Abstract
Short regularly spaced repeats (SRSRs) occur in multiple large clusters in archaeal chromosomes and as smaller clusters in some archaeal conjugative plasmids and bacterial chromosomes. The sequence, size, and spacing of the repeats are generally constant within a cluster but vary between clusters. For the crenarchaeon Sulfolobus solfataricus P2, the repeats in the genome fall mainly into two closely related sequence families that are arranged in seven clusters containing a total of 441 repeats which constitute ca. 1% of the genome. The Sulfolobus conjugative plasmid pNOB8 contains a small cluster of six repeats that are identical in sequence to one of the repeat variants in the S. solfataricus chromosome. Repeats from the pNOB8 cluster were amplified and tested for protein binding with cell extracts from S. solfataricus. A 17.5-kDa SRSR-binding protein was purified from the cell extracts and sequenced. The protein is N terminally modified and corresponds to SSO454, an open reading frame of previously unassigned function. It binds specifically to DNA fragments carrying double and single repeat sequences, binding on one side of the repeat structure, and producing an opening of the opposite side of the DNA structure. It also recognizes both main families of repeat sequences in S. solfataricus. The recombinant protein, expressed in Escherichia coli, showed the same binding properties to the SRSR repeat as the native one. The SSO454 protein exhibits a tripartite internal repeat structure which yields a good sequence match with a helix-turn-helix DNA-binding motif. Although this putative motif is shared by other archaeal proteins, orthologs of SSO454 were only detected in species within the Sulfolobus genus and in the closely related Acidianus genus. We infer that the genus-specific protein induces an opening of the structure at the center of each DNA repeat and thereby produces a binding site for another protein, possibly a more conserved one, in a process that may be essential for higher-order stucturing of the SRSR clusters.
Repetitive DNA sequences play important roles in DNA replication, DNA recombination, and gene regulation, as well as more generally in genome reorganization. Recently, comparative genomics has provided a strong basis for examining the locations and frequency of such repeats and their functional significance and has also led to the discovery of new classes of repeats. One such class consists of clusters of short regularly spaced repeats (SRSRs) of 20 to 37 bp that are interspaced with nonconserved sequences of almost constant length in the range 32 to 69 bp in Archaea. Such clusters were first observed for the haloarchaeal genus Haloferax (9, 10) and were subsequently found in a conjugative plasmid pNOB8 of the archaeal genus Sulfolobus (18). They are now known to be present in almost all of the sequenced archaeal genomes, and smaller clusters are present in some bacterial genomes (11, 13).
For archaeal genomes a few large uninterrupted SRSR clusters can be present constituting up to 1% of the genome. For example, the genomes of Sulfolobus solfataricus P2 (19) and Sulfolobus tokodaii (7) contain seven and five clusters, with a total of 441 and 455 repeat copies, respectively, falling mainly into two closely related sequence families. Moreover, the sequence within one of the repeat families in S. solfataricus is identical to that of the Sulfolobus conjugative plasmid, pNOB8 (18). In general, the repeat sequences show a smaller variation between different clusters within the same genome than between those in different genomes (11).
To date, the only insight into the function of the SRSRs is that they have been implicated in DNA segregation. In an experimental study, an additional SRSR cluster was transformed into Haloferax volcanii on a bacterial-archaeal shuttle vector (10). The vector recombined into the host chromosome and produced a decrease in both cellular growth rate and the fraction of viable cells, as well as a difference in the distribution of DNA in daughter cells (10). An SRSR cluster with six repeats was also found in an archaeal conjugative plasmid pNOB8 and was considered to be a cis element which, together with the encoded ParA and ParB homologs, ensured stable plasmid maintenance in its natural host Sulfolobus NOB8H2 (18), by analogy to a bacterial partitioning mechanism (see, for example, reference 12).
The conserved structures and large sizes of many SRSR clusters in archaea, as well as their widespread occurrence, testify to an important biological function. This is reinforced for S. solfataricus P2, in which none of the 349 putatively mobile elements in the genome interrupt SRSR clusters (4, 19). A biological function such as chromosome segregation would require that the repeats are recognized by specific DNA-binding protein(s). To date, two chromosomal binding proteins have been characterized for Sulfolobus species: SSO7d is involved in the folding and condensing of chromosomal DNA (1), and Alba (SSO10b) binds in clusters along short DNA regions (∼60 bp) and is involved in transcriptional regulation (2, 23). We describe here the purification and characterization of a protein from S. solfataricus P2 that binds specifically to the SRSR-DNA.
MATERIALS AND METHODS
Cell growth and preparation of cell extracts.
Sulfolobus sp. strain NOB8H2 was provided by W. Zillig, and S. solfataricus P2 was purchased from Deutsche Samlung Mikroorganismer, Darmstadt, Germany. Cells were grown in complex medium containing 2% tryptone at 80°C (17). Conjugation was initiated by mixing Sulfolobus sp. strain NOB8H2 with S. solfataricus cells at a ratio of 1:10,000 (17). Cells were harvested at an A600 of 0.3 by centrifugation at 4,000 rpm at 4°C. The cell pellet was suspended in buffer (10 mM Tris-HCl, pH 7.6; 150 mM KCl; 2 mM dithiothreitol [DTT]; 1 mM EDTA; 0.1 mM ZnCl2; 0.5 mM phenylmethylsulfonyl fluoride; 15% glycerol) and sonicated with a Soniprep 150 (Sanyo, Toky, Japan) at an amplitude of 8 μm for 40 s. This treatment was performed 12 times, interspersed with 40-s cooling periods in ice water. The sample was centrifuged at 106 × g for 1 h at 4°C, and aliquots of the supernatant were stored at −80°C.
DNA preparation.
DNA fragments containing the repeat sequences were amplified by PCR with pairs of oligonucleotide primers as follows. SRSR-1p (103 bp; biotin-5′-TAGTACTATCTCATCCCAT-3′ and 5′-CGAACTTACTAATCCAAATAG-3′) and SRSR-2p (158 bp; biotin-5′-TAGTACTATCTCATCCCAT-3′ and 5′-CTACCACCCGCGATAATC-3′) were both amplified from pNOB8 (18) (sequence accession no. AJ010405). SRSR-2c (151 bp; 5′-CTCCGCAACTTCATCAATAGTG-3′ and 5′-CTCCAAAAGGAATGCAAAAAGT-3′) was amplified from S. solfataricus P2 chromosomal DNA (19) (sequence accession no. NC002754). The 325-bp DNA fragment used in the competition assay was amplified by PCR from the terminator region of the putative parAB operon from pNOB8 (18) with the primers 5′-ACCTCTATGCCTTCTTCTCC-3′ and 5′-TCGTCATCCTCAACTTCCAG-3′. Oligonucleotides were purchased from DNA Technology and TAG Copenhagen (Denmark). PCR products were purified by QIAquick PCR purification kit (Qiagen, Westburg, Germany), and DNA concentrations were estimated in agarose gels by visual comparison with DNA markers.
For the DNA affinity chromatography, SRSR-2p was purified in a 5% polyacrylamide gel to remove the biotinylated primer, and ca. 140 pmol SRSR-2p was extracted from the gel and bound to magnetic streptavidin beads under standard conditions (Dynal, Oslo, Norway). pUC18 DNA was prepared by the alkaline sodium dodecyl sulfate (SDS) lysis method (16), and the DNA concentration was measured spectroscopically.
Bandshift binding assay.
A total of 1 to 5 ng of SRSR-2p or SRSR-1p was 32P end labeled on the free 5′ end by using T4 polynucleotide kinase (Amersham) and then dissolved in 10 μl of DNA-binding buffer (10 mM Tris-HCl, pH 7.6; 150 mM KCl; 2 mM DTT; 10% glycerol). Increasing concentrations of cell extract or purified SSO454 protein were added, and the components were allowed to react at 50°C for 20 min. For binding experiments with cell extracts, 0.5 μg of carrier pUC18 DNA was added as a nonspecific competitor. After the mixture cooled to room temperature, 1.5 μl of loading buffer (10 mM Tris-HCl, pH 7.6; 50% glycerol; 1 mM EDTA; 0.5% bromophenol blue) was added, and the samples were run in a prerun 7% polyacrylamide gel (14 by 20 cm) in glycerol-tolerant gel buffer (89 mM Tris-HCl, 25 mM taurine, 0.5 mM EDTA; pH 8.9) and electrophoresed at 12 mA and 90 V for 90 min. Gels were autoradiographed with Fuji RX film by using an intensifying screen at −80°C.
To compare the SRSR-binding activities of the native and recombinant SSO454 proteins, unlabeled SRSR-DNA was used, and the volume of the binding mixture was increased to 15 μl. The SRSR-DNA concentration was increased fivefold, and the protein concentration was scaled up proportionally. After electrophoresis, the polyacrylamide gel was stained with ethidium bromide for 45 min and destained in water for 10 min before photographing it by using the Biometra Ti3 illuminating system (Whatman) and a EDAS290 digital camera (Kodak).
Protein purification and sequencing.
A total of 30 ml of cell extract was obtained from 6 liters of S. solfataricus culture that had been conjugated with pNOB8. The occurrence of conjugation was established by digesting the total DNA extract with BamHI; gel electrophoresis revealed a strong pNOB8 fragment pattern indicative of a high copy number of plasmid in the conjugants (17). After treatment with 1% streptomycin sulfate (Sigma) for 15 min on ice and centrifugation at 106 × g for 10 min, three 10-ml aliquots of supernatant were applied to a ResourceQ column (Bio-Rad). Fast-performance liquid chromatography (FPLC) was performed with a gradient from 0.15 to 1.0 M KCl in 10 mM Tris-HCl (pH 7.6)-1 mM EDTA-2 mM DTT at 4°C. Eluted fractions were checked for binding to the DNA repeats, and positive fractions (containing ca. 0.42 M KCl) were pooled and concentrated with a centrifugal filter (Millipore) to a final volume of 1.8 ml. The fractions were then mixed with SRSR-2p that was bound to streptavidin-magnetic beads (Dynal), and the binding reaction was performed at 50°C for 20 min after an adjustment of the KCl concentration to 0.15 M and the addition of 250 μg of pUC18 and 10 A260 U of poly(dI-dC) (Sigma). After the reactions cooled to room temperature, the beads were absorbed on a magnetic stand (Dynal). The supernatant was removed, and the beads were resuspended in 500 μl of binding buffer containing 100 μg of pUC18 to wash the DNA-protein complex. The washing step was repeated three times under the same conditions as for binding. Washing was then performed four times without pUC18, and the protein was eluted with 150 μl of buffer (10 mM Tris-HCl, pH 7.6; 1 M KCl; 2 mM DTT; 10% glycerol). The purified protein was electrophoresed in a 12% polyacrylamide-SDS gel (8) and visualized by silver staining (Bio-Rad). Aliquots were stored at −20°C. Protein sequences were determined by Edman degradation, peptide fingerprinting, and matrix-assisted laser desorption ionization-mass spectrometry analyses of the peptides (6).
DNase I footprinting.
To ensure that only one 5′ end was labeled, 32P-end-labeled SRSR-2p and SRSR-1p were digested with RsaI, which cut just downstream from the biotin-labeled 5′ end (Fig. 1). 32P-5′-end-labeled fragments of SRSR-2c were prepared by digesting an end-labeled SRSR-4c fragment in the central spacer with HinfI. The downstream fragment is illustrated in Fig. 1. Binding reactions were performed with 30 to 72 ng of DNA and increasing concentrations of purified SSO454 protein under the same conditions as for the band shift assays, except that the reaction volume was 15 μl. Subsequently, 2 μl of DNA-binding buffer containing 85 mM MgCl2 and 0.1 U of RNase-free DNase I (Boehringer Mannheim) was added. The mixture was incubated at room temperature for 10 min, and then 17 μl of stop solution (10 mM Tris-HCl, pH 7.6; 20 mM EDTA) and 5 μg of yeast tRNA were added before the DNA was precipitated with ethanol and dissolved in 3 μl of loading buffer (5 mM EDTA [pH 7.5] and 50% deionized formamide with 0.15% each of bromophenol blue and xylene cyanol FF). After being boiled for 3 min, samples were loaded onto a 10% polyacrylamide gel in 89 mM Tris-borate (pH 8.0)-2 mM EDTA-8 M urea. Gel electrophoresis was performed, together with sequencing ladder samples, at 45 W for 2 h, and gels were autoradiographed at −80°C.
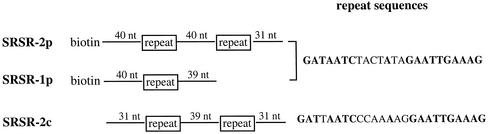
FIG. 1.
PCR fragments of SRSR-2p and SRSR-1p were amplified from pNOB8, and SRSR-2c was amplified from the S. solfataricus P2 chromosome. Sequence identities between the repeats are indicated by boldface letters. Spacer sequences can be obtained from the sequence accession numbers given in Materials and Methods. Biotin is attached to the 5′ end of the DNA strand carrying the sequences given for SRSR-2p and SRSR-1p in order to facilitate protein purification.
Cloning and expression of SSO454 in Escherichia coli.
A one-step protein purification system, IMPACT T7 (New England Biolabs), was used to express SSO454 in E. coli. Briefly, SSO454 was amplified from S. solfataricus P2 DNA by PCR with the primers 5′-GGGAATTCCATATGAGCGAGGAAGAAAACA-3′ and 5′-AAGATATGCTCTTCCGCAAGCAGATGTGGGAGAAGA-3′ and cloned into pTYB1. The insert was sequenced, and the gene was expressed in E. coli BL21(DE3)RI952 (21). The purification was performed according to a standard method (5) that was slightly modified so that, after elution from chitin beads, the recombinant protein was further purified by heating at 70°C for 15 min and any minor E. coli contaminants were removed by centrifuging at 18,000 rpm for 30 min.
Genome sequence analyses.
The algorithm LUNA (locating uniform poly-nucleotide areas; K. Brügger, unpublished data) was used (http://dac.molbio.ku.dk/bioinformatics/luna/); this algorithm can rapidly detect perfect and/or degenerate repeats, direct or inverted, in microbial genomes by using a desktop computer. It also allows a high level of control over the repeats reported. Repeats can be filtered by using several different parameters including length, distance, and level of conservation.
RESULTS
SRSRs in archaeal genomes.
A sequence repeat search was performed on the sequenced archaeal genomes by using the program LUNA. SRSRs were generally detected as multiple clusters for all archaea except Halobacterium sp. strain NRC-1, although they are present in other haloarchaeal genomes (10). Although the structure within a given SRSR cluster is fairly conserved, the sizes of the repeat and spacer regions vary for different clusters within a given genome. These size ranges are listed for the available crenarchaeal genomes and a few representative euryarchaeal genomes in Table 1. The repeat sequence within an SRSR cluster is also almost invariably conserved, whereas the sequences differ between clusters. Moreover, some repeats contain central imperfect palindromic sequences, as reported earlier for the Sulfolobus plasmid pNOB8 (18) or inverted terminal repeat sequences, as exemplified in Table 1. The spacer regions are generally A+T-rich and show no sequence conservation; occasionally, however, individual spacers are G+C-rich, suggesting that they may encode small RNA molecules. The general picture emerges that there is a considerable diversity in the number and sizes of the SRSR clusters and in the sizes and sequences of the SRSR repeats.
TABLE 1.
Characteristics of archaeal SRSR clustersa
| Archaea | Repeat size (bp) | Spacer (bp) | No. of clusters | No. of repeat units | Selected repeat sequence |
|---|---|---|---|---|---|
| Crenarchaea | |||||
| pNOB8 | 24 | 39-42 | 1 | 6 | CTTTCAATTCTATAGTAGATTATC |
| S. solfataricus | 20-25 | 35-41 | 7 | 441 | CTTTCAATTCCTTTTGGGATTAATC |
| S. tokodaii | 24-25 | 39-41 | 5 | 455 | GATGAATCCCAAAAAGGATTGAAAG |
| S. acidocaldarius | 24-25 | 37-39 | >3 | >225 | CTTTCAATTCCATTAAGGATTATC |
| A. brierleyi | 24-30 | ∼41 | >3 | >394 | CTTTCAATTCCTTTTTGGATGAAAC |
| A. pemix | 20-25 | 40-44 | 4 | 73 | CTTTCTATTCCCTTTAGGGATATGC |
| P. aerophilum | 20-25 | 40-45 | 5 | 131 | CTTTCAATCCTCTTTTTGAGATTC |
| Euryarchaea | |||||
| M. kandleri | 24-36 | 53-59 | 4 | 27 | GTTTCATTACCCGTATTATTACGGGTTAATTGCGAG |
| M. jannaschii | 24-32 | 34-47 | 15 | 170 | AATTAAAATCAGACCGTTTCGGAATGGAAA |
| P. horikoshii | 23-30 | 37-46 | 7 | 145 | GTTTCAATTCTATTTTAGTCTTATTGGAAC |
Data are included for available crenarchaeal genomes together with representative examples from the euryarchaea. The genome accession numbers are as follows: S. solfataricus P2, NC002754; S. tokodaii, NC003106; Aeropyrum pernix, NC000854; Pyrobaculum aerophilum, NC003364; Methanopyrus kandleri AV19, NC003551; Methanococcus jannaschii, NC000909; and Pyrococcus horikoshii, NC000961. Data from incomplete genomes are S. acidocaldarius (L. Chen, A. Zibat, H.P. Klenk, and R. A. Garrett, unpublished data) and Acidianus brierleyi (Q. She, unpublished data). The LUNA program was used with parameters that would detect perfect direct repeats, where the space between a set of repeats is between 20 and 80 bp and the length of the repeat is between 20 and 40 bp. Selected repeat sequences found in archaeal SRSR clusters illustrate the degree of diversity internal structure, length, and sequence. Boldface sequence letters indicate perfect or imperfect palindromes or inverted repeats.
SRSR-protein-binding activity in Sulfolobus.
For protein-binding studies we selected the repeat sequence found in the SRSR cluster of the Sulfolobus conjugative plasmid pNOB8 and amplified fragments containing double (SRSR-2p) and single (SRSR-1p) repeat structures by PCR. Fragments containing two repeats, with a different sequence, were also amplified from a large SRSR cluster present in the S. solfataricus P2 genome (SRSR-2c). These fragments are illustrated in Fig. 1. Biotin was added to the ends of SRSR.2p and SRSR-1p in order to facilitate protein purification.
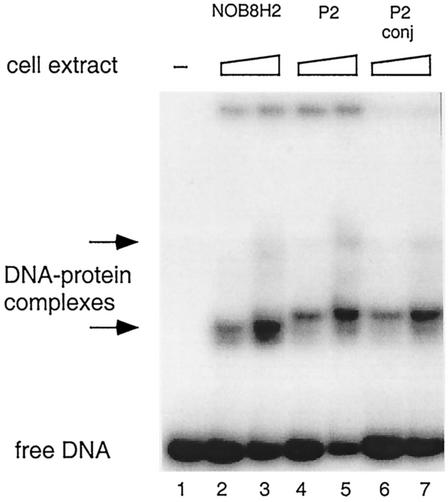
The copy number of pNOB8 increases dramatically after conjugation into S. solfataricus P2 (17), and we therefore examined the binding of SRSR-2p to cell extract proteins from both conjugated and nonconjugated S. solfataricus P2, as well as from Sulfolobus sp. strain NOB8H2, the natural host of pNOB8, which is closely related phylogenetically to the former strain (98 to 99% 16S RNA sequence identity [data not shown]). Protein binding was observed for each cell extract by using a band shift assay (Fig. 2). The two S. solfataricus samples produced similar gel patterns, but the band shift was slightly less for the NOB8H2 strain. Each of the main complex bands was a doublet, and a weak higher band was discernible at the highest protein concentrations (Fig. 2).
FIG. 2.
SRSR-2p was 32P end labeled at the free 5′ end, incubated with Sulfolobus cell extracts, and run in a polyacrylamide gel (3 ng of DNA/lane). Lane 1, no cell extract protein; lanes 2 and 3, 3.5 and 7 μg of cell extract protein, respectively, from the Sulfolobus sp. strain NOB8H2; lanes 4 and 5, 3.2 and 6.4 μg; lanes 6 and 7, 2.0 and 4.0 μg of cell extract protein from nonconjugated (P2) and conjugated (P2 conj) S. solfataricus, respectively. Positions of the protein-DNA complexes are indicated by arrows.
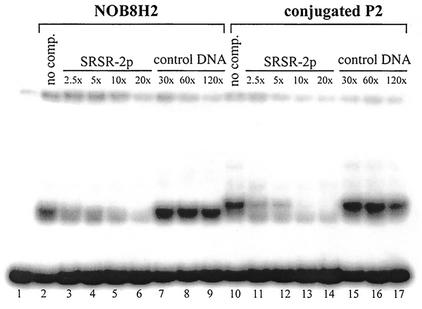
The specificity of protein binding to SRSR-2p was checked for cell extracts of Sulfolobus sp. strain NOB8H2 and conjugated S. solfataricus in a competition assay (Fig. 3). The competitor DNA was either unlabeled SRSR-2p or control DNA amplified from the putative terminator region of the parAB operon in pNOB8 (see Materials and Methods). Although unlabeled SRSR-2p competed strongly at molar excesses of 2.5- to 10-fold (lanes 3 to 6 for Sulfolobus sp. strain NOB8H2 and lanes 11 to 14 for conjugated S. solfataricus), large molar excesses of the unspecific competitor DNA had little effect (lanes 7 to 9 and lanes 15 to 17, respectively). For the specific competitor DNA, a stepwise transition from the upper doublet band to the lower is discernible. The results established that cell extracts from both Sulfolobus strains contained specific SRSR-binding protein(s).
FIG. 3.
Testing the binding specificity of the cell extract protein to SRSR-2p. SRSR-2p was 23P end labeled and a band shift binding assay was performed in the presence of unlabeled SRSR-2p or control DNA amplified from another region of pNOB8 (see text). We used 1.4 μg of cell extract protein from Sulfolobus sp. strain NOB8H2 (lanes 2 to 9) and 1 μg of cell extract protein from conjugated S. solfataricus P2 (lanes 10 to 17). The approximate molar ratios of unlabeled SRSR-2p to 32P-end-labeled SRSR-2p (3 ng) are given above the lane. “No comp.” indicates that no competing DNA was added.
Purification and sequencing of an SRSR-binding protein from S. solfataricus.
S. solfataricus was selected for protein extraction rather than Sulfolobus sp. strain NOB8H2 for two reasons. (i) Conjugation in S. solfataricus produces a high cellular level of pNOB8 and, therefore, of SRSR repeat sequences, which in turn probably enhances the expression of SRSR-binding protein(s). (ii) The availability of the complete genome sequence (19) would facilitate protein identification. Cell extracts of conjugated S. solfataricus were treated with streptomycin sulfate to remove chromosomal DNA before being loaded onto an anion-exchange column (see Materials and Methods). Fractions eluted from the column were checked for SRSR-binding activity by a band shift assay (see above). Most fractions showed DNA-binding activity, but only fractions eluted in the range of from 0.378 to 0.472 M KCl exhibited specific SRSR-binding activity, as shown by the competition assay (see above). These fractions were collected and concentrated before we purified the protein by DNA affinity chromatography with SRSR-2p as the binding substrate (Fig. 4A). Six liters of conjugated S. solfataricus P2 cells harvested at an A600 of 0.3 yielded 7.5 μg of highly purified protein, which corresponds to ca. 110 protein copies per cell. The protein had a molecular mass of ∼18 kDa and was the only visible band in a silver-stained SDS-polyacrylamide gel (Fig. 4A).
FIG. 4.
Characterization of the protein that binds to SRSR-2p from cell extracts of S. solfataricus P2 (A) and of the recombinant protein expressed in E. coli (B) in SDS-polyacrylamide gels. Lanes in panel A: 1, cell extract; 2, protein size marker; 3, FPLC fraction containing SRSR-2p-binding activity; 4, proteins partially purified with DNA affinity chromatography when the washing buffer contained insufficient pUC18 as nonspecific competitor; 5, purified protein. The gel was silver stained. Lanes in panel B: 1, recombinant SSO454 purified from E. coli; 2, protein size marker. The gel was stained with Coomassie brilliant blue. The SRSR-binding protein is indicated by a double arrow. The protein that was gradually removed by extensive washing with pUC18-containing buffer is indicated by an open arrow.
Initial attempts at N-terminal sequencing of the purified protein by Edman degradation failed, and we concluded that the N terminus was modified. Therefore, the protein band from the polyacrylamide gel (Fig. 4A, lane 5) was subjected to peptide fingerprinting. Seven tryptic peptides covering 75 amino acids of sequence were analyzed, and they all yielded perfect matches with SSO454 in the S. solfataricus P2 genome with the Mascot Search program (15). This constitutes a 150-amino-acid basic protein of unknown function (19). Compatible with this assignment, mass spectrometry produced a sharp, major peak corresponding to 17,591.57 Da (data not shown), 14 Da larger than the calculated mass of SSO454. Therefore, we inferred that a posttranslational methylation occurs at the N terminus. Secondary structure analyses of the sequence revealed an α-helix content of 70 to 80% in short segments regularly spaced along the protein. Moreover, the protein exhibits a tripartite internal repeat structure (see below).
A small doublet peak at ca. 11 kDa was also observed in the mass spectrum of the partially purified binding protein. Moreover, although most nonspecific DNA-binding proteins were removed during the streptomycin sulfate precipitation step and by FPLC fractionation (see above), an 11-kDa protein copurified with the 18-kDa protein during small-scale protein purification by using DNA affinity chromatography. It is indicated in the partially purified protein preparation by an open arrowhead (Fig. 4A). This protein was gradually removed by more extensive washing in the presence of nonspecific competitor pUC18 DNA. N-terminal sequencing yielded the sequence AKKRSKLEIIQAILE, which corresponds to the N-terminal sequence of open reading frame SSO10449 (95 amino acids, 11,072 Da), another protein of unknown function (19).
Specificity of DNA binding.
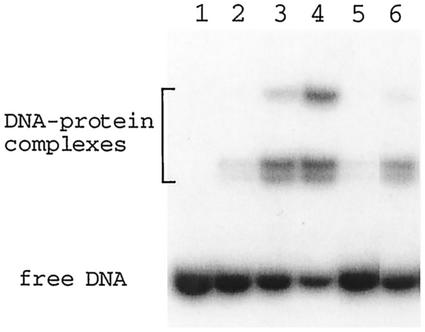
The specificity of binding of the SSO454 protein to SRSR-2p was tested in a competitive band shift assay. Protein was added to 32P-end-labeled SRSR-2p at 0.5-, 1-, and 2-fold molar protein/DNA ratios, and increasing yields of complex were produced (Fig. 5). Evidence supporting the formation of a specific complex was obtained from a competition experiment in which a twofold molar excess of unlabeled SRSR-2p disrupted the complex (Fig. 5, lane 5), whereas competition with a twofold molar excess of nonspecific control DNA (also used in Fig. 3) had little effect on the complex yield (Fig. 5, lane 6).
FIG. 5.
Binding specificity of purified SSO454 to SRSR-2p. A band shift assay was performed with 3 ng of 32P-end-labeled SRSR-2p and purified native SSO454 with no carrier DNA. Lanes: 1, no protein; lanes 2, 3, and 4, protein was added at 0.5-, 1-, and 2-fold protein-DNA molar ratios, respectively; lanes 5 and 6, protein was added at a 1:1 molar ratio. A twofold molar excess of unlabeled SRSR-2p and control DNA (see Fig. 3) was included in lanes 5 and 6, respectively.
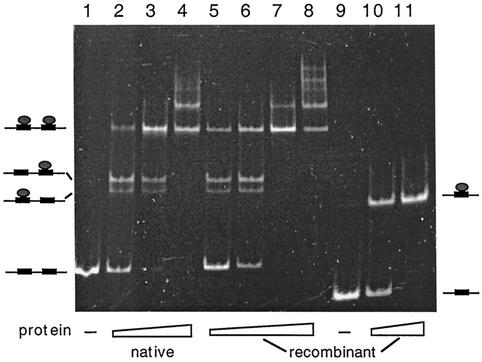
To establish that no minor Sulfolobus protein components were contributing to the complex formation in Fig. 5, the SSO454 gene was cloned into a TYB1 vector, expressed in E. coli, and purified (Fig. 4B). A band shift binding assay was performed with the recombinant protein and both SRSR-2p and SRSR-1p, and the native protein complex with SRSR-2p was included as a control. The native and recombinant proteins produced similar complex band patterns with SRSR-2p (Fig. 6, lanes 1 to 8), confirming that no other Sulfolobus proteins were involved. Moreover, the recombinant protein produced only a single DNA-protein band with SRSR-1p, indicating that the protein can recognizes a single copy of the repeat sequence.
FIG. 6.
Binding of the native and recombinant SSO454 to SRSR-2p (lanes 1 to 8) and of recombinant SSO454 to SRSR-1p (lanes 9 to 11). Protein-DNA molar ratios of mixing were as follows: lanes 1 and 9, no protein; lanes 2 to 4, 2.7, 6.8, and 13.5, respectively; lanes 5 to 8, 1.6, 4.2, 8.4, and 16, respectively; and lanes 10 to 11, 2.8 and 5.6, respectively. Models for the protein-DNA complexes are drawn adjacent to the gel bands.
The doublet bands observed for the protein-SRSR-2p complex are also discernible in the experiments performed with Sulfolobus cell extracts (Fig. 2 and 3). Therefore, these bands do not arise, as first suspected, from the binding of an additional different protein. Thus, we infer that the doublet corresponds to the two single protein-DNA complexes, whereas the single upper band corresponds to the DNA fragment with two bound proteins, as illustrated in Fig. 6. The multiple slow-moving bands observed at higher protein concentrations (Fig. 6, lanes 4 and 8) suggest that the protein can also induce some higher-order structuring of the DNA fragments.
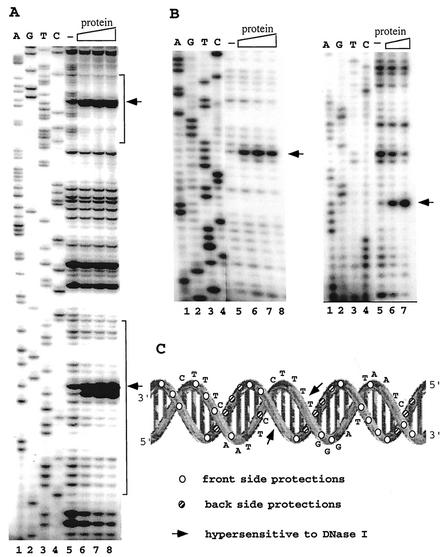
DNase I footprinting of the purified SSO454 protein-SRSR complex.
To localize the DNA-binding site, DNase I footprinting was performed on the complex of the native SSO454 protein and SRSR-2p, 32P labeled at one 5′ end, and SRSR-2c, 32P 5′ end labeled at both ends (see Materials and Methods). Strongly enhanced cutting was observed at the center of each repeat structure, and protection effects were observed along most of the repeat sequence (Fig. 7A). Moreover, the same DNase I cutting pattern was observed for both repeats in SRSR-2p (Fig. 7A). There are no effects within the spacer regions except for weak protection at the first one or two nucleotides adjacent to the repeat (see Fig. 7A, lower repeat), which may reflect steric hindrance of the DNase I by the bound protein. For the SRSR-2c that exhibits seven repeat sequence differences from SRSR-2p concentrated within the central region (Fig. 1), fragment footprinting was performed on both DNA strands. Again, strongly enhanced cutting was observed at the center of the repeat, and protection effects occur over most of the repeat sequence. The results are shown for each strand of one repeat in Fig. 7B. The footprinting results for the protein-SRSR-2c complex are superimposed on the DNA secondary structure in Fig. 7C. The two hypersensitive DNase I sites lie adjacent across the minor groove at a site that is central and outward facing, whereas multiple protection effects are concentrated in the three rear-facing major groove sites. This suggests that the protein binds primarily along the back of the repeat structure and causes a distortion, possibly involving unwinding and/or bending of the double helix, at the center. The footprinting data for one strand of SRSR-2p (Fig. 7A) yield a similar pattern of enhancement and protection when superimposed on the DNA secondary structure. For the latter, we infer that the lower band of the strongly enhanced doublet (Fig. 7A) probably derives from a secondary cutting after formation of the upper band. The strongest protection effects for both SRSR-2p and SRSR-2c occur in the first one-third of the repeat sequence (Fig. 7C), where the sequence is also most highly conserved (see Discussion), and this may constitute the primary binding site of the native SSO454.
FIG. 7.
DNase I footprinting analysis of the native SSO454-SRSR complexes. (A) SRSR-2p was sequenced (lanes 1 to 4) and footprinted (lanes 5 to 8). Lane 5, no protein; lanes 6, 7, and 8, a two-, four-, and sixfold molar excess of purified protein, respectively, was incubated with 30 ng of DNA, 32P end labeled at the free 5′ end in the presence of 600 ng of pUC18 DNA, and digested with DNase I (see Materials and Methods). The positions of the two repeat units are indicated by brackets. (B) Both strands of SRSR-2c were sequenced (lanes 1 to 4) and footprinted (lanes 5 to 8). In the left gel, 72 ng of labeled DNA was complexed with no protein (lane 5) or with a 0.6-, 1.2-, or 2.4 molar protein-DNA ratio (lanes 6, 7, and 8, respectively). In the right gel, 36 ng of labeled DNA was incubated with no protein (lane 5) or with a 1.6- or 6.4-fold molar excess of protein (lanes 6 and 7, respectively). All of the footprinting samples were digested with DNase I in the presence of 600 ng of pUC18 DNA. Strongly enhanced DNase I cuts in panels A and B are indicated by arrows. (C) Data from panel B are superimposed on the secondary structure of SRSR-2c.
DNase I treatment of the native protein-SRSR-1p complex yielded the same cutting pattern as illustrated in Fig. 7A for SRSR-2p (data not shown), which indicates, in combination with the binding assay (Fig. 6), that the purified protein can bind specifically to a single copy of the repeat sequence.
DISCUSSION
The SRSR-binding protein SSO454 constitutes another chromosomal binding protein from Sulfolobus. It binds to one side of each SRSR and produces an opening of the DNA structure on the other side. This may mean that it produces a binding site for another protein that is, possibly, required for higher-order structuring of the SRSR clusters. At present, we have no further insight into such a process, and the genome context of SSO454 yields no insight because the protein is encoded in a typical single gene, with a promoter and no Shine-Dalgarno sequence (22), in a conserved region of the Sulfolobus genome (19).
Large SRSR clusters seem to be a general feature of archaeal genomes (Table 1). The small clusters observed in some mesophilic bacteria show insignificant sequence similarity with archaea (11) and, although there are a few extremely thermophilic bacteria, including Thermatoga maritima, that contain larger clusters, with repeat sequences that partially align with those of archaea, these organisms occupy environments in which the genetic exchange with archaea has been shown to occur (14). For the archaea, there appears to be a fairly high level of repeat sequence conservation at the genus level. In addition to Sulfolobus, this has also been shown for the genera Haloferax (10) and Pyrococcus (24). These results are all compatible with the finding of a genus-specific protein, SSO454, binding to the Sulfolobus repeats.
Three archaeal orthologs of SSO454 were identified, with confidence, that are confined to the closely related genera Sulfolobus and Acidianus, and all four proteins show sequence similarity in the range 51 to 59% identity and 58 to 74% similarity. Moreover, all exhibited the tripartite repeat structure that we found in SSO454, and three carry a conserved hydrophobic C-terminal tail. This is illustrated for the aligned sequences (Fig. 8), for which a consensus sequence for the protein repeat is given. The protein repeat sequences give good matches with helix-turn-helix motifs that are implicated in DNA binding (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/). They also give good matches with motifs in many diverse archaeal proteins, to some of which have been attributed DNA-binding functions. However, we could detect no obvious paralogs of SSO454 in the Sulfolobus genome sequences which might, for example, recognize other SRSR sequences.
FIG. 8.
Tripartite structure of SSO454 and its homologs. The repeat structure in the S. solfataricus protein was identified by using the program Radar (European Bioinformatics Institute, Hinxton, United Kingdom). The three repeat regions of SSO454 are aligned together with the corresponding regions of the homologous proteins from S. tokodaii (identity and similarity, 55 and 73%), S. acidocaldarius (51 and 58%), and Acidianus brierleyi (59 and 74%). A consensus sequence for the repeat is given below. The sequences were aligned manually.
There is a positive sequence correlation between SSO454, and its orthologs, and the SRSRs, which suggests that the binding protein and repeats have coevolved. The SRSR alignment (Fig. 9) demonstrates that, with one exception, all of the main clusters from Sulfolobus and Acidianus species show closely similar sequence conservation patterns, whereas the other crenarchaea show marked differences, especially in the second half of their repeats. This raises the possibility that different proteins recognize the families of DNA repeats occurring in different genera (Table 1). In the present study, we demonstrate that SSO454 can bind specifically to the two major groups of repeat sequence found in S. solfataricus, groups which only exhibit ca. 70% sequence identity (Fig. 1). However, the observation that Sulfolobus acidocaldarius, exceptionally, carries two major SRSR clusters with quite divergent repeat sequences (54% identity; Fig. 9) suggests that some organisms may contain more than one repeat binding protein.
FIG. 9.
Alignment of the available crenarchaeal SRSR sequences. The closely similar Sulfolobus and Acidianus sequences were aligned manually in a group, and the more divergent crenarchaeal sequences were aligned separately. Conserved nucleotides are blackened. The asterisks denote the sequences used in the present study. The positions of the strongly enhanced DNase I cuts on the two DNA are indicated by arrows above the alignment. The figure was prepared by using the BOX shade program (http://www.ch.embnet.org/software/BOX_form.html).
We still know little about the biological function of SRSRs. The sheer size and regularity of the clusters in most archaeal genomes testifies to their importance. This is reinforced further by their integrity; even in the S. solfataricus genome, which contains ca. 350 potentially mobile elements and several integrated regions, there are no insertions (3, 19, 20). The evidence for a role in DNA segregation is based on the detrimental effect on cell division of inserting vector-borne SRSR clusters into haloarchaeal chromosomes (10). Although the evidence is relatively weak, it receives circumstantial support from analyses of the SRSR-containing Sulfolobus plasmid pNOB8 (18) and from our observation that the copy of the pNOB8-like plasmid that has integrated irreversibly into the S. tokodaii chromosome has lost its SRSR cluster. Further indirect evidence arises from the observation that SRSR clusters tend to be located near the origins, or termini, of DNA replication in Pyrococcus chromosomes (24) and probably also in S. solfataricus chromosomes (19). If they play a role in DNA segregation, then it is likely that the SRSR clusters are condensed in the cell and that other proteins are involved.
Acknowledgments
We thank the following people for help with protein sequencing: Susanne Jacobsen, Danish Technical University, who performed the Edman degradation; Grete Koch, Rigshospitet, who carried out the peptide fingerprinting; and Gerhard Saalbach, Risø National Laboratory, who performed matrix-assisted laser desorption ionization-mass spectrometry analyses on the peptides.
This research was supported by a postdoctoral stipendium (X.P.) and a talent stipendium (Q.S.), both from the Danish Technical Science Research Council. B.S. received a DANIDA fellowship, and R.A.G. was awarded a Archaeal Centre Grant from the Danish Natural Science Research Council.
REFERENCES
- 1.Agback, P., H. Baumann, S. Knapp, R. Ladenstein, and T. Hard. 1998. Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nat. Struct. Biol. 5:579-584. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 3.Brügger, K., P. Redder, Q. She, F. Confalonieri, Y. Zivanovic, and R. A. Garrett. 2002. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206:131-141. [DOI] [PubMed] [Google Scholar]
- 4.Charlebois, R. L., Q. She, D. P. Sprott, C. W. Sensen, and R. A. Garrett. 1998. Sulfolobus genome: from genomics to biology. Curr. Opin. Microbiol. 1:581-584. [DOI] [PubMed] [Google Scholar]
- 5.Chong, S., F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus, and M.-Q. Xu. 1997. Single column purification of free recombinant proteins using a self-cleavable affinity tag from a protein splicing element. Gene 192:277-281. [DOI] [PubMed] [Google Scholar]
- 6.Gobom, J., E. Nordhoff, E. Mirgorodskaya, R. Ekman, and P. Roepstorff. 1999. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34:105-116. [DOI] [PubMed] [Google Scholar]
- 7.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Mojica, F. J., G. Juez, and F. Rodriguez-Valera. 1993. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 9:13-21. [DOI] [PubMed] [Google Scholar]
- 10.Mojica, F. J., C. Ferrer, G. Juez, and F. Rodriguez-Valera. 1995. Long stretches of short tandem repeats are present in the largest replicons of the archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 17:85-93. [DOI] [PubMed] [Google Scholar]
- 11.Mojica, F. J., C. Diez-Villasenor, E. Soria, and G. Juez. 2000. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36:244-246. [DOI] [PubMed] [Google Scholar]
- 12.Mori, H., A. Kondo, A. Ohshima, T. Ogura, and S. Hiraga. 1986. Structure and function of the F plasmid genes essential for partitioning. J. Mol. Biol. 192:1-15. [DOI] [PubMed] [Google Scholar]
- 13.Nakata, A., M. Amemura, and K. Makino. 1989. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J. Bacteriol. 171:3553-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesbø, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermatoga maritima. J. Bacteriol. 184:4457-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567 [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Schleper, C., I. Holz, D. Janekovic, J. Murphy, and W. Zillig. 1995. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177:4417-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.She, Q., H. Phan, R. A. Garrett, S. V. Albers, K. M. Stedman, and W. Zillig. 1998. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2:417-425. [DOI] [PubMed] [Google Scholar]
- 19.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, P. Gordon, G. Allard, M. J. Awayez, C.-Y. Chan-Weiher, I. G. Clausen, B. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. Heidekamp de Jong, A. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. Phan Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. M. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.She, Q., K. Brügger, and L. Chen. 2002. Archaeal integrative genetic elements and their impact on genome evolution. Res. Microbiol. 153:25-332. [DOI] [PubMed]
- 21.Tito, B. J. D., Ward, J., Hodgson, C. J. L. Gershater, H. Edwards, L. A. Wysocki, F. A. Watson, G. Sathe, and J. F. Kane. 1995. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol. 177:7086-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolstrup, N., C. W. Sensen, R. A. Garrett, and I. G. Clausen. 2000. Two different and highly organised mechanisms of translation initiation in the archaeon S. solfataricus. Extremophiles 4:175-179. [DOI] [PubMed] [Google Scholar]
- 23.Xue, H., R. Guo, Y. Wen, D. Lin, and L. Huang. 2000. An abundant DNA-binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 182:3929-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zivanovic, Y. P., P. Lopez, H. Philippe, and P. Forterre. 2002. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 30:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]