Abstract
The conversion of ketomethiobutyrate to methionine has been previously examined in a number of organisms, wherein the aminotransferases responsible for the reaction have been found to be members of the Ia subfamily (L. C. Berger, J. Wilson, P. Wood, and B. J. Berger, J. Bacteriol. 183:4421-4434, 2001). The genome of Bacillus subtilis has been found to contain no subfamily Ia aminotransferase sequences. Instead, the analogous enzymes in B. subtilis were found to be members of the If subfamily. These putative aspartate aminotransferases, the yugH, ywfG, ykrV, aspB, and patA gene products, have been cloned, expressed, and characterized for methionine regeneration activity. Only YkrV was able to convert ketomethiobutyrate to methionine, and it catalyzed the reaction only when glutamine was used as amino donor. In contrast, subcellular homogenates of B. subtilis and Bacillus cereus utilized leucine, isoleucine, valine, alanine, phenylalanine, and tyrosine as effective amino donors. The two putative branched-chain aminotransferase genes in B. subtilis, ybgE and ywaA, were also cloned, expressed, and characterized. Both gene products effectively transaminated branched-chain amino acids and ketoglutarate, but only YbgE converted ketomethiobutyrate to methionine. The amino donor preference for methionine regeneration by YbgE was found to be leucine, isoleucine, valine, phenylalanine, and tyrosine. The B. subtilis ybgE gene is a member of the family III of aminotransferases and falls in a subfamily designated here IIIa. Examination of B. cereus and Bacillus anthracis genome data found that there were no subfamily IIIa homologues in these organisms. In both B. cereus and B. anthracis, two putative branched-chain aminotransferases and two putative d-amino acid aminotransferases were discovered as members of subfamily IIIb. These four sequences were cloned from B. cereus, expressed, and characterized. Only the gene product from the sequence designated Bc-BCAT2 was found to convert ketomethiobutyrate to methionine, with an amino donor preference of leucine, isoleucine, valine, phenylalanine, and tyrosine. The B. anthracis homologue of Bc-BCAT2 was also cloned, expressed, and characterized and was found to be identical in activity. The aminooxy compound canaline was found to be an uncompetitive inhibitor of B. subtilis YbgE and also inhibited growth of B. subtilis and B. cereus in culture.
Methionine (Met) is a key amino acid required for protein synthesis, biological methylation, and polyamine biosynthesis. This latter function is particularly important in rapidly growing cells, such as cancer cells and most bacteria and parasites, which must continually synthesize polyamines in order to replicate their DNA (31). The de novo production of Met is energetically expensive and requires aspartate, ATP, NADPH, succinyl-coenzyme A or acetyl-coenzyme A, cysteine or H2S, and 5-methyltetrahydrofolate. In addition, many organisms lack the ability to make Met and rely on exogenous sources. For these reasons, Met bioavailability is limiting and tightly controlled. Almost all organisms examined to date, including those that can synthesize Met, have recycling pathways that can regenerate the amino acid from metabolic by-products.
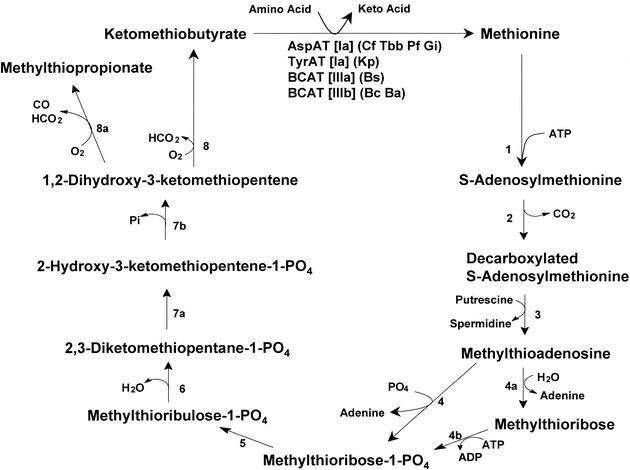
In the production of spermidine from putrescine or spermine from spermidine, Met is consumed (in the form of decarboxylated S-adenosylmethionine) in a one-to-one stoichiometry (Fig. 1). The end product of this reaction is methylthioadenosine, which can be recycled to adenine and Met via a unique pathway, which has been found in organisms ranging from bacteria to mammals (Fig. 1) (5, 17, 30, 48, 51). Studies with cancer cells, bacteria, malaria, and trypanosomes have demonstrated that interference with this Met regeneration pathway leads to the death of rapidly growing cell types. In these studies, the target enzyme was methylthioadenosine phosphorylase or methylthioadenosine nucleosidase, which represents the first step in the bioconversion to Met (4, 18, 41, 48).
FIG. 1.
The Met regeneration pathway. The labeled enzymes are as follows: 1, S-adenosylmethionine synthetase; 2, S-adenosylmethionine decarboxylase; 3, spermidine/spermine synthetase; 4, methylthioadenosine phosphorylase; 4a, methylthioadenosine nucleosidase; 4b, methylthioribose kinase; 5, unidentified isomerase; 6, unidentified dehydratase; 7, enolase-phosphatase; 8, nonenzymatic or dioxygenase; 8a, dioxygenase. The specific aminotransferases that catalyze the final step are shown, with the subfamily membership in square brackets. The organism abbreviations are as follows: Cf, Crithidia fasciculata; Tbb, Trypanosoma brucei brucei; Pf, Plasmodium falciparum; Gi, Giardia intestinalis; Kp, Klebsiella pneumoniae; Bs, Bacillus subtilis; Bc, Bacillus cereus; Ba, Bacillus anthracis.
This laboratory has been investigating the final step of the Met regeneration pathway, where ketomethiobutyrate (KMTB) is converted to Met via an aminotransferase. The exact enzyme catalyzing this final step has been examined in a number of organisms, including Trypanosoma brucei brucei, Crithidia fasciculata, Giardia intestinalis, and Plasmodium falciparum (8, 9). In these organisms, aspartate aminotransferase (AspAT) was found to be the enzyme responsible. In vitro growth inhibition studies have also shown that inhibitors of the parasite AspAT have a cytocidal effect on the cells. In the case of P. falciparum, inhibitors that had a 50% inhibitory concentration (IC50) in the 200 to 500 nM range were uncovered (7). In studies on the gram-negative bacterium Klebsiella pneumoniae, tyrosine aminotransferase (TyrAT) was found to catalyze the reaction (22). Both the parasite AspATs and the bacterial TyrAT were found to be members of the same subfamily of aminotransferases (subfamily Ia; Fig. 2). This subfamily consists of eukaryotic AspATs, gram-negative bacterial AspATs, and bacterial TyrATs, and its constituents tend to have fairly broad substrate specificities (27).
FIG. 2.
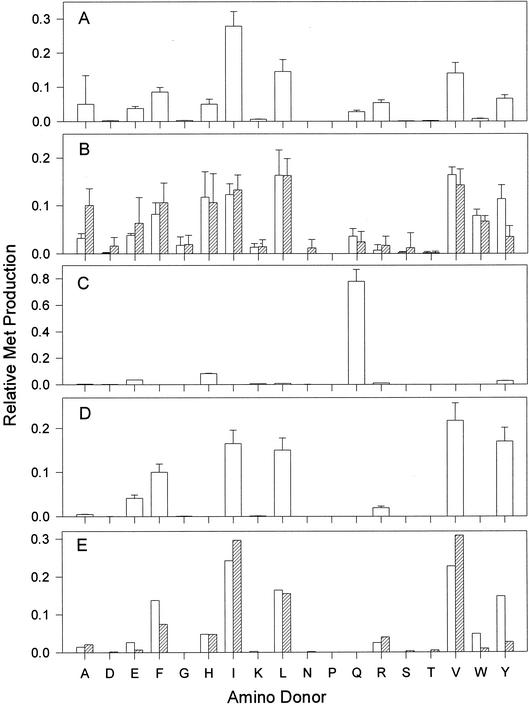
The amino donor range for Met regeneration in Bacillus spp. An enzyme source was mixed with 1.0 mM KMTB, a 2.0 mM concentration of a single amino acid, and pyridoxal phosphate for 30 min at 37°C before analysis of Met production by HPLC. The enzyme sources are as follows: B. subtilis homogenate from cells grown in nutrient broth (A), B. cereus homogenates from cells grown in nutrient broth (open bars) or minimal medium (hatched bars) (B), recombinant B. subtilis YkrV (C), recombinant B. subtilis YbgE (D), or recombinant B. cereus BCAT2 (open bars) or recombinant B. anthracis BCAT2 (hatched bars) (E).
During these previous studies it became apparent, by using aminotransferase sequences available from completed genome projects, that gram-positive bacteria and archaebacteria have no members of the aminotransferase Ia subfamily (see below). The few AspATs that have been characterized for these organisms, such as one from Bacillus subtilis (32) and one from Sulfolobus solfataricus (13), are members of the If subfamily (Fig. 2). To date, members of this subfamily have not been examined for their capacity to transaminate KMTB.
In this study, we have determined the phylogenic relationship of the AspATs from B. subtilis and have cloned, functionally expressed, and characterized these enzymes in regard to Met regeneration. In addition, based on the amino acid preference shown by bacterial homogenates in catalyzing the conversion of KMTB to Met, we have also cloned, expressed, and characterized selected members of the family III of aminotransferases from B. subtilis, B. cereus, and B. anthracis. It was discovered that the final step of Met recycling in these organisms was catalyzed by a single branched-chain amino acid aminotransferase (BCAT).
MATERIALS AND METHODS
Cells and homogenates.
Bacillus subtilis 168 (ATCC 23857) and Bacillus cereus 14579 were obtained from the American Type Culture Collection (Manassas, Va.) and were routinely cultured in nutrient broth at 30°C with agitation at 250 rpm. For selected experiments, B. subtilis was grown in a liquid minimal methionine-free medium as described by Anagnostopoulos and Spizizen (2). For similar experiments, B. cereus was grown in a minimal methionine-free medium as described by Milner et al. (34). Bacillus anthracis Ames was obtained from Defence R&D Canada-Suffield reference stocks and was originally acquired from the U.S. Army Medical Research Institute for Infectious Diseases (USAMRIID) (Frederick, Md.). Growth of B. anthracis was under the same conditions as for B. cereus, except that growth temperature was 37°C.
Cells in late log or stationary phase were harvested by centrifugation at 3,500 × g for 20 min and 4°C. The cell pellets were resuspended in a minimal volume of 25 mM PO4 (pH 7.8)-120 mM KCl-2.5 mM α-ketoglutarate (KG)-1.0 mM dithiothreitol (DTT)-0.2 mM pyridoxal phosphate-protease inhibitors (Complete Tablets; Roche Biomedical, Laval, Quebec, Canada), lysozyme was added to a 0.3% final concentration (wt/vol), and then the mixtures were incubated on ice for 60 min. The samples were then sonicated on ice and dialyzed against 10 mM HEPES (pH 7.4)-1.0 mM EDTA-1.0 mM DTT at 4°C. After dialysis, the samples were briefly resonicated on ice and stored at 4°C for enzyme assays. For long-term storage, glycerol was added to a final concentration of 20% (vol/vol) and the samples were kept at −20°C.
Cloning of aminotransferases.
Genomic DNA was isolated from B. subtilis 168, B. cereus 14579, or B. anthracis Ames by digestion with 0.3% (wt/vol) lysozyme for 1 h on ice, followed by incubation with an equal volume of 100 mM NaCl-10 mM Tris-HCl (pH 8.0)-25 mM EDTA-0.5% (wt/vol) sodium dodecyl sulfate-0.1 mg of proteinase K/ml at 37°C for 1 h with occasional mixing. The mixture was then subjected to extraction with phenol-chloroform-isoamyl alcohol (24:1), and the DNA was ethanol precipitated.
The nucleotide sequences of the B. subtilis aminotransferases were obtained from the SubtiList website (http://www.genolist.pasteur.fr/SubtiList/) (36) and used to design oligonucleotide primers for each enzyme. A gapped genome of B. cereus 14579 was obtained from Integrated Genomics (www.integratedgenomics.com/Public/IG_Release.html), and data from the completed genome project for B. anthracis Ames were obtained from The Institute for Genomic Research (Rockville, Md.) and USAMRIID (40). The nucleotide sequences of the B. cereus and B. anthracis aminotransferases were obtained by examination of the appropriate genome data by using the BLAST program (1) running within BioEdit (20) and used to design oligonucleotide primers for each enzyme. The 5′ primers contained a 12-nucleotide ligation-independent cloning (LIC) (3) sequence and an in-frame start codon, while the 3′ primers contained a 13-nucleotide LIC sequence and an in-frame stop codon. Primer sequences are available from the authors. The target sequences were amplified from the genomic DNA and cloned into pCALnFLAG by using the LIC procedure outlined by Stratagene (La Jolla, Calif.) before transformation into Escherichia coli XL10 competent cells (Stratagene). The recombinant plasmid was purified from these cells by using the QiaSpin miniprep kit (Qiagen), and the presence of the insert was confirmed by digestion with NdeI and SacI. The insert was sequenced by using the Big-Dye cycle sequencing kit (ABI, Foster City, Calif.) and an ABI Prism 310 genetic analyzer.
Phylogenetic analysis.
Additional aminotransferase sequences were obtained from GenBank and were aligned by using the Clustal algorithm and the BLOSUM sequence substitution table in the ClustalX program (50). Aligned sequences were visualized with the Bioedit program (20). The aligned sequences were then used with the ProtDist component of PHYLIP (16) to construct a distance matrix which was the basis for tree construction using neighbor joining (42). All trees were visualized by using Treeview (39).
Expression of recombinant enzymes.
The plasmids from positive clones were transformed into E. coli BL21 DE3 CodonPlus RIL cells (Stratagene) for functional expression. The BL21 cells containing the recombinant plasmid were grown in Luria-Bertani liquid medium containing 50 μg of ampicillin/ml and 50 μg of chloramphenicol/ml at 37°C and 250 rpm until the cell density reached an A600 of 0.6 to 0.8. The culture was then cooled to 28°C, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 1.0 mM before 2 to 5 h of continued culture at 28°C and 250 rpm. The cells were then pelleted by centrifugation at 3,500 × g for 20 min at 4°C and resuspended in a minimal volume of 10 mM HEPES (pH 7.8)-150 mM NaCl-1.0 mM DTT-1.0 mM imidazole-2.0 mM CaCl2 before storage at −20°C. The sample was thawed, sonicated on ice, and centrifuged at 3,500 × g for 20 min at 4°C. The resulting supernatant was loaded onto a 1.6- by 8.0-cm calmodulin-agarose column (Stratagene) equilibrated with the resuspension buffer. The column was eluted with 10 mM HEPES (pH 7.8)-1.2 M NaCl-1.0 mM DTT-3.0 mM EGTA. The eluted enzyme was concentrated to less than 5.0 ml by using a 10-kDa molecular mass cutoff centrifugal filter (Pall Filtron; Mississauga, Ontario, Canada). The concentrated enzyme was kept at 4°C for short-term storage and at −20°C with 20% (vol/vol) glycerol for long-term storage.
Enzyme assays.
Aminotransferase activities were assayed by a high-performance liquid chromatography (HPLC) method (9). Ten microliters of subcellular homogenate or a variable volume of recombinant enzyme was added to 100 μl of substrate mix (100 mM PO4, 50 μM pyridoxal-5-phosphate, and various concentrations of amino acid and keto acid) and incubated for 30 min at 37°C. The samples were then stored at −20°C until analysis by HPLC. BCAT activity was assayed by using 2.0 mM valine, isoleucine, or leucine and 1.0 mM KG mixtures, while d-alanine aminotransferase (DAAT) activity was measured by using 2.0 mM d-alanine-1.0 mM KG. AspAT activity was measured by using 2.0 mM aspartate-1.0 mM KG in the substrate mix. Met regeneration was screened by using 2.0 mM concentrations of each of the amino acids A, D, E, F, G, H, I, K, L, N, Q, R, S, T, W, Y and 1.0 mM KMTB in the substrate mix. The range of effective amino donors for Met formation was determined by using 2.0 mM individual amino acid and 1.0 mM KMTB in the substrate mix. For the determination of Michaelis-Menten constants, the substrate mixes contained 0.1 to 10 mM concentrations of substrate and 5 or 10 mM concentrations of the cosubstrate. Similar kinetic constants were determined by using valine, isoleucine, or leucine and KG mixtures at the same concentrations as for Met formation from KMTB. The apparent Km and Vmax for each substrate pairing was determined by nonlinear curve fitting by using the Scientist software program (Micromax, Salt Lake City, Utah). All samples were analyzed by pre-column derivatization and reverse-phase HPLC by using an Agilent 1100 HPLC equipped with an autosampler, variable wavelength UV and visible spectrophotometric detector, Chemstation operating system, and a 2.1- by 200-mm ODS-AA column (Agilent; Mississauga, Ontario, Canada).
Protein concentration was determined by using the Bio-Rad dye (Bio-Rad; Mississauga, Ontario, Canada). Recombinant protein samples were examined by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gels followed by Coomassie brilliant blue R250 staining.
Inhibition studies.
The inhibition constant for canaline against recombinant B. subtilis YbgE was determined by adding 0, 1.0, 5.0, 10, 50, or 100 μM canaline to enzyme incubation mixtures containing 10 mM KMTB and 1.0, 2.5, 5.0, or 10.0 mM leucine. The resulting data were analyzed as Cornish-Bowden plots (12). For measurement of in vitro growth inhibition, 100 μl of nutrient broth or appropriate minimal medium containing 2 × 104 CFU of B. subtilis or B. cereus was added to an equal volume of medium containing 0 to 10 mM canaline. The cultures were then incubated for 16 h, and the turbidity of the samples was measured at 650 nm by using a ThermoMax microtiter plate spectrophotometer (Molecular Devices; Sunnyvale, Calif.). The IC50 was calculated by nonlinear curve fitting of the data to the Chou equation (11), and the MIC was determined by inspection of the wells for the lowest drug concentration permitting no cell growth.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been submitted to GenBank under accession numbers AF527041, AF527042, AF527043, AF527044, and AF527045.
RESULTS
Methionine regeneration in B. subtilis and B. cereus.
Subcellular homogenates of B subtilis grown in nutrient broth were examined for the range of effective amino donors for the transamination of KMTB (Fig. 2A). Alanine, isoleucine, leucine, and valine were clearly the most effective amino donors, with phenylalanine and tyrosine also able to act as donors. The maximal rate of activity ranged from 1.20 to 1.48 nmol/min/mg of protein (for Ile:KMTB) which was equivalent to that seen in K. pneumoniae homogenates (1.92 nmol/min/mg of protein for Glu:KMTB [22]).
Subcellular homogenates of B. cereus prepared from cells grown in nutrient broth were found to utilize leucine, isoleucine, valine, tyrosine, phenylalanine, tryptophan, and histidine as preferred amino donors for the reaction (Fig. 2B). Alanine, glutamate and glutamine could also act as amino donors to a lesser degree. Cells grown in a minimal medium with sulfate as the only exogenous sulfur source presented a nearly identical amino donor spectrum, with the exception of a greater utilization of alanine and glutamate and a lower use of tyrosine.
Relationships of B. subtilis family I aminotransferases.
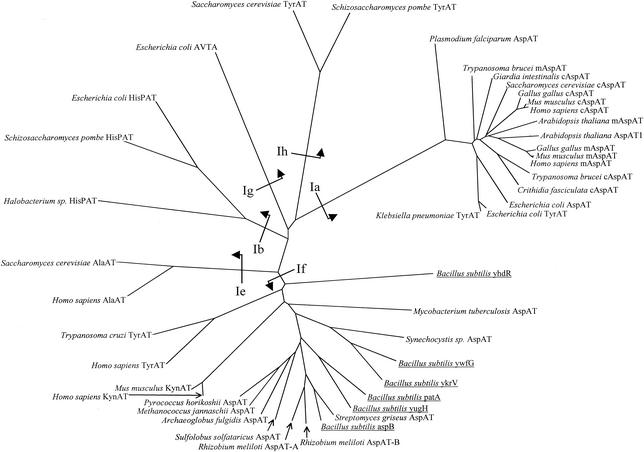
Previous research into the final step of Met recycling in the lower eukaryotes C. fasciculata, T. brucei brucei, G. intestinalis, and P. falciparum and in the gram-negative bacterium K. pneumoniae has demonstrated that the reaction in these organisms is catalyzed by AspATs and TyrAT that belong to the Ia subfamily of the enzymes (9, 22). However, tree construction of the complete aminotransferase I family clearly showed that gram-positive bacteria and archaebacteria had no enzymes in the Ia subfamily (22). The previously assembled phylogenies have suggested that gram-positive bacterial AspATs are members of the aminotransferase If subfamily. The B. subtilis genome has six open reading frames (aspB, patA, ykrV, yugH, ywfG, and yhdR) with significant homology to AspATs from diverse organisms (28). One of these putative AspATs, the aspB gene product, corresponds to an AspAT which had been previously purified and characterized from cell homogenates (32). Alignment of the six B. subtilis sequences with selected members of the aminotransferase I family and construction of a phylogeny via neighbor joining clearly showed that the B. subtilis sequences clustered with other enzymes of the If subfamily (Fig. 3). Therefore, the putative AspATs in B. subtilis are all members of the If subfamily.
FIG. 3.
Family I aminotransferases. Selected sequences were aligned via the clustal algorithm and utilized for tree construction with the neighbor-joining method. The putative B. subtilis AspAT sequences are underlined, and the subfamily designations (as defined by Iraqui et al. [25a]) are shown on the base of the appropriate branches. The enzymes are abbreviated as follows: AspAT, aspartate aminotransferase; TyrAT, tyrosine aminotransferase; AVTA, alanine:valine aminotransferase; HisPAT, histidinol-phosphate aminotransferase; AlaAT, alanine aminotransferase; KynAT, kynurenine aminotransferase.
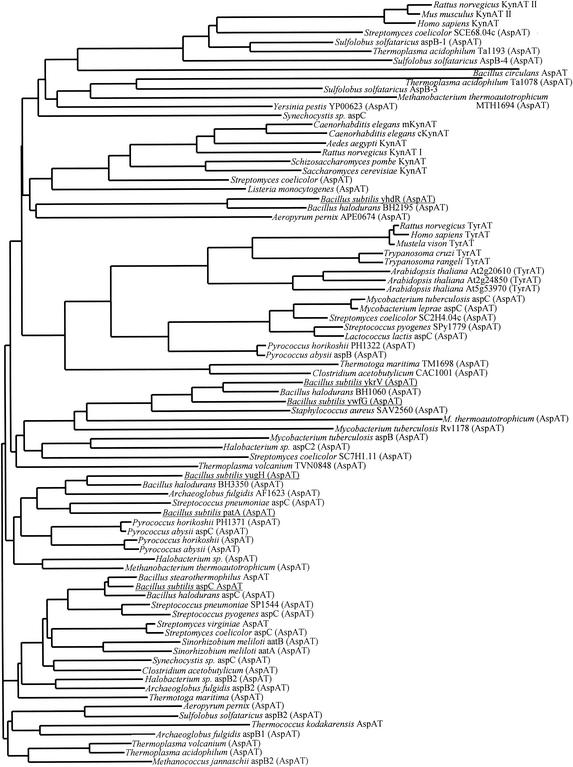
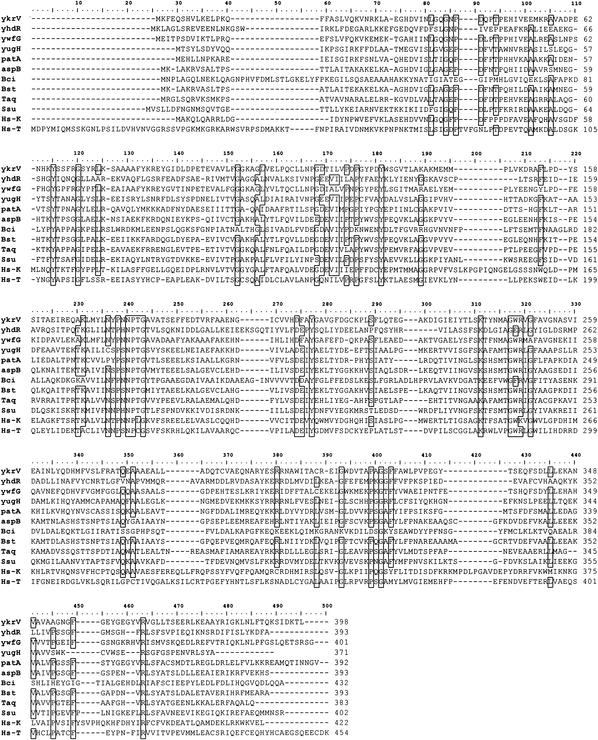
Sequence alignment and tree construction of all the existing members of the If subfamily available in public databases showed that the B. subtilis sequences are broadly dispersed among several divisions of the subfamily (Fig. 4). The YhdR sequence was 50% identical to that of the Bacillus halodurans BH2195 gene product and had a low identity to the other subfamily If enzymes. The YkrV and YwfG sequences were 49% identical to each other and had 60 and 48% identity, respectively, to the sequence of the B. halodurans BH1060 gene product. The YugH sequence was 53% identical to that of the B. halodurans BH3350 gene product, and the PatA sequence was 45% identical to the Streptococcus pneumoniae aspC gene product. Finally, the B. subtilis AspB sequence was 73% identical to the B. halodurans aspC gene product. It is interesting that, aside from the YkrV and YwfG sequences, the putative B. subtilis AspATs bear little resemblance to each other and are highly divergent within subfamily If.
FIG. 4.
The If subfamily of aminotransferases. The sequences were aligned with the clustal algorithm and used for tree construction with the neighbor-joining method. The putative B. subtilis AspATs are underlined.
Characterization of the B. subtilis subfamily If aminotransferases.
The alignment of the six B. subtilis sequences with selected subfamily If enzymes that have been fully characterized in the literature highlighted the lack of sequence conservation within subfamily If (Fig. 5). In the alignment, only Y115, G120, Y181, P238, N240, P241, G243, D274, Y277, K311, G317, R401, and R463 are conserved across the 12 enzymes. Of these residues, G243(G197), D274(D222), K311(K258), and R463(R386) are reported by Mehta et al. to be conserved across all four aminotransferase families (33). By the convention of Mehta et al. (33), conserved residues are labeled in parentheses with the corresponding residue number from pig cytosolic AspAT. Residues Y115(Y70), G120(G110), N240(N194), P241(P195), Y277(Y225), and G317(G268) are reported by Jensen and Gu to be conserved across all family I aminotransferases (27). Therefore, unlike subfamily Ia aminotransferases, the subfamily If enzymes do not present a recognizable unique sequence motif.
FIG. 5.
Alignment of the putative B. subtilis AspATs. The following sequences were aligned by using the clustal algorithm: B. subtilis YkrV, YhdR, YwfG, YugH, PatA, and AspB, B. circulans (Bci) AspAT (6), Bacillus sp. (Bst) AspAT (49), Thermus aquaticus AspAT (Taq) (38), Sulfolobus solfataricus (Ssu) AspAT (13), human KynAT (Hs-K) (35), and human TyrAT (Hs-T) (47). Residues conserved by 75% of the sequences are boxed.
The aspB, patA, yugH, ykrV, and ywfG genes were cloned and functionally expressed as calmodulin-binding peptide fusion proteins. Each of the putative enzymes was screened for Met regeneration activity by using a mixture containing KMTB as the amino acceptor and an amino acid mixture as potential amino donors. YugH, YwfG, PatA, and AspB catalyzed little detectable Met regeneration under these conditions. Only YkrV was able to produce a moderate amount of Met (approximately 40 nmol/min/mg of protein) by using the mixture of amino donors. The enzyme was rescreened by using KMTB and individual amino acids as amino donors (Fig. 2C). The enzyme was found to have an almost exclusive preference for Gln as an amino donor for the transamination of KMTB. The enzyme was also found to be similarly active in catalyzing Gln:KG aminotransfer (data not shown). In this regard, YkrV has the characteristics of a glutamine aminotransferase (KynAT). Of the putative B. subtilis AspATs, only YkrV was effective at transaminating KMTB. However, the amino donor specificity for recombinant YkrV did not match the major activity seen above in B. subtilis cell homogenates grown in nutrient broth or minimal medium. Another aminotransferase(s) is clearly responsible for the cellular activity seen by using branched-chain and aromatic amino acids as amino donors.
Characterization of B. subtilis family III aminotransferases.
The strong preference for branched-chain amino acids as amino donors suggested the possibility that a BCAT was responsible for this Met regeneration activity in B. subtilis. In a previous publication, Hall et al. (21) reported that rat brain BCAT could catalyze Met:KG aminotransfer at a rate approximately 10% that of Leu:KG, but this reaction has not been further examined, particularly in the reverse reaction. Similarly, bacterial BCATs are known to play a role in the conversion of Met to odor compounds in cheese bacteria (10, 10, 54), but the use of KMTB in the reverse reaction has not been studied.
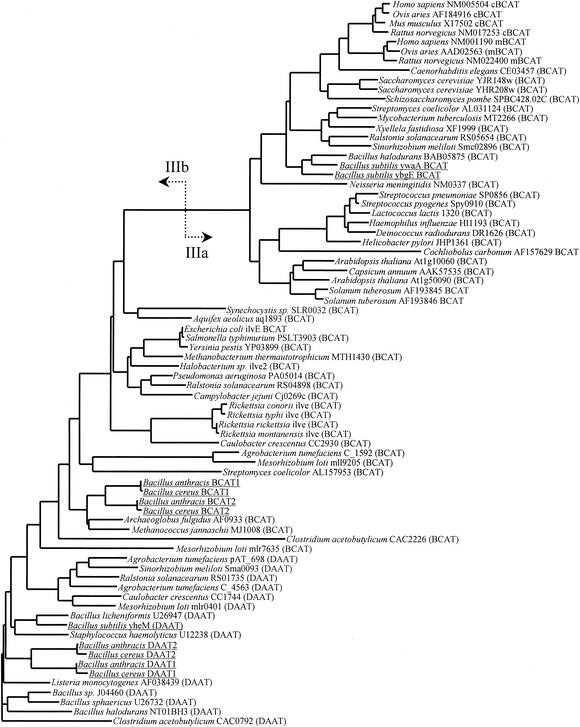
BCATs are members of family III of the aminotransferase superfamily, which appears to be evolutionarily unrelated to the family I and II aminotransferases (26). Members of family III have not been subjected to the same level of phylogenic analysis as family I, and there is no information available on potential subfamilies within family III. Clustal alignment of all the available family III aminotransferase sequences followed by tree construction using a variety of algorithms demonstrated a clear division into two subfamilies (Fig. 6). The first, designated subfamily IIIa, contained BCATs from eukaryotic and bacterial sources. The second, designated subfamily IIIb, contained BCATs from archaeal and bacterial sources, as well as DAATs. Analysis of the B. subtilis genome data (28) has uncovered three sequences with homology to existing members of family III: ywaA, ybgE, and yheM (Fig. 7). The YheM sequence clearly localized among the DAAT sequences in subfamily IIIb and was 62% identical to the Bacillus licheniformis DAAT. The YbgE and YwaA sequences were found in the IIIa subfamily and were 59% identical to each other and 60 and 62% identical, respectively, to the sequence of the B. halodurans BAB05575 gene product.
FIG. 6.
Family III aminotransferases. The sequences were aligned with the Clustal algorithm and used for tree construction with the neighbor-joining method. The division between subfamilies IIIa and IIIb is shown by arrows. The B. subtilis, B. cereus, and B. anthracis sequences are underlined.
FIG. 7.
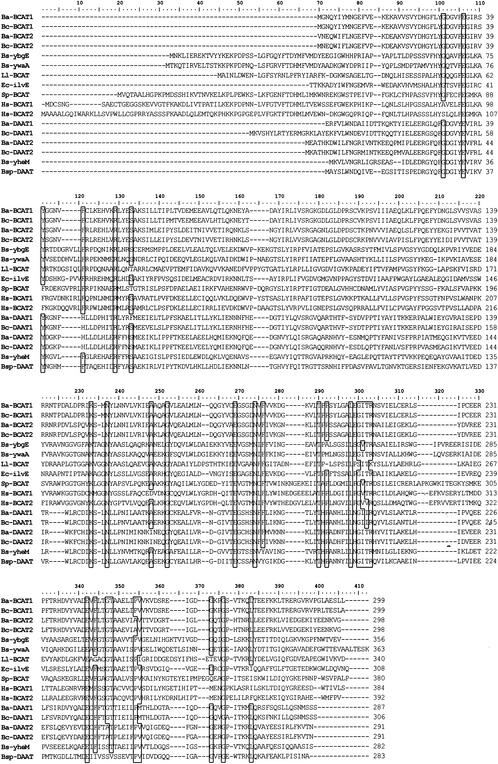
Alignment of the B. subtilis, B. cereus, and B. anthracis family III enzymes. The following sequences were aligned with the Clustal algorithm: B. subtilis YbgE, YwaA, and YheM; B. cereus and B. anthracis BCAT1 and -2 and DAAT1 and -2; Ll-BCAT, Lactococcus lactis BCAT (54); Ec-ilvE, E. coli BCAT (29); Sp-BCAT, Schizosaccharomyces pombe BCAT (14); Bsp-DAAT, B. sphaericus DAAT (53); Hs-BCAT1, human BCAT1 (25); Hs-BCAT2, human BCAT2 (15). Residues conserved by 75% of the sequences are boxed.
Alignment of YheM, YbgE, and YwaA sequences with six other family III sequences that have been fully characterized in the literature and with the B. cereus and B. anthracis family III sequences (see below) demonstrated a low number of conserved residues across the family (Fig. 7). As the family III aminotransferases are now thought of as unrelated to the family I and II enzymes (26), it would be inappropriate to continue using pig cytosolic AspAT as the reference sequence for family III enzymes. The residues in parentheses reflect the corresponding residue in the E. coli ilvE gene product, which has been well studied (29). Residues E106(E37), R129(R59), K233(K159), N237(N163), G252(G178), E269(E193), N274(N198), T290(T209), L298(L217), E342(E251), and G376(G278) are conserved across the sequences shown in Fig. 7, with K233(K159) as the pyridoxal-phosphate binding site. Even among this small selection of family III enzymes, it is clear that there is no retention of D/E222 (in pig cytosolic AspAT), which was postulated by Mehta et al. (33) as an invariant residue across all aminotransferases. While completely conserved across family I and II enzymes, this residue appears to be unnecessary for activity in family III enzymes.
The two putative BCATs were cloned and functionally expressed as calmodulin-binding peptide fusion proteins. Both recombinant enzymes were screened by using KMTB and single amino acids as amino sources. YwaA catalyzed little Met production, whereas YbgE readily formed Met using leucine, isoleucine, valine, phenylalanine, and tyrosine as amino donors (Fig. 2D). Both enzymes were active in leucine:KG aminotransfer (data not shown). As the amino donor preference for Met regeneration with the recombinant YbgE closely mirrored that seen in B. subtilis homogenates and in the semipurified fraction, more-detailed kinetic studies were performed with the BCATs (Table 1). The Km values for Leu, Val, and Ile in the presence of KMTB or KG were very similar for both YbgE and YwaA and ranged from 2.11 to 5.68 mM. Both YbgE and YwaA catalyzed KMTB transamination at a rate approximately 10-fold lower than that of the corresponding reaction with KG. In addition, YwaA was 100-fold less active than YbgE for all of the reactions examined. Therefore, it would appear that YbgE is responsible for the majority of the Met regeneration seen in the B. subtilis homogenates.
TABLE 1.
Kinetic characterization of the Bacillus BCATsa
| Gene product | Substrate | Cosubstrate | Apparent Km (mM) | Apparent Vmax (μmol/min/mg of protein) |
|---|---|---|---|---|
| B. subtilis YbgE | Leu | KG | 3.99 ± 0.75 | 14.58 ± 0.12 |
| Val | KG | 2.82 ± 1.55 | 13.93 ± 0.29 | |
| Ile | KG | 3.15 ± 0.93 | 16.61 ± 0.20 | |
| Leu | KMTB | 2.78 ± 0.76 | 1.87 ± 0.14 | |
| Val | KMTB | 3.20 ± 0.58 | 2.03 ± 0.09 | |
| Ile | KMTB | 2.36 ± 0.59 | 1.84 ± 0.12 | |
| B. subtilis YwaA | Leu | KG | 3.04 ± 0.33 | 0.10 ± 0.02 |
| Val | KG | 3.34 ± 1.22 | 0.05 ± 0.01 | |
| Ile | KG | 4.83 ± 1.33 | 0.23 ± 0.03 | |
| Leu | KMTB | 5.68 ± 0.39 | 0.02 ± 0.00 | |
| Val | KMTB | NDb | ND | |
| Ile | KMTB | 2.11 ± 0.94 | 0.02 ± 0.00 | |
| B. cereus BCAT2 | Leu | KG | 0.48 ± 0.29 | 0.54 ± 0.07 |
| Val | KG | 0.91 ± 0.45 | 0.74 ± 0.10 | |
| Ile | KG | 0.59 ± 0.25 | 0.63 ± 0.06 | |
| KG | Leu | 4.34 ± 1.21 | 0.80 ± 0.10 | |
| Leu | KMTB | 2.09 ± 0.72 | 0.52 ± 0.06 | |
| Val | KMTB | 3.09 ± 1.26 | 0.90 ± 0.13 | |
| Ile | KMTB | 1.37 ± 0.57 | 1.16 ± 0.15 | |
| KMTB | Leu | 2.84 ± 1.01 | 1.44 ± 0.20 | |
| B. anthracis BCAT2 | Leu | KG | 0.41 ± 0.06 | 0.25 ± 0.01 |
| Val | KG | 0.71 ± 0.33 | 0.34 ± 0.04 | |
| Ile | KG | 0.67 ± 0.28 | 0.33 ± 0.04 | |
| KG | Leu | 0.83 ± 0.22 | 0.13 ± 0.01 | |
| Leu | KMTB | 1.75 ± 0.58 | 0.44 ± 0.06 | |
| Val | KMTB | 3.23 ± 1.18 | 0.43 ± 0.06 | |
| Ile | KMTB | 1.66 ± 0.52 | 0.45 ± 0.05 | |
| KMTB | Leu | 0.95 ± 0.20 | 0.42 ± 0.02 |
The enzymes were incubated with various amounts of substrate and 10 mM cosubstrate before analysis by HPLC as described in Materials and Methods.
ND, no detectable Met produced.
Characterization of the B. cereus family III aminotransferases.
As the primary catalyst of KMTB transamination in B. subtilis proved to be the ybgE gene product and was a member of the family III of aminotransferases, B. cereus and B. anthracis genome data were examined for potential homologues. Both B. cereus and B. anthracis were found to contain four sequences with high identity to known members of family III aminotransferases. Two of these sequences were similar to those of BCATs, and two were similar to those of DAATs, and these were given the names Bc-BCAT1 (GenBank AF527041), Bc-BCAT2 (GenBank AF527043), Bc-DAAT1 (GenBank AF527045), Bc-DAAT2 (GenBank AF527042), Ba-BCAT1, Ba-BCAT2 (GenBank AF527044), Ba-DAAT1, and Ba-DAAT2.
Despite their relatively close relationship to B. subtilis, neither B. cereus nor B. anthracis had any aminotransferases with homology to subfamily IIIa. All four B. cereus and B. anthracis family III aminotransferases are members of subfamily IIIb and are not closely related to the B. subtilis ybgE gene product (Fig. 6). Given the extreme similarity of the B. cereus and B. anthracis genomes, it is not surprising that each of the family III enzymes from B. cereus is most identical to its B. anthracis counterpart, with identities in excess of 96%. The BCAT1s were also found to be 61% identical to the BCAT2s, and all four BCATs were 56% identical to the Archaeoglobus fulgidus AF0933 gene. The B. subtilis ybgE gene product, which is responsible for Met regeneration in that organism and is a member of subfamily IIIa, was only 17% identical to the B. cereus and B. anthracis BCATs.
As with the BCATs, the putative DAAT sequences from B. cereus were most identical to their B. anthracis counterparts, with an identity in excess of 86%. The two DAAT1 sequences were found to be 47% identical to the DAAT2 sequences, and all four DAAT sequences were 42% identical to the B. subtilis yheM gene product sequence.
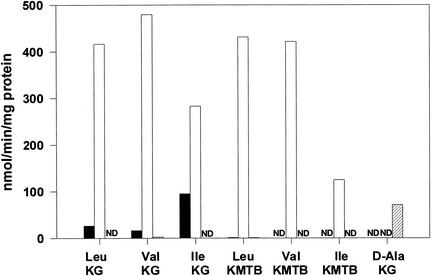
The four putative family III aminotransferase sequences in B. cereus were cloned and functionally expressed as calmodulin-binding peptide fusion proteins in E. coli. Bc-DAAT2 was expressed completely as insoluble, inactive inclusion bodies. Bc-BCAT1, Bc-BCAT2, and BC-DAAT1 were screened with branched-chain amino acids and KG or KMTB, as well as d-alanine and KG, in order to determine their capacity for transamination of these substrates (Fig. 8). As would be expected, only Bc-DAAT1 catalyzed the d-alanine:KG aminotransfer, confirming its identity as a DAAT. Both Bc-BCAT1 and Bc-BCAT2 were active with branched-chain amino acids when KG was used as an amino acceptor, confirming their identities as BCATs. However, Bc-BCAT2 catalyzed these reactions three- to fourfold better than Bc-BCAT1. Only Bc-BCAT2 had any appreciable activity with branched-chain amino acids and KMTB as an amino donor. Therefore, while both BCATs were capable of transaminating KG and branched-chain amino acids, only Bc-BCAT2 had the ability to produce Met from KMTB. Therefore, Bc-BCAT2 is likely the analogue of YbgE in B. subtilis and acts as the primary catalyst of Met regeneration in B. cereus. The recombinant Bc-BCAT2 was screened with single amino acids and KMTB in order to define the amino donor preference of the enzyme (Fig. 2E). The enzyme used isoleucine, leucine, valine, tyrosine, phenylalanine, and, to a lesser degree, histidine and arginine. Therefore, with the exception of alanine and glutamine as amino donors, Bc-BCAT2 would appear to be responsible for almost all the Met regeneration activity seen in the B. cereus homogenates (Fig. 2B).
FIG. 8.
Substrate preference for the recombinant B. cereus family III aminotransferases. B. cereus BCAT1 (black bars), BCAT2 (white bars), or DAAT1 (hatched bars) was incubated with 2.0 mM amino acid-1.0 mM keto acid-pyridoxal phosphate before HPLC analysis for the production of Met from KMTB or glutamate from KG. ND, no detectable activity.
Selected substrate pairings were examined in more detail in order to determine the kinetic parameters of the enzyme (Table 1). The Km and Vmax values were found to be similar regardless of whether KG or KMTB was used as the amino acceptor, with Km values ranging from 0.48 to 4.34 mM and Vmax values ranging from 0.54 to 1.44 μmol/min/mg of protein. Therefore, the enzyme appears to be equally active in using branched-chain amino acids for producing glutamate or Met. This result is in contrast to those obtained with YbgE in B. subtilis, where KMTB transamination had a Vmax 10-fold lower than that of KG transamination. The B. cereus enzyme is thus better adapted for Met production than that in B. subtilis and may be a reflection of membership in different subfamilies within family III.
B. anthracis BCAT2 and methionine regeneration.
As Bc-BCAT2 was identified as the sole family III enzyme responsible for Met regeneration in B. cereus, the B. anthracis homologue was cloned and expressed as a calmodulin-binding peptide fusion protein in E. coli. Ba-BCAT2 was screened by using single amino acids and KMTB as an amino donor, with results nearly identical to that seen with Bc-BCAT2 (Fig. 2E). The only difference seen between the two enzymes was a lower preference for aromatic amino acids as amino donor with Ba-BCAT2. Again, selected substrate pairs were further examined in order to define the kinetic properties of the enzyme (Table 1). As with Bc-BCAT2, Ba-BCAT2 had Km and Vmax values that were similar regardless of the amino acceptor. The Km values were 0.41 to 0.83 mM for KG and 0.95 to 3.23 mM for KMTB, while the Vmax values were 0.13 to 0.34 μmol/min/mg of protein for KG and 0.42 to 0.44 μmol/min/mg of protein for KMTB. Therefore, Ba-BCAT2 catalyzes the formation of Met or glutamate equally well and likely acts as the primary source of Met regeneration in B. anthracis.
Growth inhibition by canaline.
The production of Met from KMTB and leucine by the recombinant B. subtilis YbgE was examined in the presence of the aminotransferase inhibitor canaline, which has been demonstrated to be an effective inhibitor of Met regeneration (7, 8, 9, 22). Canaline was found to interfere with the reaction in an uncompetitive manner, with a calculated Ki of 48.41 ± 19.64 μM (mean ± standard deviation).
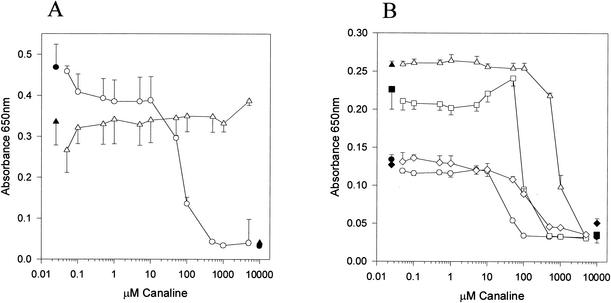
Canaline was also examined as an inhibitor of B. subtilis growth in vitro. When 2 × 104 CFU of B. subtilis was used as the inoculum in a final volume of 200 μl of nutrient broth, up to 5.0 mM canaline was found to have little effect on cell growth over 16 h (Fig. 9A). When the experiment was repeated with a minimal medium, canaline was found to be an effective growth inhibitor, with an IC50 of 36.69 ± 9.62 μM and an MIC of 500 μM. These experiments were repeated with B. cereus in nutrient broth and in a defined minimal medium (Fig. 9B). Since B. cereus and B. anthracis are the same species, differing only in plasmid content (23), the former organism makes a convenient, lower pathogenicity model for the latter. As was seen with B. subtilis, canaline is much more effective as an antibacterial when utilized in minimal medium. However, the degree of difference between these two media was not as marked as with B. subtilis. Canaline killed B. cereus with an IC50 of 38 μM in minimal medium and 759 μM in nutrient broth and MICs of 100 and 5,000 μM, respectively. Total B. cereus growth in minimal medium (which has no exogenous protein and uses sulfate as a sulfur source) was approximately half of that seen in nutrient broth (which contains 30 mg/ml of protein and uses methionine and cysteine as sulfur sources). Addition of 1 or 10 mM methionine to the minimal medium reversed most of this inherent growth inhibition but had little effect on the IC50 of canaline. Supplementation of minimal medium with 30 mg of bovine serum albumin (BSA)/ml had no effect on inherent growth rate and also had little effect on the IC50 of canaline.
FIG. 9.
In vitro inhibition of Bacillus sp. growth by canaline. (A) B. subtilis early log cells were inoculated into nutrient broth (triangles) or minimal medium (circles) in the presence of various concentrations of canaline. Growth after incubation overnight at 30°C was measured by turbidity at 650 nm. (B) B. cereus early-log-phase cells were inoculated into nutrient broth (triangles) or minimal medium (circles) in the presence of various amounts of canaline. Growth after incubation overnight at 30°C was measured by turbidity at 650 nm. The squares and diamonds represent minimal medium supplemented with 10 mM Met and 30 mg of BSA/ml, respectively. The dark symbols are the appropriate values for growth with no inhibitor and for medium without cells.
DISCUSSION
Polyamine biosynthesis and its associated Met regeneration pathway have been the subject of a number of studies on experimental chemotherapeutics with bacteria, protozoa, and cancer cells (31). To date, none of the previous reports have examined this chemotherapeutic potential in gram-positive bacteria. Many of the enzymes shown in Fig. 1 have been characterized for B. subtilis, including S-adenosylmethionine synthase (52), S-adenosylmethionine decarboxylase (43), methylthioadenosine nucleosidase (44), and methylthioribose kinase (46), but have not been considered as drug targets. Previous work with parasitic protozoa and K. pneumoniae had identified the regeneration of Met from KMTB by aspartate or tyrosine aminotransferase as a potential target for therapeutic development (7-9, 22). In this study, we have identified branched-chain amino acid aminotransferase as the primary source of Met recycling in B. subtilis, B. cereus, and B. anthracis.
Unlike the situation seen with the TyrAT in K. pneumoniae, where tyrosine:KMTB and tyrosine:KG aminotransfer occurred at equal rates (22), the B. subtilis YbgE catalyzed leucine:KG aminotransfer approximately 10-fold better than leucine:KMTB aminotransfer. When growing in nutrient broth, B. subtilis has access to large amounts of exogenous Met and might repress the expression of an aminotransferase required for Met regeneration. However, we have examined subcellular homogenates made from B. subtilis grown in minimal medium with no exogenous Met and sulfate as the sulfur source and have found no substantial increase in the formation of Met from KMTB (data not shown).
As was the case with B. subtilis, B. cereus homogenates preferentially catalyzed KMTB transamination by using branched-chain amino acids. However, while this activity is catalyzed by YbgE in B. subtilis, neither B. cereus nor B. anthracis was found to contain any aminotransferase with a high identity to YbgE. The sequence identity between YbgE and the B. cereus and B. anthracis aminotransferases is very low (approximately 17%). Given that the B. cereus complex and B. subtilis are fairly closely related as judged by 16S ribosomal DNA sequences (data not shown), this lack of homology between BCAT sequences is quite striking. It seems likely that acquisition or loss of BCATs occurred after the split from a Bacillus progenitor to pre-subtilis and pre-cereus complexes. As B. halodurans also contains a BCAT within subfamily IIIa and is much more distantly related to B. subtilis and B. cereus, the most likely explanation is that the B. cereus complex lost any subfamily IIIa sequence(s) and evolved BCAT functionality from DAAT members of subfamily IIIb. These results highlight the perils of extrapolating biochemical findings from one bacterial species to another regardless of the appearance of a close evolutionary relationship.
That family III aminotransferases might catalyze Met regeneration in the place of subfamily Ia aminotransferases is interesting. Family I and family III enzymes appear to be unrelated and the result of convergent, rather than divergent, evolution. Structural studies have shown that family I and family III aminotransferases consist of different folds and do not have a spatial resemblance (26). In addition, family III enzymes are substantially smaller than family I aminotransferases and lack one of the four universally conserved residues found in the other aminotransferase families (33). The evolution of this functionality in family III aminotransferases in Bacillus spp. suggests that this family of enzymes should be examined in other gram-positive bacteria and archaebacteria, which lack subfamily Ia enzymes, and in mammals, where the subfamily Ia enzymes have been found not to catalyze Met regeneration (8). The work of Hutson (24) has shown that rat and human BCAT can catalyze Met:KG aminotransfer, but this finding has not been further developed.
Among the subfamily If enzymes examined in this study, only one, YkrV, was found to have any appreciable Met regeneration activity. However, this enzyme only produced Met from KMTB using glutamine as an amino donor, did not have any aspartate:KG activity, and had substantial glutamine:KG activity. This profile did not match with that seen in subcellular homogenates. Recently, Sekowska and Danchin (45) and Murphy et al. (37) identified members of the ykrTS and ykrWXYZ operons as enzymes involved in Met regeneration. These operons were found to be members of the S-box regulon (19), which is up-regulated during sulfur limitation. In particular, the authors have unequivocally identified the ykrT gene product as the B. subtilis methylthioribose kinase (see Fig. 1). By gene deletion the ykrS, ykrY, ykrZ, ykrU, and ykrW genes have been shown to be involved in the methionine regeneration pathway, but their enzymatic activity has not yet been demonstrated (45). The ykrV gene lies immediately between the ykrTS and ykrWXYZ operons and was hypothesized to act as the catalyst for the last step in the cycle. However, ykrV itself is not S-box regulated, the gene product has not been previously examined for its enzymatic properties, and Sekowska and Danchin have found that deletion of the gene has no effect on methionine regeneration in B. subtilis (45). While we have shown that YkrV will convert KMTB to Met by using glutamine as an amino donor, it does not act in this capacity under the conditions we have examined. Perhaps YkrV plays a role in Met recycling under specific growth conditions not replicated by planktonic growth in nutrient broth or minimal medium. This aspect of YkrV function deserves further attention.
As seen with the other enzymes previously investigated (7-9, 22), canaline acts as an efficient inhibitor of Met regeneration by YbgE with a Ki of 48 μM. However, in nutrient broth, B. subtilis growth in the presence of 5.0 mM canaline showed little inhibition. In a minimal medium containing no exogenous Met, cysteine, or protein, canaline was an effective inhibitor of B. subtilis growth with an IC50 of 37 μM and an MIC of 500 μM. It is interesting that the IC50 for canaline in this minimal medium is very similar to the Ki for canaline against YbgE, suggesting that inhibition of this enzyme plays a role in the toxic effect of the drug. Unlike B. subtilis, where canaline had no effect on cell growth in nutrient broth, B. cereus in nutrient broth was inhibited by canaline with an IC50 of 760 μM and an MIC of 5.0 mM. The basis for the differential sensitivity to canaline between rich and minimal media was examined from two potential mechanisms: exogenous methionine as an antagonist and drug binding to exogenous protein. Addition of methionine to minimal medium had little effect on the IC50 of canaline, which suggested that methionine neither interfered with canaline transport nor rescued metabolically starved cells. The addition of 30 mg of BSA/ml to the minimal medium also had little effect on the IC50 of canaline. Therefore, the compound does not bind well to BSA. As canaline is known to bind to pyridoxal phosphate-dependent enzymes and pyridoxal phosphate itself, a logical step for the future would be the examination of nutrient broth for the levels of pyridoxine, pyridoxal, and pyridoxal phosphate. Addition of a similar amount of cofactor to the minimal medium would then test for loss of canaline activity.
The effectiveness of canaline against B. cereus in in vitro growth inhibition tests suggests that the compound should be examined against B. anthracis in vitro and against B. cereus and B. anthracis in vivo. These experiments and the screening of further aminooxy analogues against the bacilli are planned for the immediate future. In addition, potential synergy with other inhibitors of enzymes involved in polyamine biosynthesis and Met regeneration is to be examined.
Acknowledgments
This work was funded in part by a Defence R&D Canada Technology Investment Fund award. Shane English was supported by the Defence R&D Canada Summer Research Associate program.
We gratefully acknowledge the assistance of Mark Dertzbaugh, USAMRIID, in obtaining prepublication access to the Bacillus anthracis genome data.
Genomic data for Bacillus cereus 14579 were made available by Integrated Genomics (Chicago, Ill.) and were funded by DARPA. Genomic data for Bacillus anthracis Ames were made available by USAMRIID (Frederick, Md.) and The Institute for Genomic Research (Rockville, Md.) and were funded by DARPA.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslanidis, C., and P. J. de Jong. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18:6069-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacchi, C. J., J. R. Sufrin, H. C. Nathan, A. J. Spiess, T. Hannan, J. Garofalo, K. Alecia, L. Katz, and N. Yarlett. 1991. 5′-Alkyl-substituted analogs of 5′-methylthioadenosine as trypanocides. Antimicrob. Agents Chemother. 35:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backlund, P. S., Jr., and R. A. Smith. 1981. Methionine synthesis from 5′-methylthioadenosine in rat liver. J. Biol. Chem. 256:1533-1535. [PubMed] [Google Scholar]
- 6.Battchikova, N., M. Koivulehto, A. Denesyuk, L. Ptitsyn, Y. Boretsky, J. Hellman, and T. Korpela. 1996. Aspartate aminotransferase from an alkalophilic Bacillus contains an additional 20-amino acid extension at its functionally important N-terminus. J. Biochem. (Tokyo) 120:425-432. [DOI] [PubMed] [Google Scholar]
- 7.Berger, B. J. 2000. Antimalarial activities of aminooxy compounds. Antimicrob. Agents Chemother. 44:2540-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger, B. J., W. W. Dai, and J. Wilson. 1998. Methionine formation from alpha-ketomethiobutyrate in the trypanosomatid Crithidia fasciculata. FEMS Microbiol. Lett. 165:305-312. [DOI] [PubMed] [Google Scholar]
- 9.Berger, L. C., J. Wilson, P. Wood, and B. J. Berger. 2001. Methionine regeneration and aspartate aminotransferase in parasitic protozoa. J. Bacteriol. 183:4421-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnarme, P., L. Psoni, and H. E. Spinnler. 2000. Diversity of l-methionine catabolism pathways in cheese-ripening bacteria. Appl. Environ. Microbiol. 66:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, T. C. 1976. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J. Theor. Biol. 59:253-276. [DOI] [PubMed] [Google Scholar]
- 12.Cornish-Bowden, A. 1974. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 137:143-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cubellis, M. V., C. Rozzo, G. Nitti, M. I. Arnone, G. Marino, and G. Sannia. 1989. Cloning and sequencing of the gene coding for aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. Eur. J. Biochem. 186:375-381. [DOI] [PubMed] [Google Scholar]
- 14.Eden, A., and N. Benvenisty. 1998. Characterization of a branched-chain amino-acid aminotransferase from Schizosaccharomyces pombe. Yeast 14:189-194. [DOI] [PubMed] [Google Scholar]
- 15.Faure, M., F. Glomot, R. Bledsoe, S. Hutson, and I. Papet. 1999. Purification and cloning of the mitochondrial branched-chain amino acid aminotransferase from sheep placenta. Eur. J. Biochem. 259:104-111. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Ghoda, L. Y., T. M. Savarese, D. L. Dexter, R. E. Parks, Jr., P. C. Trackman, and R. H. Abeles. 1984. Characterization of a defect in the pathway for converting 5′-deoxy-5′-methylthioadenosine to methionine in a subline of a cultured heterogeneous human colon carcinoma. J. Biol. Chem. 259:6715-6719. [PubMed] [Google Scholar]
- 18.Gianotti, A. J., P. A. Tower, J. H. Sheley, P. A. Conte, C. Spiro, A. J. Ferro, J. H. Fitchen, and M. K. Riscoe. 1990. Selective killing of Klebsiella pneumoniae by 5-trifluoromethylthioribose. Chemotherapeutic exploitation of the enzyme 5-methylthioribose kinase. J. Biol. Chem. 265:831-837. [PubMed] [Google Scholar]
- 19.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 20.Hall, T. A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 21.Hall, T. R., R. Wallin, G. D. Reinhart, and S. M. Hutson. 1993. Branched chain aminotransferase isoenzymes. Purification and characterization of the rat brain isoenzyme. J. Biol. Chem. 268:3092-3098. [PubMed] [Google Scholar]
- 22.Heilbronn, J., J. Wilson, and B. J. Berger. 1999. Tyrosine aminotransferase catalyzes the final step of methionine recycling in Klebsiella pneumoniae. J. Bacteriol. 181:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutson, S. 2001. Structure and function of branched chain aminotransferases. Prog. Nucleic Acid Res. Mol. Biol. 70:175-206. [DOI] [PubMed] [Google Scholar]
- 25.Hutson, S. M., R. K. Bledsoe, T. R. Hall, and P. A. Dawson. 1995. Cloning and expression of the mammalian cytosolic branched chain aminotransferase isoenzyme. J. Biol. Chem. 270:30344-30352. [DOI] [PubMed] [Google Scholar]
- 25a.Iraqui, I., S. Vissers, M. Cartiaux, and A. Urrestarazu. 1998. Characterisation of Saccharomyces cerevisiae AR08 and AR09 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:238-248. [DOI] [PubMed] [Google Scholar]
- 26.Jansonius, J. N. 1998. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 8:759-769. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, R. A., and W. Gu. 1996. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 178:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 29.Kuramitsu, S., T. Ogawa, H. Ogawa, and H. Kagamiyama. 1985. Branched-chain amino acid aminotransferase of Escherichia coli: nucleotide sequence of the ilvE gene and the deduced amino acid sequence. J. Biochem. (Tokyo) 97:993-999. [DOI] [PubMed] [Google Scholar]
- 30.Marchitto, K. S., and A. J. Ferro. 1985. The metabolism of 5′-methylthioadenosine and 5-methylthioribose 1-phosphate in Saccharomyces cerevisiae. J. Gen. Microbiol. 131:2153-2164. [DOI] [PubMed] [Google Scholar]
- 31.Marton, L. J., and A. E. Pegg. 1995. Polyamines as targets for therapeutic intervention. Annu. Rev. Pharmacol. Toxicol. 35:55-91. [DOI] [PubMed] [Google Scholar]
- 32.Mavrides, C., and M. Comerton. 1978. Aminotransferases for aromatic amino acids and aspartate in Bacillus subtilis. Biochim. Biophys. Acta 524:60-67. [DOI] [PubMed] [Google Scholar]
- 33.Mehta, P. K., T. I. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214:549-561. [DOI] [PubMed] [Google Scholar]
- 34.Milner, J. L., S. J. Raffel, B. J. Lethbridge, and J. Handelsman. 1995. Culture conditions that influence accumulation of zwittermicin A by Bacillus cereus UW85. Appl. Microbiol. Biotechnol. 43:685-691. [DOI] [PubMed] [Google Scholar]
- 35.Mosca, M., L. Cozzi, J. Breton, N. Avanzi, S. Toma, E. Okuno, R. Schwarcz, C. Speciale, S. Magagnin, M. Mostardini, and L. Benatti. 1996. Cloning of rat and human kynurenine aminotransferase. Adv. Exp. Med. Biol. 398:449-454. [DOI] [PubMed] [Google Scholar]
- 36.Moszer, I., L. M. Jones, S. Moreira, C. Fabry, and A. Danchin. 2002. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 30:62-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy, B. A., F. J. Grundy, and T. M. Henkin. 2002. Prediction of gene function in methylthioadenosine recycling from regulatory signals. J. Bacteriol. 184:2314-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Farrell, P. A., G. Sannia, J. M. Walker, and S. Doonan. 1997. Cloning and sequencing of aspartate aminotransferase from Thermus aquaticus YT1. Biochem. Biophys. Res. Commun. 239:810-815. [DOI] [PubMed] [Google Scholar]
- 39.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 40.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 41.Riscoe, M. K., A. J. Ferro, and J. H. Fitchen. 1988. Analogs of 5-methylthioribose, a novel class of antiprotozoal agents. Antimicrob. Agents Chemother. 32:1904-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Sekowska, A., J. Y. Coppee, J. P. Le Caer, I. Martin-Verstraete, and A. Danchin. 2000. S-adenosylmethionine decarboxylase of Bacillus subtilis is closely related to archaebacterial counterparts. Mol. Microbiol. 36:1135-1147. [DOI] [PubMed] [Google Scholar]
- 44.Sekowska, A., and A. Danchin. 1999. Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis. DNA Res. 6:255-264. [DOI] [PubMed] [Google Scholar]
- 45.Sekowska, A., and A. Danchin. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekowska, A., L. Mulard, S. Krogh, J. K. Tse, and A. Danchin. 2001. MtnK, methylthioribose kinase, is a starvation-induced protein in Bacillus subtilis. BMC Microbiol. 1:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seralini, G. E., V. Luu-The, and F. Labrie. 1995. Cloning and expression of human tyrosine aminotransferase cDNA. Biochim. Biophys. Acta 1260:97-101. [DOI] [PubMed] [Google Scholar]
- 48.Sufrin, J. R., S. R. Meshnick, A. J. Spiess, J. Garofalo-Hannan, X. Q. Pan, and C. J. Bacchi. 1995. Methionine recycling pathways and antimalarial drug design. Antimicrob. Agents Chemother. 39:2511-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung, M. H., K. Tanizawa, H. Tanaka, S. Kuramitsu, H. Kagamiyama, and K. Soda. 1990. Purification and characterization of thermostable aspartate aminotransferase from a thermophilic Bacillus species. J. Bacteriol. 172:1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, S. Y., D. O. Adams, and M. Lieberman. 1982. Recycling of 5′-methylthioadenosine-ribose carbon atoms into methionine in tomato tissue in relation to ethylene production. Plant Physiol. 70:117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yocum, R. R., J. B. Perkins, C. L. Howitt, and J. Pero. 1996. Cloning and characterization of the metE gene encoding S-adenosylmethionine synthetase from Bacillus subtilis. J. Bacteriol. 178:4604-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yonaha, K., H. Misono, T. Yamamoto, and K. Soda. 1975. D-amino acid aminotransferase of Bacillus sphaericus. Enzymologic and spectrometric properties. J. Biol. Chem. 250:6983-6989. [PubMed] [Google Scholar]
- 54.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]