Abstract
Methanococcus maripaludis is a mesophilic species of Archaea capable of producing methane from two substrates: hydrogen plus carbon dioxide and formate. To study the latter, we identified the formate dehydrogenase genes of M. maripaludis and found that the genome contains two gene clusters important for formate utilization. Phylogenetic analysis suggested that the two formate dehydrogenase gene sets arose from duplication events within the methanococcal lineage. The first gene cluster encodes homologs of formate dehydrogenase α (FdhA) and β (FdhB) subunits and a putative formate transporter (FdhC) as well as a carbonic anhydrase analog. The second gene cluster encodes only FdhA and FdhB homologs. Mutants lacking either fdhA gene exhibited a partial growth defect on formate, whereas a double mutant was completely unable to grow on formate as a sole methanogenic substrate. Investigation of fdh gene expression revealed that transcription of both gene clusters is controlled by the presence of H2 and not by the presence of formate.
Hydrogen and carbon dioxide are common substrates for methanogenic Archaea. Many species of hydrogenotrophic methanogens can use formate in place of H2, while others cannot. The ability to use formate can be attributed to the enzyme formate dehydrogenase, which in methanogens catalyzes the reaction that produces reduced coenzyme F420 from the oxidation of formate (9, 10, 18). The enzyme consists of two protein subunits (17), α and β, encoded by fdhA and fdhB, respectively (15, 19).
The presence or absence of formate dehydrogenase genes does not predict the ability to grow on formate. Species known to contain contiguously encoded homologs of fdhA and fdhB include Methanococcus maripaludis, Methanococcus voltae, Methanobacterium formicicum, Methanococcus jannaschii, and Methanopyrus kandleri. Of these species, only the first three can use formate (4). In the species Methanothermobacter thermautotrophicus, strain Z-245 can use formate but strain ΔH cannot. This difference is attributed to the presence in the former strain of fdh genes that are absent from the same genomic context in the latter (15). Nevertheless, fdhA and fdhB homologs are present in the genome of strain ΔH, albeit not closely related to those known in strain Z-245. Furthermore, at least one species that is unable to grow on formate, M. jannaschii, possesses formate dehydrogenase enzyme activity (11).
The organization and regulation of formate dehydrogenase genes have been studied in M. formicicum (23) and M. thermoautotrophicus strain Z-245 (15), both of which use formate. In each species a gene designated fdhC precedes fdhA and fdhB. FdhC contains several potential membrane-spanning regions (23) and may encode a formate transporter. Northern blots in both species indicated the presence of three mRNA types, one containing fdhC, fdhA, and fdhB, another containing fdhC alone, and the third containing fdhA and fdhB. An apparent transcription start site was identified upstream of fdhC in both species. The smaller mRNAs could be formed by termination or processing between fdhC and fdhA, although a separate initiation of transcription in this region could not be ruled out. In M. thermoautotrophicus Z-245 the levels of fdh mRNA increased when the availability of hydrogen was decreased (15).
We have studied the formate dehydrogenase genes of M. maripaludis. Examination of the genome sequence of M. maripaludis indicated the presence of genes encoding two distinct formate dehydrogenases. Biochemical studies had shown that two distinct formate dehydrogenases exist in the related species Methanococcus vannielii (9, 10), but genetic studies were not carried out. Due to the development of genetic methods for M. maripaludis (21), we have been able to produce mutants in both sets of fdh genes and to determine their roles in growth on formate. In addition, we used lacZ fusions to determine factors that regulated the expression of both gene sets.
MATERIALS AND METHODS
Strains and growth conditions.
Strict anaerobic growth conditions were used when culturing M. maripaludis as previously described (2). The wild-type strain LL is also designated strain S2 (13, 24). LL and its derivatives were grown in McC (24) or in nitrogen-free liquid medium (3) supplemented with 10 mM ammonium chloride. Tubes contained 5 ml of medium and 20 ml of headspace pressurized to 40 lb/in2 with either H2:CO2 (80%:20%) or N2:CO2 (80%:20%); 20% CO2 was always necessary, since a CO2-bicarbonate equilibrium buffers the pH of the medium. Cultures were supplemented with 100 mM sodium formate where appropriate, and Tris-HCl (100 mM, pH 7) was added to buffer the pH during formic acid utilization. Antibiotics were used at the following concentrations (in micrograms per milliliter) where appropriate: neomycin sulfate, 500 (solid medium) or 1,000 (liquid medium); puromycin, 2.5; kanamycin sulfate, 25; ampicillin, 100.
Construction of mutants.
The fdhA1::neo mutant, MM707, was constructed as follows. A 2,064-bp PCR product containing the fdhA1 gene was amplified from LL chromosomal DNA by using primers FdhA1 and FdhA2 (Table 1). The PCR product was TA-cloned into pCR2.1Topo (Invitrogen), producing pCRfdhA303. A promoterless, terminatorless neomycin resistance cassette was amplified from pRCN230 (pRCN115 [5] with the Pur cassette replaced by a Neo cassette [1]) by using primers NeoForCla and NeoRevNde and then were cloned into the ClaI and NdeI sites of pCRfdhA303. The resulting plasmid, pCRfdhA303neo, was used to transform LL as described below.
TABLE 1.
Primers used in this study
| Target | Plasmid | Primer name and sequence (5′-3′) |
|---|---|---|
| fdhA1 | pCRfdhA303 | FdhA1: CCTTTCAAAAGACACCCTG |
| FdhA2: ATGCAGTAACTGCCCCACCAC | ||
| neo | pCRfdhA303neo | NeoForCla: CCATCGATGGATCTGATCAAGAGACAGG |
| NeoRevRINde: GAATTCCATATGCGAACCCCAGAGTCCCGC | ||
| fdhA2 | pBSfdh336-11 | Fdh/dhySXba: GGTCTAGAATCTGGGCTGACGCAACG |
| FdhB62Xho: GGCTCGAGGATCTGAATTTCTTGGAACTTTCAAT | ||
| Pmcr-pac-Tmcr | pBSfdh336-11pac | Pmcr2MluI: GGACGCGTTCTAGAGGATGATTAATTTTAAG |
| Tmcr2PpuRI: GAATTCAGGTCCCTAAAAATATATAAAAAAAGGAAACC | ||
| fdhA2 upstream region | pFL10 | Fdh336Nsi: CCCATGCATTATTCCACCTTTTAATTTTATACTTATTTTTAACC |
| Fdh336Spe4: GGACTAGTCATTCAAATTTTTGAGACTAGG | ||
| fdhA2 upstream region | pFL11 | Fdh336Nsi: CCCATGCATTATTCCACCTTTTAATTTTATACTTATTTTTAACC |
| Fdh336Spe3: GGACTAGTCGATATAGGTATTATTTAACAGG | ||
| fdhA2 upstream region | pFL12 | Fdh336Nsi: CCCATGCATTATTCCACCTTTTAATTTTATACTTATTTTTAACC |
| Fdh336Spe2: CCACTAGTGAAAAGATATTTCTTGATATCTCTC | ||
| fdhA2 upstream region | pFL13 | Fdh336Nsi: CCCATGCATTATTCCACCTTTTAATTTTATACTTATTTTTAACC |
| Fdh336Spe1: GGACTAGTCGATTGTATTTCCAATGCTCG | ||
| fdhA1 upstream region | pFL20 | Fdh303Nsi: GGGATGCATATTTCGATCACCATTTTATAA |
| Fdh303Spe3: GGACTAGTGCACAAATTTGAATGGTGTG | ||
| fdhA1 upstream region | pFL21 | Fdh303Nsi: GGGATGCATATTTCGATCACCATTTTATAA |
| Fdh303Spe2: CCACTAGTGCCTACAATAATATACTTAACTATATC | ||
| fdhA1 upstream region | pFL22 | Fdh303Nsi: GGGATGCATATTTCGATCACCATTTTATAA |
| Fdh303Spe1: GGACTAGTGCCTAAAAAAGGGTAATGTGG | ||
| fdhC1 upstream region | pFL30 | FdhCNsi: GGGATGCATGATATTACCTCCATGTGAATATTG |
| Fdh303Spe4: GGACTAGTCCACTTGATGTTAATTACTAC | ||
| fdhC1 upstream region | pFL31 | FdhCNsi: GGGATGCATGATATTACCTCCATGTGAATATTG |
| Fdh303Spe5: GGACTAGTGTAGACGCTATCGTTGAAGC | ||
| fdhC1 upstream region | pFL32 | FdhCNsi: GGGATGCATGATATTACCTCCATGTGAATATTG |
| Fdh303Spe6: GGACTAGTGGGGCGCACATATATAAGAGG | ||
| fdhC1 upstream region | pFL33 | FdhCNsi: GGGATGCATGATATTACCTCCATGTGAATATTG |
| Fdh303Spe7: GGACTAGTAAAGGAAAAGTGACATTTTACAC |
To make the fdhA2::pac mutant, MM708, fdhA2 and surrounding DNA was amplified from LL chromosomal DNA by using primers Fdh/dhySXba and FdhB62Xho. The resulting 3,327-bp product was cloned into the XhoI and XbaI sites of pBluescript KS(+) (Stratagene), forming pBSfdh336-11. A puromycin resistance cassette, in which pac expression is controlled by the promoter (Pmcr) and terminator (Tmcr) of the M. voltae mcr operon (8), was amplified from pJK3 (14) by using primers Pmcr2MluI and Tmcr2PpuRI. This PCR product was then cloned into the EcoRI and MluI sites of pBSfdh336-11 to form pBSfdh336-11pac.
Allelic exchange was accomplished by transforming (20) LL with pCRfdh303neo or pBSfdh336-11pac and selecting on McC agar plates containing neomycin or puromycin, respectively, under 16 lb of H2:CO2/in2. PCR and Southern blots confirmed that the wild-type allele had been replaced by the disrupted allele in these mutants and that no vector sequences remained (data not shown). MM708 was then transformed with pBSfdh336-11pac to produce the double mutant MM709.
Growth curves.
Overnight McC-H2:CO2-grown cultures were diluted 1:50 into duplicate tubes containing 5 ml of the appropriate fresh medium. Tubes were pressurized with H2:CO2 or N2:CO2 to 40 lb/in2. The optical density at 600 nm was determined immediately and at intervals throughout the growth curve. Readings were taken from duplicate cultures and were averaged.
Construction of lacZ reporter plasmids.
Various regions upstream of fdhA1, fdhC1, and fdhA2 (see Fig. 4) were PCR amplified and then were cloned into the SpeI and NsiI sites of pWLG40+lacZ (6) such that the ATG was fused in frame with the ATG of lacZ. Each construct used the native ribosome binding site present upstream of the respective gene. The primers used to construct each plasmid are shown in Table 1. Each plasmid was sequenced to confirm that no errors had been introduced by PCR and then were used to transform LL. Plasmid pWLG40+lacZ was used as a positive control because it contains the hmv promoter, which is constitutively expressed (6). Plasmid pWLGΔhmv is identical to pWLG40+lacZ except that the DNA between the SpeI and NsiI sites was deleted, thus removing the hmv promoter; this plasmid served as a promoterless negative control.
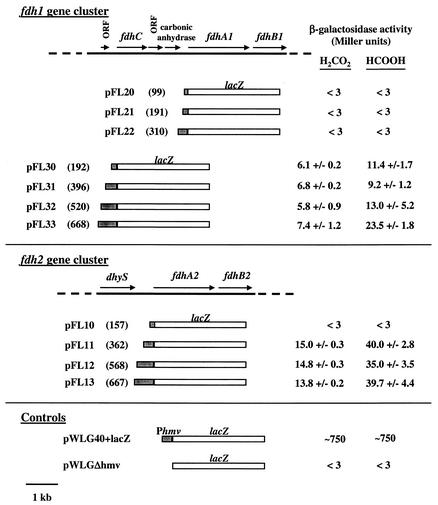
FIG. 4.
Regulation mediated by regions upstream of fdh genes. Strains containing plasmids with lacZ fused to the indicated upstream regions were grown on H2:CO2 or on formate. Numbers in parentheses indicate the number of base pairs upstream of the putative start codon. β-Galactosidase activities are means ± standard deviations of duplicate measurements from a single culture after 9 h of growth on the indicated substrate. The medium used was McC plus Tris.
β-Galactosidase assays.
Overnight McC-H2:CO2-grown cultures were diluted 1:25 in fresh medium as appropriate and then were pressurized to 40 lb/in2 with H2:CO2 or N2:CO2. The optical density at 600 nm of the cultures was determined immediately (0 h), an aliquot (0.3 ml) was removed to determine β-galactosidase activity, and the cultures were placed in a 37°C shaker. Samples for the 3-, 6-, and 9-h time points were collected similarly. β-Galactosidase activities were determined by using standard procedures in duplicate, and the means and standard deviations were determined.
Sequence analysis and nucleotide sequence accession numbers.
The genome sequence of M. maripaludis strain LL is an ongoing collaboration of J. A. Leigh with M. Olson (University of Washington Genome Center). M. maripaludis fdh sequences are deposited in GenBank (accession numbers AY236515 and AY236516).
RESULTS
M. maripaludis strain LL uses formate as a substrate for methanogenesis.
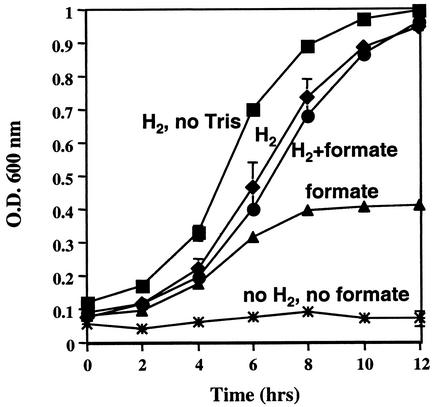
It has previously been reported that M. maripaludis strain JJ can grow on formate as a methanogenic substrate (12). To determine if this is also true for strain LL we compared its growth with either H2:CO2 or sodium formate (in the presence of N2:CO2) as methanogenic substrates. As shown in Fig. 1, LL grew on both H2:CO2 and formate, with maximal growth on H2:CO2. A higher final culture density was achieved on H2:CO2, presumably because 40 lb of H2:CO2/in2 provides approximately five times more growth substrate than 100 mM formate, assuming that all available substrate is consumed. Cultures were inhibited somewhat by the Tris buffer that was necessary to stabilize the pH of cultures utilizing formic acid. Strain LL did not grow when both H2 and formate were absent, confirming the lack of growth-supporting substrate.
FIG. 1.
Growth of M. maripaludis LL on H2:CO2 or formate. Squares, H2:CO2; circles, H2:CO2 plus Tris plus formate; triangles, N2:CO2 plus Tris plus formate; diamonds, H2:CO2 plus Tris; asterisks, N2CO2 plus Tris. The medium used was McC. O.D. 600 nm, optical density at 600 nm.
M. maripaludis LL contains two sets of formate dehydrogenase genes.
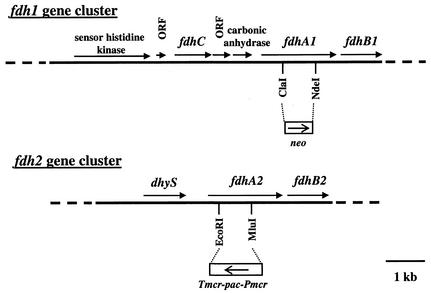
We searched the genome sequence of M. maripaludis strain LL for formate dehydrogenase α subunit genes and found two separate gene clusters, which we named fdh1 and fdh2 (Fig. 2). We found no other formate dehydrogenases in the genome; distant homologs to FdhA appeared to be molybdopterin-containing proteins with different functions. The fdhA1 gene lay immediately upstream of an fdhB homolog, fdhB1. An fdhC homolog, encoding a putative formate transporter, was found upstream of fdhA1 separated by an open reading frame (ORF) of unknown function and a carbonic anhydrase homolog. In contrast, in M. formicicum (23) and M. thermoautotrophicus strain Z-245 (15) fdhC is immediately upstream of fdhA, and genes corresponding to the ORF of unknown function and the carbonic anhydrase homolog are not present. In addition, a small ORF of unknown function lay upstream of fdhC in M. maripaludis. The entire fdh1 cluster was preceded by a sensor histidine kinase homolog. The fdhA2 gene lay immediately upstream of an fdhB homolog (fdhB2); no fdhC homolog was found near fdhA2, nor was a carbonic anhydrase gene or a homolog of either unknown ORF present. The fdh2 cluster was preceded by dhyS encoding a homolog of deoxyhypusine synthase. The two fdh clusters of M. maripaludis encoded closely related proteins; FdhA1 and FdhA2 were 65% identical and FdhB1 and FdhB2 were 62% identical. Each gene was preceded by a sequence similar to that of a ribosome binding site (data not shown). Both fdhA alleles contained a TGA codon at amino acid 132 (fdhA1) or 133 (fdhA2) which presumably encodes a selenocysteine residue. The presence of selenocysteine is not uncommon in formate dehydrogenases (8, 10).
FIG. 2.
The fdh gene clusters of M. maripaludis LL. Intergenic distances are as follows (in base pairs, between genes): sensor histidine kinase-213-ORF-247-fdhC1-40-ORF-23-carbonic anhydrase-304-fdhA1-38-fdhB1; dhyS-639-fdhA2-23-fdhB2. Mutations constructed in fdhA1 and fdhA2 are shown. neo, neomycin resistance gene; pac, puromycin resistance gene; Pmcr and Tmcr, promoter and terminator derived from the M. voltae methylreductase gene (7).
In an effort to determine the origin of the two formate dehydrogenases of M. maripaludis, we carried out a phylogenetic analysis of FdhA and FdhB in methanogens. The results (not shown) indicated that the duplications leading to the presence of two fdhA-fdhB gene clusters in M. maripaludis occurred within the methanococcal lineage.
Both fdhA genes contribute to growth on formate.
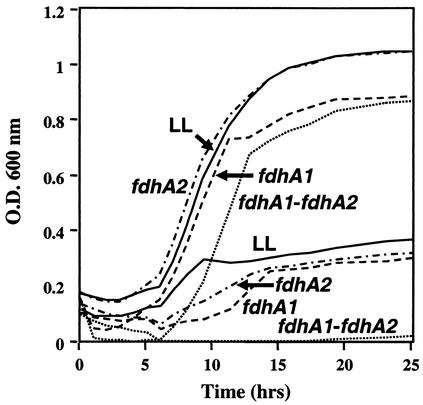
The relative importance of the two fdh gene clusters in growth on formate was assessed by constructing mutants in either or both fdhA genes. Internal sections of each gene were deleted and replaced by neomycin or puromycin resistance markers (Fig. 2). The mutants were then analyzed for growth on formate, and their growth was compared to that of wild-type strain LL (Fig. 3). Mutants lacking either fdhA1 or fdhA2 showed a significant growth defect on formate. When both fdhA alleles were inactivated, M. maripaludis was completely unable to grow with formate as sole methanogenic substrate. All strains grew well on H2:CO2. The entire experiment was repeated, with similar results. These results suggest that both fdhA genes contribute to growth on formate as the methanogenic substrate.
FIG. 3.
Growth of M. maripaludis fdh mutants on H2:CO2 (upper curves) or N2:CO2 plus formate (lower curves). Solid lines, LL; dashed lines, fdhA1 mutant; dashed and dotted lines, fdhA2 mutant; dotted lines, double mutant. Lines join readings that were taken every 1 to 2 h. Readings from duplicate cultures were averaged. The medium used was McC plus Tris. O.D. 600 nm, optical density at 600 nm.
White (22) found that free formate, not formyl-tetrahydromethanopterin, is the substrate for the in vitro synthesis of purines in M. thermoautotrophicus and Sulfolobus solfataricus. We hypothesized that in addition to functioning in growth on formate, the fdh genes may be required for the synthesis of formate and hence of purines during growth on H2:CO2 in M. maripaludis. To test this hypothesis we performed growth experiments in chemically defined medium that lacked exogenous purines (nitrogen-free medium supplemented with ammonium chloride). Both single mutants and the double mutant grew as well as LL, suggesting that neither fdhA allele is required to produce formate for purine biosynthesis (data not shown).
Regulation of fdh expression.
We studied the expression of each fdh gene cluster by constructing a series of plasmid-encoded lacZ fusions to possible promoter region DNAs. Intergenic regions upstream of fdhC1, fdhA1, and fdhA2 were judged to be of sufficient length to contain possible promoter regions (see Fig. 2). Therefore, regions extending various distances upstream of fdhA1, fdhC1, and fdhA2 were PCR amplified and fused in frame with the ATG of lacZ on a low-copy replicative plasmid. Thus, each construct contained the native ribosome binding site and start codon fused to lacZ. The extent of each upstream region is shown in Fig. 4. M. maripaludis LL was transformed with these plasmids, and β-galactosidase assays were done with McC medium with either H2:CO2 or formate (with N2:CO2) as methanogenic substrate. Controls included a plasmid (pWLG40+lacZ) in which lacZ is constitutively expressed (6) and a plasmid (pWLGΔhmv) which lacks a promoter upstream of lacZ. As shown in Fig. 4, fusions to DNA immediately upstream of fdhA1 (pFL20 to pFL22) were not expressed on either substrate, suggesting that no promoter lies in this region. Fusions made to DNA upstream of fdhC1 (pFL30 to pFL33) had β-galactosidase activity, suggesting that a promoter lies in this region. In all cases fdhC-lacZ expression was higher when cultures were grown on formate in the presence of N2:CO2 than when grown on H2:CO2. Plasmids pFL30 to pFL32, collectively containing DNA ranging up to 520 bp upstream of fdhC, gave similar results. In contrast, higher expression and greater regulation appeared to occur when DNA between 520 and 668 bp upstream of fdhC was included in the region fused to lacZ (pFL33). This result suggests that the regions between the start codon of fdhC and 520 bp and between 520 and 668 bp both contain separate sequences involved in the activation of fdhC expression with formate plus N2:CO2. Notably, 520 bp upstream of fdhC is only 30 bp upstream of a small ORF of unknown function (see Fig. 2). Therefore, activation involving the region upstream of 520 bp might be due not only to DNA that acts cis to fdhC but also to the expression of this ORF.
β-Galactosidase fusions were also made by using DNA upstream of fdhA2 (pFL10 to pFL13, Fig. 4). No β-galactosidase was expressed from plasmid pFL10, suggesting that no promoter is present in the 157 bp immediately upstream of the start codon of fdhA2. Fusions containing ≥362 bp of DNA were all expressed, indicating the presence of a promoter in this region. Likewise, all three fusions were upregulated two- to threefold in formate-containing medium with N2:CO2, suggesting the presence of a regulatory site.
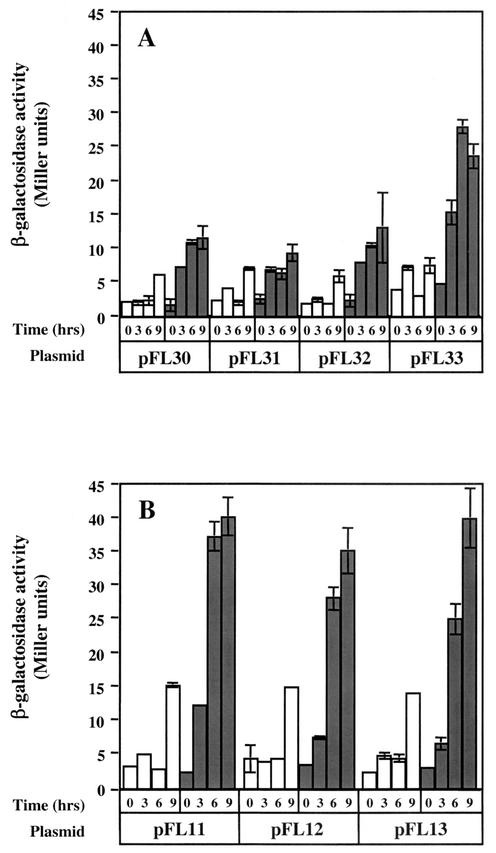
A time course experiment was performed to extend the above observations. In all cases, enhancement of fdhC1 (Fig. 5A) and fdhA2 (Fig. 5B) expression in the presence of formate and N2:CO2 was observed starting 3 h after inoculation. As before, the highest expression of fdhC1 occurred only with pFL33, which contains 668 bp of upstream DNA. An increase in expression of both fdhC1 and fdhA2 was consistently seen after 9 h of growth on H2:CO2. One possible explanation is that H2:CO2 pressure was decreasing and that, as shown below, fdh expression is controlled by the presence of H2.
FIG. 5.
Time course of fdh regulation. Strains containing plasmids as shown in Fig. 4 were grown with H2:CO2 and then were used to inoculate medium with H2:CO2 (open bars) or formate plus N2:CO2 (filled bars). The medium used was McC plus Tris.
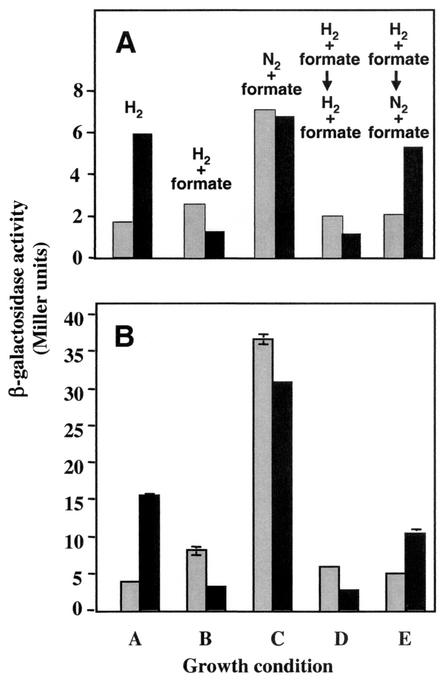
To test the hypothesis that fdh expression is regulated by the presence of H2 we repeated the time course experiment, this time including cultures containing formate with an H2:CO2 atmosphere. Thus, three conditions were compared: H2 alone, H2 plus formate, and formate alone (N2 replacing H2). Comparison of these three conditions allowed us to separately assess the effects of hydrogen and of formate (Fig. 6). Constructs containing the maximum upstream regions of fdhC1 (Fig. 6A) and fdhA2 (Fig. 6B) were used. When both H2:CO2 and formate were present (growth condition B), fdhC1-lacZ and fdhA2-lacZ expression were both low compared to that of formate with an N2:CO2 atmosphere (growth condition C). These results suggested that the presence of H2 resulted in decreased gene expression. To further test the effect of H2, cultures were grown for 6 h in tubes containing both H2:CO2 and formate, β-galactosidase activity was measured, and the H2:CO2 was flushed away and replaced with 40 lb of N2:CO2/in2 (growth condition E). Control cultures were flushed with N2:CO2 and were repressurized with 40 lb of H2:CO2/in2 (growth condition D). β-Galactosidase activities increased at 9 h due to the removal of H2. Furthermore, repeat experiments with each construct revealed the same pattern. These results confirm that both fdhC1 expression and fdhA2 expression are controlled by the presence or absence of H2. In contrast, formate had no marked effect.
FIG. 6.
Effects of H2 and formate on fdhC1 and fdhA2 regulation. LL containing pFL33 (A) or pFL13 (B) was grown under the following conditions: lanes A, H2:CO2; lanes B, H2:CO2 plus formate; lanes C, N2:CO2 plus formate; lanes D, H2:CO2 plus formate flushed with H2:CO2 at 6 h; lanes E, H2:CO2 plus formate flushed with N2:CO2 at 6 h. Grey bars, 6 h; black bars, 9 h. The medium used was McC plus Tris.
DISCUSSION
Apparent gene duplication within the methanococcal lineage has resulted in M. maripaludis containing two sets of formate dehydrogenase genes. We considered two alternative hypotheses for the function of these genes. One hypothesis was that duplication of function has occurred: both formate dehydrogenases convert formic acid to carbon dioxide and reduce F420 for the purpose of methanogenesis. The other hypothesis was that only one formate dehydrogenase has this catabolic function while the other functions biosynthetically, producing from H2 and CO2 the formate that has been suggested as a substrate for purine biosynthesis in other methanogens (22). Our results support the first hypothesis: both FdhA proteins appeared to contribute to the ability to grow on formate as the catabolic substrate, and together the two formate dehydrogenases accounted fully for this ability. Our findings are in concurrence with those of a recent study in which the selB gene encoding the archaeal translation factor specialized for selenocysteine insertion was inactivated in M. maripaludis strain JJ (16). The resulting mutant was unable to grow on formate, presumably because both fdhA genes encode selenocysteine-containing proteins. We found no evidence that either formate dehydrogenase is required for biosynthesis, since the double mutant grew well in minimal medium on H2 and CO2.
Maximum expression of both fdh clusters depended on the absence of H2 and not the presence of formate. This observation parallels that of Nolling and Reeve (15), who noted that in M. thermoformicicum Z-245, fdh expression was regulated by the availability of H2. Whether formate had any regulatory effect was not addressed in that study. In M. maripaludis, growth on H2 was superior to growth on formate (Fig. 1), and downregulation of fdh by H2 would be in accordance with the organism's tendency to grow on formate only when H2 is absent. However, why significant fdh expression still occurs when hydrogen is present is unknown; a biosynthetic function would be one explanation, but as stated above we were unable to find evidence to support this idea. Besides regulation by H2, fdh expression in M. formicicum is stimulated by molybdate (23), a phenomenon we did not investigate with M. maripaludis.
By fusing different segments of fdh promoter region DNA to lacZ we were able to localize the sites necessary for expression and regulation. In the case of fdh2 both expression and regulation were mediated by DNA between 157 and 362 bp upstream of the fdhA coding sequence. In the case of fdh1 transcription was initiated within 192 bp upstream of fdhC, and no transcription was initiated upstream of fdhA. These results are consistent with those of studies with M. formicicum (23) and M. thermoautorophicus strain Z-245 (15) which suggested that transcription occurred from a promoter upstream of fdhC with processing of some transcripts between fdhC and fdhA. Regulation of fdh1 appeared to be more complex than that of fdh2, being mediated partially by DNA within 192 bp of fdhC but with additional activation depending on DNA between 520 and 668 bp upstream of fdhC. Future experiments will characterize the regulatory roles of these upstream regions in more detail, including the possible function of the small ORF upstream of fdhC.
Acknowledgments
This work was supported by grants GM-55255 and GM-60403 from the National Institutes of Health and grant DE-FG03-01ER15252 from the Department of Energy's Microbial Cell Program.
REFERENCES
- 1.Argyle, J. L., D. L. Tumbula, and J. A. Leigh. 1996. Neomycin resistance as a selectable marker in Methanococcus maripaludis. Appl. Environ. Microbiol. 62:4233-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blank, C. E., P. S. Kessler, and J. A. Leigh. 1995. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J. Bacteriol. 177:5773-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boone, D. R., and R. W. Castenholz (ed.). 2001. The Archaea and the deeply branching phototrophic bacteria, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 5.Cohen-Kupiec, R., C. Blank, and J. A. Leigh. 1997. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. USA 94:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner, W. L., and W. B. Whitman. 1999. Expression vectors for Methanococcus maripaludis: overexpression of acetohydroxyacid synthase and beta-galactosidase. Genetics 152:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gernhardt, P., O. Possot, M. Foglino, L. Sibold, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221:273-279. [DOI] [PubMed] [Google Scholar]
- 8.Jones, J. B., G. L. Dilworth, and T. C. Stadtman. 1979. Occurrence of selenocysteine in the selenium-dependent formate dehydrogenase of Methanococcus vannielii. Arch. Biochem. Biophys. 195:255-260. [DOI] [PubMed] [Google Scholar]
- 9.Jones, J. B., and T. C. Stadtman. 1980. Reconstitution of a formate-NADP+ oxidoreductase from formate dehydrogenase and a 5-deazaflavin-linked NADP+ reductase isolated from Methanococcus vannielii. J. Biol. Chem. 255:1049-1053. [PubMed] [Google Scholar]
- 10.Jones, J. B., and T. C. Stadtman. 1981. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J. Biol. Chem. 256:656-663. [PubMed] [Google Scholar]
- 11.Jones, W. J., J. A. Leigh, F. Mayer, C. R. Woese, and R. S. Wolfe. 1983. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136:254-261. [Google Scholar]
- 12.Jones, W. J., M. J. B. Paynter, and R. Gupta. 1983. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 135:91-97. [Google Scholar]
- 13.Lie, T. J., and J. A. Leigh. 2002. Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source. J. Bacteriol. 184:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolling, J., and J. N. Reeve. 1997. Growth- and substrate-dependent transcription of the formate dehydrogenase (fdhCAB) operon in Methanobacterium thermoformicicum Z-245. J. Bacteriol. 179:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rother, M., I. Mathes, F. Lottspeich, and A. Bock. 2003. Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J. Bacteriol. 185:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schauer, N. L., and J. G. Ferry. 1986. Composition of the coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum. J. Bacteriol. 165:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauer, N. L., and J. G. Ferry. 1982. Properties of formate dehydrogenase in Methanobacterium formicicum. J. Bacteriol. 150:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuber, A. P., E. C. Orr, M. A. Recny, P. F. Schendel, H. D. May, N. L. Schauer, and J. G. Ferry. 1986. Cloning, expression, and nucleotide sequence of the formate dehydrogenase genes from Methanobacterium formicicum. J. Biol. Chem. 261:12942-12947. [PubMed] [Google Scholar]
- 20.Tumbula, D. L., T. L. Bowen, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 21.Tumbula, D. L., and W. B. Whitman. 1999. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 22.White, R. H. 1887. Purine biosynthesis in the domain Archaea without folates or modified folates. J. Bacteriol. 179:3374-3377. [DOI] [PMC free article] [PubMed]
- 23.White, W. B., and J. G. Ferry. 1992. Identification of formate dehydrogenase-specific mRNA species and nucleotide sequence of the fdhC gene of Methanobacterium formicicum. J. Bacteriol. 174:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]