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohnishi, K., F. Fan, G. J. Schoenhals, M. Kihara, and R. M. Macnab. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 179:6092-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohnishi, K., Y. Ohto, S.-I. Aizawa, R. M. Macnab, and T. Iino. 1994. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Page, A.-L., P. Sansonetti, and C. Parsot. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533-1542. [DOI] [PubMed] [Google Scholar]
- 35.Ryu, J., and R. J. Hartin. 1990. Quick transformation in Salmonella typhimurium LT2. BioTechniques 8:43-44. [PubMed] [Google Scholar]
- 36.Silva-Herzog, E., and G. Dreyfus. 1999. Interaction of FliI, a component of the flagellar export apparatus, with flagellin and hook protein. Biochim. Biophys. Acta 1431:374-383. [DOI] [PubMed] [Google Scholar]
- 37.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11998-12002. [DOI] [PMC free article] [PubMed]
- 38.Suzuki, T., T. Iino, T. Horiguchi, and S. Yamaguchi. 1978. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J. Bacteriol. 133:904-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, T., and Y. Komeda. 1981. Incomplete flagellar structures in Escherichia coli mutants. J. Bacteriol. 145:1036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, A. W., S. Yamaguchi, F. Togashi, S.-I. Aizawa, I. Kawagishi, and R. M. Macnab. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255-265. [DOI] [PubMed] [Google Scholar]
- 42.Yokoseki, T., and K. Kutsukake. 1994. Role of the FliS protein on the flagellar filament formation in Salmonella typhimurium. Jpn. J. Genet. 69:778.