Abstract
Recent studies have shown that expression of the Staphylococcus aureus lrgAB operon inhibits murein hydrolase activity and decreases sensitivity to penicillin-induced killing. It was proposed that the lrgAB gene products function in a manner analogous to an antiholin, inhibiting a putative holin from transporting murein hydrolases out of the cell. In the present study the cidAB operon was identified and characterized based on the similarity of the cidA and cidB gene products to the products of the lrgAB operon. Zymographic and quantitative analyses of murein hydrolase activity revealed that mutation of the cidA gene results in decreased extracellular murein hydrolase activity compared to that of S. aureus RN6390, the parental strain. Complementation of cidA expression restored the wild-type phenotype, indicating that expression of the cidAB operon has a positive influence on extracellular murein hydrolase activity. The cidA mutant also displayed a significant decrease in sensitivity to the killing effects of penicillin. However, complementation of the cidA defect did not restore penicillin sensitivity to wild-type levels. Reverse transcriptase PCR also revealed that cidAB is maximally expressed during early exponential growth, opposite of what was previously observed for lrgAB expression. Based on these results, we propose that the cidAB operon encodes the holin-like counterpart of the lrgAB operon and acts in a manner opposite from that of lrgAB by increasing extracellular murein hydrolase activity and increasing sensitivity to penicillin-induced killing.
Bacterial murein hydrolases, or autolysins, comprise a broad and diverse family of enzymes that specifically cleave structural components of the bacterial cell wall. These enzymes have been shown previously to participate in a number of important biological functions during cell growth and division, including daughter cell separation, cell wall growth, and peptidoglycan recycling and turnover (14, 15, 17, 23, 38, 42, 55). Murein hydrolases also appear to play an important role in several bacterial developmental processes, such as spore formation, swarming motility, and competence (10, 24, 29, 40). Because of their potentially lethal capacity to hydrolyze the cell wall, it is critical that tight control be exercised over the expression and activity of murein hydrolases. For example, compartment-specific and temporal expression of the complement of murein hydrolases involved during Bacillus subtilis sporulation is dependent on several sporulation-specific sigma factors (11, 30, 32, 51), as well as the late-growth regulator Sin (30, 47). At the posttranscriptional level, murein hydrolase activity has been shown elsewhere to be modulated by several mechanisms, such as substrate modification, selective transport, interactions with lipoteichoic acids and cationic peptides, and cleavage by proteolytic enzymes (7, 13, 17, 23, 34, 36, 49, 54). Recent evidence has suggested that the proton motive force (PMF) and its effect on the cell wall pH also regulate murein hydrolase activity in B. subtilis (4, 5).
In the case of Staphylococcus aureus, Mani et al. (31) have provided initial evidence for a murein hydrolase regulatory locus. Two Tn917-lacZ mutants were identified that displayed negligible rates of autolysis and exhibited normal cell division (31). Furthermore, our group had previously identified a second regulatory locus, designated lytSR, whose mutation resulted in a significantly increased rate of autolysis (2). The lytSR gene products are members of the bacterial two-component regulatory family of proteins and positively regulate expression of the lrgAB operon that is located immediately downstream of lytSR (3). Recent studies in our laboratory have demonstrated that lrgAB expression inhibits extracellular murein hydrolase activity and promotes penicillin tolerance (19). Interestingly, the lrgA gene product shares many structural similarities to the bacteriophage holin family of proteins (19). These small membrane proteins control the timing of bacteriophage-induced lysis by regulating access of the bacteriophage-encoded murein hydrolase, or endolysin, to the cell wall peptidoglycan (53). The activity of a holin is usually inhibited by the presence of a homologous protein, called the antiholin (53). Since the lrgAB locus inhibits murein hydrolase activity, it was hypothesized that LrgA functions in a manner analogous to an antiholin that, accordingly, functions by inhibiting an unidentified holin (19).
In the present study, we have identified an operon homologous to lrgAB, designated cidAB, within the S. aureus genome. Like LrgA, the cidA gene product shared many holin-like characteristics. Analysis of a cidA mutant indicated that this gene enhances extracellular murein hydrolase activity and promotes sensitivity to penicillin. Therefore, we propose that the proteins encoded by the cidAB locus may indeed function in a holin-like manner that is inhibited by the products of the lrgAB operon.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. aureus was grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, Mich.) or filter-sterilized NZY broth (3% [wt/vol] N-Z Amine A [Sigma Chemical Co., St. Louis, Mo.], 1% [wt/vol] yeast extract [Fisher Scientific, Fair Lawn, N.J.] adjusted to pH 7.5), supplemented as necessary with 1.5% (wt/vol) granulated agar (Difco). Escherichia coli DH5α was grown in Luria-Bertani medium (Fisher Scientific). All cultures were grown with shaking (250 rpm) at 37°C in volumes that did not exceed 20% of the flask volume. Antibiotics were purchased from either Sigma Chemical Co. or Fisher Scientific and were used at the indicated concentrations: erythromycin (ERY; 2 μg · ml−1), tetracycline (5 μg · ml−1), ampicillin (100 μg · ml−1), and spectinomycin (50 μg · ml−1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Highly transformable strain; restriction deficient | 28 |
| RN6390 | Parental laboratory strain | 37 |
| KB350 | RN6390 cidA::Erm; Emr | This study |
| E. coli DH5α | Host strain for construction of recombinant plasmids | 22 |
| Plasmids | ||
| pDG647 | Source of Emr cassette; Emr Apr | 21 |
| pCL52.2 | Temperature-sensitive shuttle vector; Tcr Spr | 45 |
| pRN5548 | Gram-positive bacterial expression vector; Cmr | 35 |
| pRN-cidA | CidA ORF cloned into EcoRI and BamHI sites of pRN5548; Cmr | This study |
DNA manipulations.
Chromosomal DNA was isolated from S. aureus by the method of Dyer and Iandolo (9). Plasmid DNA was purified by using either the QIAprep Spin miniprep kit from Qiagen, Inc. (Chatsworth, Calif.), or the WizardPlus SV DNA purification kit from Promega, Inc. (Madison, Wis.). Enzymes used in the manipulation of DNA in this study were purchased from either New England BioLabs (Beverly, Mass.) or Invitrogen Life Technologies (Carlsbad, Calif.). Preparation and transformation of E. coli were accomplished by the procedure described by Inoue et al. (25). Electroporation and φ11-mediated transduction were carried out by previously described methods (27, 48).
RT-PCR analysis of RNA.
Temporal expression of the cidAB operon was examined by reverse transcriptase PCR (RT-PCR) analysis. An overnight culture of RN6390 was used to inoculate 400 ml of NZY in a 2-liter flask to an optical density at 600 nm (OD600) of 0.1 and was grown for 12 h at 37°C with shaking at 250 rpm. Growth was monitored by measuring the OD600 at regular intervals, and corresponding samples of RNA were isolated as previously described (6, 41). Contaminating DNA was removed from each sample by DNase treatment with the DNA-Free kit (Ambion, Inc., Austin, Tex.) according to the manufacturer's protocols. The primers, cidB-R (5′-CCCCTCGAGATAGAATAATAAAATTAGAACAGG-3′) and gyrA-R (5′-TAACTGGCGTACGTTTACCATAACC-3′), were used to generate cDNA templates by standard protocols (43). Briefly, RT reactions were carried out in 10-μl volumes, containing 1.0 μg of total RNA template (denatured at 65°C for 5 min), 1 μM reverse primer, 1 mM deoxynucleoside triphosphates (dNTPs), 10 U of Moloney murine leukemia virus RT (Ambion, Inc.), and 1 μl of the 10× buffer supplied with the RT enzyme. The RT reaction mixtures were incubated at 42°C for 60 min, followed by inactivation at 70°C for 5 min. Each reaction mixture was subsequently treated with 1 U of RNase H (Ambion, Inc.) at 37°C for 20 min. As a control for genomic DNA contamination, duplicate reactions were performed as described above for each RNA sample, except that no RT enzyme was included in the reaction mixture. The cidAB and gyrA transcripts were then detected by PCR with the primer pairs cidA-F (5′-CCCCATATGCACAAAGTCCAATTA-3′)-cidB-R and gyrA-F (5′-CGTGAAGGTGACGAAGTTGTAGG-3′)-gyrA-R, respectively. The cidA-F-cidB-R primer pair generates a 1.1-kb amplicon corresponding to nucleotides (nt) 2626687 to 2625613 of the S. aureus 8325 genome (http://www.genome.ou.edu/staph.html), while the gyrA-F-gyrA-R primer pair amplifies a 100-bp product corresponding to nt 4236 to 4339 of the S. aureus gyr locus (GenBank accession no. D10489). PCR amplification was carried out in 25-μl reaction mixtures containing 2 mM MgCl2, 1.25 U of Taq DNA polymerase (Invitrogen Life Technologies), 2.5 μl of 10× PCR buffer, 0.2 mM dNTP mix, 0.2 μM (each) forward and reverse primers, and 1.5 μl of RT template. Following an initial denaturation at 94°C for 5 min, the cycling conditions (carried out for 25 cycles) included 94°C denaturation for 30 s, 52°C annealing for 30 s, and 72°C extension for 1 min.
Primer extension reactions.
A primer extension analysis of the cidAB promoter region was carried out with the reverse primer cidA-3 (5′-GCCGGCTAAGGGAAGATG-3′), which was complementary to the 5′ end of the cidA gene (nt 2626577 to 2626594 of the S. aureus 8325 genome). One hundred nanograms of the primer was end labeled with 200 μCi of [γ-32P]ATP (6,000 Ci · mmol−1) and T4 polynucleotide kinase (Invitrogen Life Technologies). One hundred micrograms of total RNA, isolated from an S. aureus culture grown as described above in TSB plus 0.5% (wt/vol) glucose, was mixed with 106 cpm (approximately 0.5 μCi) of labeled primer, ethanol precipitated, dissolved in 25 μl of hybridization buffer (40 mM Tris-HCl [pH 8.3], 25 mM NaCl, 40 U of RNase inhibitor [MBI Fermentas, Hanover, Md.]), denatured at 70°C for 5 min, and then allowed to anneal at 42°C for 60 min. The primer extension reaction was performed by adding 21 μl of 2× extension buffer (100 mM Tris-HCl [pH 8.3], 100 mM KCl, 2 mM [each] dNTP, 8 mM dithiothreitol, 16 mM MgCl2, 50 mM NaCl, 160 mg of acetylated bovine serum albumin · ml−1, 200 μg of actinomycin D-mannitol · ml−1) containing 12 U of Omniscript RT enzyme (Qiagen, Inc.) and 40 U of RNase inhibitor (MBI Fermentas), and the reaction mixture was incubated at 42°C for 60 min. After the reaction was stopped by adding 1.0 μl of 0.5 M EDTA (pH 8.0), 1 μl of 10-mg · ml−1 RNase A was added and the reaction mixture was incubated at 37°C for 30 min. This was followed by addition of 150 μl of TEN buffer (10 mM Tris-HCl, 1 mM EDTA, 0.1 M NaCl, pH 7.6), followed by extraction with phenol-chloroform and ethanol precipitation. The precipitated cDNA extension product was resuspended in 5 μl of Sequenase stop solution provided with the Sequenase kit (United States Biochemical Corporation, Cleveland, Ohio). A DNA sequencing ladder of the cidAB promoter region was also obtained with the CidA-1 primer with the Sequenase kit, according to the manufacturer's recommendations for using an end-labeled primer in the sequencing reaction. A plasmid containing 1,010 bp upstream of the ATG translational start site of cidA was used as template in the sequencing reactions. The sequencing and primer extension products were heated at 85°C for 5 min prior to their resolution on an 8% (wt/vol) denaturing polyacrylamide gel. The bands were visualized by autoradiography.
Allele replacement of the cidA gene.
A cidA mutation was generated in RN6390 by the following strategy. First, a 486-bp DNA fragment spanning a region 3′ to cidA (nt 2626532 to 2626066 of the S. aureus 8325 genome) was PCR amplified with the primers cidA-Cla (5′-CGCGATCGATTGTACCGCTAACTTGGGTAG-3′) and cidA-Pst (5′-CCGGCTGCAGTGGGTACGCTAAACATACGACCG-3′) and ligated into the ClaI and PstI sites of the plasmid pDG647 (21) downstream of an Em cassette. This recombinant plasmid was designated pBF648. Next, a 471-bp DNA fragment spanning a region 5′ to cidA (nt 2627146 to 2626675 of the strain 8325 genome) was PCR amplified with primers cidA-Eco (5′-GGCTTTGTTCCGAATTCTGTAGCGCA-3′) and cidA-Bam (5′-GGACTTTGTGCATGGCGGGATCCCTTTCTAA-3′) and ligated into the EcoRI and BamHI sites of pBF648, upstream of the Em cassette. This plasmid, designated pBF649, was subsequently digested with EcoRI and HindIII to liberate the Em cassette along with the flanking cidA sequences. The resulting 2.56-kb fragment was ligated into the EcoRI and HindIII sites of pCL52.2 (45) to generate pBF650. This plasmid was then transformed into S. aureus strain RN4220 by electroporation, spread onto tryptic soy agar (TSA) plates containing ERY, and incubated at 37°C overnight. The plasmid was then transferred into RN6390 by bacteriophage-mediated transduction, followed by growth at the nonpermissive temperature (44°C) in the presence of ERY to select for cells in which the plasmid had integrated into the chromosome via homologous recombination. To promote a second recombination event, a single colony was inoculated into antibiotic-free TSB medium and grown at 30°C for 5 days, with 1:1,000 dilutions into fresh antibiotic-free medium each day. After the fifth day, dilutions of the culture were spread on TSA medium to yield isolated colonies, which were subsequently screened for Emr and Tcs. Verification that 142 bp (nt 2626675 to 2626533 of the 8325 genome) had been deleted from the 5′ end of the cidA gene was carried out by PCR amplification and Southern blot analyses. The confirmed mutant strain was designated KB350 (Table 1).
Complementation of the cidA mutation in KB350 was achieved by PCR amplifying the cidA open reading frame (ORF) (nt 2626708 to 2626271 of the strain 8325 genome) with primers cidA-1 (5′-CGCGGATCCTATTTAGAAAGGGATGGCGCCATG-3′) and cidA-2 (5′-CCGGAATTCATTAATAAGGCTTGCACGTAATCA-3′). The resulting fragment was then cloned into the BamHI and EcoRI sites of the gram-positive bacterial expression vector pRN5548 (35). The ability of cidAB to complement KB350 was also determined by an identical strategy, with PCR primers cidA-1 and cidB-Eco (5′-TCCCTTTCTGTCTAGAATTCTAAATATCTAAA-3′) to amplify the cidAB ORF (nt 2626708 to 2625578 of the strain 8325 genome). Overexpression of cidA and cidAB from these plasmid constructs was confirmed by Northern blot analysis of strains harboring pRN-cidA and pRN-cidAB (data not shown).
Penicillin sensitivity assays.
Penicillin-induced killing of S. aureus strains was assessed by dilution plating as described previously (19), with the following modifications: overnight cultures of RN6390 and KB350 were diluted 1:100,000 in TSB and grown to early exponential phase (2.5 h) prior to the addition of 20× MIC of penicillin (0.04 μg/ml). In addition, flow cytometry analysis of penicillin-treated cultures was performed. In this assay, overnight cultures of RN6390 and KB350 were diluted 1:100 in 10 ml of TSB and grown to early exponential phase, and 20× MIC of penicillin (0.04 μg/ml) was added. After 2 h, cells from each culture were collected by centrifugation, washed once with sterile double-distilled H2O, and resuspended in 2 ml of sterile double-distilled H2O. Cell viability was assessed by using the Live/Dead BacLight bacterial viability kit for microscopy and quantitative assays (Molecular Probes, Inc., Eugene, Oreg.) according to the manufacturer's protocols. Briefly, SYTO9 (green fluorescent stain) and propidium iodide (red fluorescent stain) were mixed at a 1:1 ratio, and 6-μl aliquots were added to each sample of cells. The samples were then incubated at room temperature in the dark for 15 min and analyzed on a Becton Dickinson FACScalibur flow cytometer equipped with a 15 mW air-cooled 488-nm argon-ion laser. Green fluorescence, indicating the population of live, or undamaged, cells, was detected by parameter FL1, which measures emitted light with wavelengths between 515 and 545 nm. Red fluorescence, indicating the population of dead, or damaged, cells, was detected by parameter FL3, which measures emitted light greater than 650 nm. Both detectors were set on logarithmic amplification. The sample flow rate was set at 35 μl/min, and the sheath pressure was 4.5 lb/in2/g. Five thousand events were collected for each sample taken. Data were collected by using CellQuest software (Becton Dickinson) and were analyzed by using the WinMidi program, version 2.8.
Murein hydrolase assays.
Overnight S. aureus cultures were diluted 1:100 in 10 ml of NZY broth and grown for 15 h at 37°C and 250 rpm. The supernatants were collected by centrifugation and concentrated three- to fivefold in a Centricon-3 concentrator (Millipore, Bedford, Mass.). Total protein concentrations in each sample were determined by the Bradford assay (Bio-Rad Laboratories, Hercules, Calif.). Zymographic analysis was carried out as described previously (19). Quantitative cell wall hydrolysis assays were also performed as described previously (19), except that the turbidity of the samples was monitored by measuring the absorbance at 580 nm (A580) with a spectrophotometer.
RESULTS
Identification of lrgAB homologs.
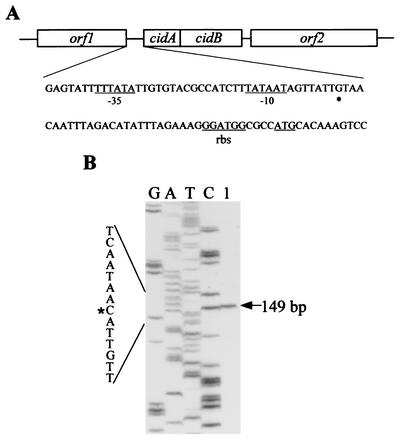
A BLAST search of the S. aureus 8325 (parent strain of RN6390) genome sequence database (http://www.genome.ou.edu/staph.html) revealed the presence of two ORFs, designated cidA and cidB (Fig. 1A), that exhibit significant similarity to the previously characterized lrgA and lrgB genes, respectively (19). The 393-nt cidA ORF overlaps by 8 nt with the 687-nt cidB ORF, suggesting that these two genes are translationally coupled. Consistent with this is the absence of a recognizable Shine-Dalgarno sequence in front of cidB. The transcription start site was mapped to a guanine residue 36 bp upstream of the cidA ATG start codon by primer extension analysis (Fig. 1B) and was preceded by a canonical −10 element (TATAAT) and near-canonical −35 element (four of six matches with the consensus sequence). In contrast to the lrgAB operon, which lies immediately downstream from the lytSR two-component regulatory system (3), the cidA and cidB genes lie downstream from an uncharacterized ORF that encodes a potential member of the LysR family of transcriptional regulators (46). Analysis of the sequence downstream of cidAB revealed the presence of a gene encoding a putative pyruvate oxidase.
FIG. 1.
Diagram of the cidAB locus (A) and primer extension analysis of the cidAB transcript (B). (A) The cidA and cidB ORFs correspond to nt 2626701 to 2625610 of the S. aureus 8325 genome. The −10 and −35 promoter elements, putative ribosomal binding site (rbs), and ATG start codon are underlined, and the +1 start site of transcription is indicated by an asterisk on the expanded cidAB sequence (nt 2626765 to 2626675 of the strain 8325 genome). (B) Primer extension of total cellular RNA (100 μg) (lane 1) from RN6390 yielded a 149-bp cDNA product, mapping the +1 site of cidAB transcription to a guanine residue located 36 bp upstream of the cidA ATG start codon. The size of the extension product was determined by comparison with the DNA sequencing ladder (shown to the left of lane 1) of the cidAB promoter region. Primer extension and sequencing reactions were performed with the same primer (see Materials and Methods).
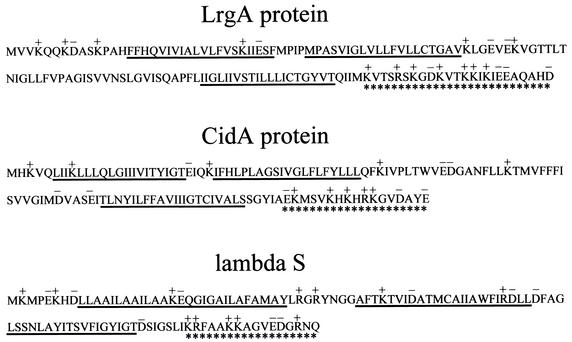
The predicted amino acid sequence of the cidA gene product (CidA) contains 131 amino acids, and the product has a deduced molecular mass of 14.7 kDa and a pI of 8.7. The cidB gene potentially encodes a 229-amino-acid protein (CidB) with a predicted molecular mass of 25.0 kDa and a pI of 9.1. The CidA and CidB proteins also contain four and six predicted membrane-spanning domains, respectively (8). CidA shares 23% amino acid sequence identity with LrgA, whereas CidB shares 31% amino acid sequence identity with LrgB. A BLAST search of other microbial genomes revealed the presence of cidAB and lrgAB homologs in many other gram-positive and gram-negative bacteria, including Staphylococcus epidermidis, B. subtilis, Bacillus cereus, Bacillus anthracis, Streptococcus mutans, E. coli, Yersinia enterocolitica, and Salmonella enterica serovar Typhimurium. Furthermore, cidAB and/or lrgAB homologs were identified in the genomes of archaeal species such as Methanosarcina mazei, Methanosarcina acetivorans, Pyrococcus abyssi, and Pyrococcus horikoshii. The apparently ubiquitous nature of these genes suggests that they may be important in bacterial physiology. Like the LrgA protein, CidA also contains several structural features that are characteristic of the bacteriophage holin family of proteins (Fig. 2). These include a relatively small size, two or more putative membrane-spanning domains, a polar N-terminal sequence, and a charge-rich C-terminal domain (53, 57). Despite having these structural features in common with holins, CidA is likely not a remnant of a bacteriophage genome, since no other bacteriophage genes are located in this region of the chromosome.
FIG. 2.
Structural similarities among LrgA, CidA, and the prototypical bacteriophage lambda S holin. The symbols used are as previously defined by Young and Blasi (57) as follows: the predicted charge pattern is shown above the amino acid sequence with K and R residues shown as positively charged and E and D residues shown as negatively charged. Potential membrane-spanning domains (8) are underlined, and the highly charged carboxyl termini are underscored with asterisks.
Effect of the cidA mutation on extracellular murein hydrolase activity.
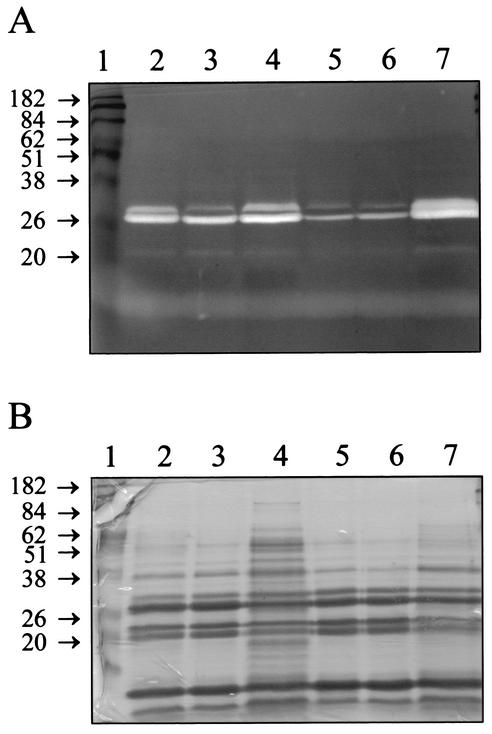
A cidA mutant derivative of RN6390 (designated KB350) was generated by replacing 140 bp from the 5′ end of this gene with an ERY resistance cassette. To determine the effect, if any, of the cidA mutation on extracellular murein hydrolase activity, proteins were isolated from stationary-phase cultures of RN6390 and KB350 and analyzed by zymography with Micrococcus luteus cells as a substrate (Fig. 3A). Interestingly, KB350 appeared to produce decreased levels of extracellular murein hydrolases (Fig. 3A, lane 5) compared to RN6390 (Fig. 3A, lane 2). Complementation of the KB350 mutant by supplying CidA in trans resulted in an increase in the overall murein hydrolase activity produced (Fig. 3A, lane 7) compared to that of the KB350 control strains (Fig. 3A, lanes 5 and 6). These levels of murein hydrolase activity appeared to be greater than those produced by RN6390 (Fig. 3A, lane 2) and RN6390(pRN5548) (Fig. 3A, lane 3). Furthermore, overexpression of cidA in RN6390 (Fig. 3A, lane 4) also appeared to produce a detectable increase in extracellular murein hydrolase activity compared to that of the RN6390 control strains (Fig. 3A, lanes 2 and 3).
FIG. 3.
Zymographic analysis (A) and corresponding secreted protein profile (B) of RN6390 and KB350, the cidA mutant. (A) Extracellular proteins were isolated, and 15 μg of each was separated in a sodium dodecyl sulfate-polyacrylamide gel containing 1.0 mg of M. luteus cells · ml−1. Murein hydrolase activity was detected by incubation overnight at 37°C in a buffer containing Triton X-100, followed by staining with methylene blue. Lanes: 1, molecular mass markers; 2, RN6390; 3, RN6390(pRN5548); 4, RN6390(pRN-cidA); 5, KB350; 6, KB350(pRN5548); 7, KB350(pRN-cidA). (B) Fifteen micrograms of protein from each of the sample preparations described above was separated in a sodium dodecyl sulfate-polyacrylamide gel and stained with Coomassie blue to detect the total secreted protein profile. The lanes are identical to those described for panel A. Molecular mass markers are indicated in kilodaltons.
One possibility that could potentially account for the observed changes in murein hydrolase activity is that the overall pattern of secreted proteins was affected by either the cidA mutation and/or overexpression of cidA. To address this, a duplicate polyacrylamide gel containing 15 μg of each protein extract was stained with Coomassie blue to determine the secreted protein profile of each sample (Fig. 3B). It is clear that the wild-type strain RN6390 (Fig. 3B, lanes 2 and 3) and the cidA mutant KB350 (Fig. 3B, lanes 5 and 6) displayed nearly identical secreted protein profiles. The secreted protein profiles of RN6390 and KB350 overexpressing cidA (Fig. 3B, lanes 4 and 7, respectively) both appeared to contain a slight overall increase in protein bands relative to those of the other strains, possibly attributable to cell lysis. However, increased cell lysis is likely not responsible for the increased murein hydrolase activity observed in these strains, as this would lead to a decreased proportion of murein hydrolases present in these 15-μg samples and a corresponding reduction in activity. Therefore, the changes in murein hydrolase activity observed are likely a direct function of the cidA mutation and not due to changes in the overall pattern of secreted proteins.
To quantify the observations of the zymographic analysis, cell wall hydrolysis assays were performed as described previously (19). As shown in Fig. 4, RN6390(pRN5548) exoproteins degraded approximately 20% more cell wall substrate after 2 h of incubation than did exoproteins isolated from KB350(pRN5548). In contrast, the presence of pRN-cidA in KB350 or RN6390 resulted in approximately fourfold more degradation of the cell wall substrate after 2 h of incubation compared to RN6390 containing the control vector. These results, in conjunction with the zymographic analysis, demonstrate that cidA expression causes an increase in extracellular murein hydrolase activity produced by S. aureus.
FIG. 4.
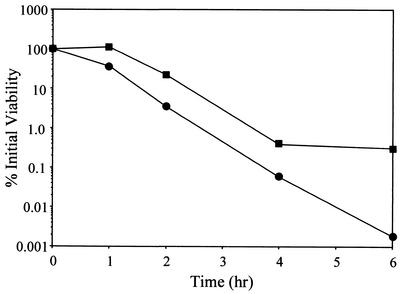
Quantitative murein hydrolase assays of the cidA mutant. One-hundred-microgram aliquots of the extracellular proteins used in the experiment the results of which are shown in Fig. 3 were added to a 1-mg · ml−1 suspension of M. luteus cells, and the turbidity was monitored for 6 h. Strains used in this analysis were RN6390(pRN5548) (closed circles), RN6390(pRN-cidA) (open circles), KB350(pRN5548) (closed squares), and KB350(pRN-cidA) (open squares). These data are from a single representative experiment and were reproduced several times. OD, optical density.
Effect of the cidA mutation on sensitivity to penicillin.
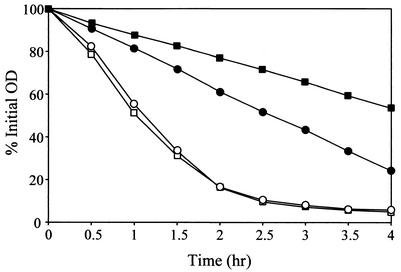
Previous work in our laboratory demonstrated that overexpression of the lrgAB operon in exponentially growing cultures of S. aureus inhibited killing of the bacteria by penicillin, while mutation of the lrgAB operon enhanced killing as the cell approached stationary phase (19). Likewise, the effect of the cidAB locus on the bacterium's response to penicillin was assessed by comparing the viability of exponential-phase KB350 (the cidA mutant) and RN6390 cultures after exposure to penicillin G (20× MIC) (Fig. 5). Specifically, the KB350 culture contained approximately 2 orders of magnitude more viable cells than did RN6390 at 6 h after the addition of penicillin. These results were not a reflection of a difference in growth rate between the two strains, as the doubling time of KB350 was comparable to that of the parental strain (data not shown). In contrast to the murein hydrolase activity observed, complementation of KB350 by introducing either pRN-cidA or pRN-cidAB did not restore its sensitivity to penicillin-induced killing to wild-type levels (data not shown).
FIG. 5.
Effect of the cidA mutation on penicillin sensitivity. Penicillin (20× MIC) was added to early-exponential-phase S. aureus RN6390 (circles) and KB350 (squares). Cultures and viable cell counts were determined by diluting aliquots of the cultures and plating them on TSA medium. These data are from a single representative experiment and were reproduced several times.
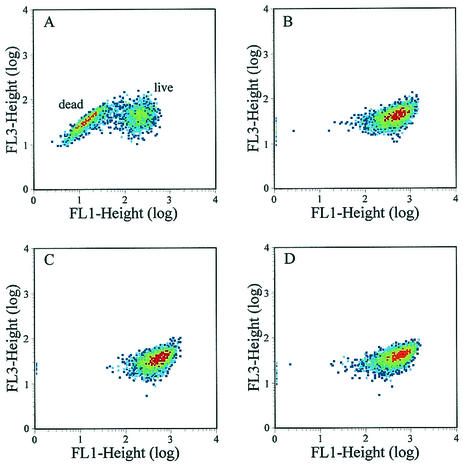
In a separate experiment, the viability of RN6390 and KB350 cultures after exposure to penicillin (20× MIC) was also measured by flow cytometry (fluorescence-activated cell sorting [FACS]) analysis with the Live/Dead BacLight bacterial viability kit (Molecular Probes, Inc.). In this assay, the dead or damaged cell population fluoresces red because the damaged cell membranes allow uptake of propidium iodide, whereas the live or undamaged population fluoresces green. After a 2-h exposure to penicillin, 48.7% of the RN6390 cell population remained intact (Fig. 6A) compared with 98% of the KB350 cell population (Fig. 6C). Furthermore, no apparent differences in viability were observed between RN6390 and KB350 cultures grown in the absence of penicillin (Fig. 6B and D).
FIG. 6.
Analysis of RN6390 and KB350 after exposure to penicillin with the Live/Dead BacLight bacterial viability kit (Molecular Probes, Inc.). Exponentially growing RN6390 and KB350 cells were treated with 20× MIC of penicillin (A and C, respectively) or left untreated (B and D, respectively). Samples of each culture were collected 2 h after the addition of penicillin and were prepared and analyzed as described in Materials and Methods. The population of dead or damaged cells in each panel is represented by a decrease in green fluorescence (FL1).
It is interesting that 48.7% of the penicillin-treated RN6390 cells stained as viable in the FACS analysis (Fig. 6A), whereas only about 5% of the RN6390 cells remained intact in the plating assays after 2 h (Fig. 5). One possibility that may account for this discrepancy is the fact that the Live/Dead stain used in the FACS analysis theoretically detects all viable cells, whereas the plating method, by its nature, is capable only of detecting those cells that are viable and able to form a colony. In other words, the cells that are still viable but damaged and unable to form a colony after penicillin treatment would be missed by the plating method. Therefore, the discrepancy in viability between these two types of assays may reflect the differing abilities of these two assays to detect a population of viable but nonculturable cells. Despite these quantitative discrepancies, both of these assays clearly demonstrate that KB350, the cidA mutant, has a reduced sensitivity to penicillin relative to that of RN6390.
Analysis of cidAB expression.
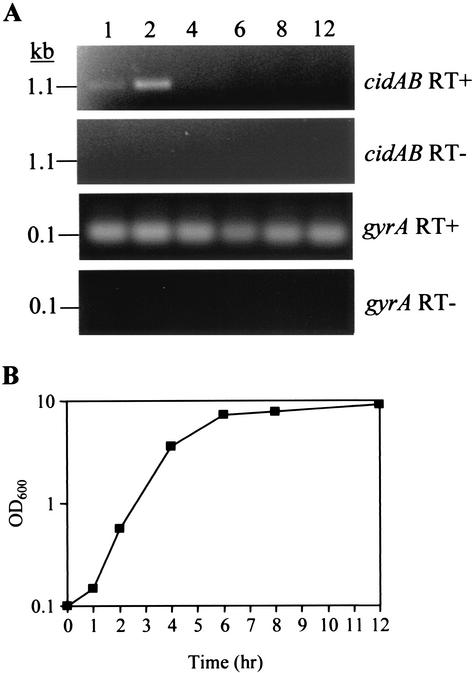
Previous studies of lrgAB transcription indicated that this operon is temporally regulated, with maximal expression occurring in the transition from exponential to stationary growth phase (19). To address the extent to which cidAB expression is growth phase dependent, we used RT-PCR to analyze cidAB expression throughout growth by collecting RNA samples from an RN6390 culture at various time points. As shown in Fig. 7A, cidAB transcription appeared to be most abundant at 2 h of growth, early in exponential growth (Fig. 7B). These results support the idea that the cid and lrg gene products function in a diametrically opposing manner, as lrgAB transcription occurs at low levels in early exponential growth phase and is maximal as cells enter stationary phase (19). In agreement with a previous report (18), transcription of the gene encoding gyrase A (gyrA) was constitutively expressed at all time points examined. Therefore, the pattern of cidAB transcription observed was due to temporal regulation and not due to differences in the amount of initial cDNA template in each RT reaction. This experiment indicates that, under these growth conditions, the cidAB genes are cotranscribed and expressed in early exponential growth phase.
FIG. 7.
RT-PCR analysis of cidAB expression (A) and corresponding growth curve (B). (A) RNA was isolated from S. aureus over 12 h of growth and subjected to RT-PCR (see Materials and Methods) to detect transcription of cidAB and gyrA. The corresponding gels are labeled “cidAB RT+” and “gyrA RT+,” respectively. Control reactions without RT enzyme were also performed, and the corresponding gels are labeled “cidAB RT−” and “gyrA RT−.” The numbered lanes on the figure represent the time points (in hours) at which RNA samples were collected. (B) Growth of the RN6390 culture analyzed for panel A was monitored by measuring its OD600 with a spectrophotometer.
DISCUSSION
Previous investigation by our laboratory has demonstrated that the S. aureus lrgAB operon inhibited extracellular murein hydrolase activity and penicillin-induced killing (19). It was hypothesized that this operon encodes an antiholin-like protein that inhibits murein hydrolase activity by interacting with a putative holin, analogous to what is thought to occur during bacteriophage infection (1, 19). In this study, we identified and investigated the role of a second S. aureus operon that encodes the potential holin counterpart of this system. Because of their involvement in regulating murein hydrolase activity and the bactericidal response to penicillin, we have designated the two genes of this operon cidA and cidB. The absence of any remnant bacteriophage sequences surrounding the cid and lrg operons argues against the possibility that these operons belong to a bacteriophage.
In contrast to the lrg operon, expression of the cid operon was shown to increase extracellular murein hydrolase activity (Fig. 3 and 4), suggesting that one or both of the cidAB gene products encode a holin counterpart to lrgAB. Indeed, the CidA protein also shares several characteristics in common with bacteriophage holin proteins (Fig. 2), indicating that it might possess a holin-like function. It is interesting that, in previous studies, hyperexpression of the λS holin resulted in a loss of host cell viability of more than 5 orders of magnitude within 5 min (50). However, in our study expression of cidA or cidAB from a plasmid did not appear to be detrimental to S. aureus viability. Although CidA displays several structural holin-like features, it may be divergent enough that it is not lethal when overexpressed. Alternatively, the level of overexpression achieved with these plasmids may not have been sufficient to be lethal to the cell, or the potential lethal action of CidA may be dependent on other as-yet-unidentified proteins or regulatory elements.
Holins and antiholins play a crucial role in the life cycle of most lytic bacteriophages by providing a timing mechanism for release of newly formed bacteriophage particles. The prototypical holin, λS, allows the bacteriophage-encoded murein hydrolases to gain access to their substrate, the cell wall peptidoglycan (57). This function is thought to be carried out by small nonspecific channels, or “holes,” formed by the oligomerization of individual holin subunits in the cytoplasmic membrane of the host. The murein hydrolases (endolysins) of many bacteriophages, including λS, lack a signal peptide, and therefore their transport is Sec independent. Without an active holin, these murein hydrolases would simply accumulate within the cytoplasm of the host cell (16). However, the murein hydrolases encoded by the atl and lytM genes of S. aureus appear to possess an N-terminal signal peptide, indicating that their transport is dependent on the Sec machinery of the cell (12, 36, 39). Therefore, if the cidA and lrgA genes do encode holin and antiholin-like proteins, it is unlikely that the sole function of these proteins is to mediate murein hydrolase export via hole formation.
Recently, it has been shown that the Oenococcus oeni bacteriophage fOg44 (Lys44) lysin contains an N-terminal signal peptide and is synthesized as a precursor whose export is dependent on the host cell general secretion pathway (44). Sec-dependent secretion and subsequent holin-independent lysis by the coliphage P1 and 21 endolysins have also been reported elsewhere (M. Xu, I. N. Wang, J. Deaton, and R. F. Young, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. M-35, p. 303, 2002). It should be emphasized that, in both of these latter cases, the corresponding holin was required for proper timing of the lytic event (Xu et al., Abstr. 102nd Gen. Meet. Am. Soc. Microbiol.). Although the exact mechanism by which holins precisely mediate the timing of host cell lysis during bacteriophage infection is unknown, the charged state of the bacterial membrane appears to play an important role in holin function, as a variety of energy poisons that collapse the PMF can instantly trigger premature lysis (56). Furthermore, holin accumulation results in collapse of the PMF several seconds prior to host cell lysis (20). In this respect, it has been demonstrated elsewhere that activity of murein hydrolases and regulation of autolysis in B. subtilis are dependent on the energized state of the membrane and the resulting pH of the local cell wall environment (4, 5, 26). Collectively, these observations suggest the intriguing possibility that the lrgAB and cidAB gene products regulate murein hydrolase activity in S. aureus by affecting the PMF. The effects of lrgAB and cidAB expression on the membrane potential and PMF are currently under investigation.
In agreement with previous studies of the lrgAB operon, mutation of the cidA gene was also shown to affect penicillin-induced killing, albeit in an opposing manner from that of lrgAB. The cidA mutant displayed increased tolerance to the bactericidal effects of penicillin (Fig. 5), whereas the lrgAB mutant had previously exhibited decreased tolerance to similar concentrations of this antibiotic (19). However, this phenotype could not be complemented in the cidA mutant when either CidA or CidA and CidB was expressed in trans from a plasmid (data not shown). The ability of these plasmids to complement murein hydrolase activity and not the level of tolerance to penicillin-induced killing in the cidA mutant is unknown. Northern blot analysis of strains harboring these plasmids has demonstrated that cidA and cidAB are highly transcribed relative to the wild-type strain RN6390 lacking either of these plasmids (data not shown). Furthermore, the ability of these plasmids to complement the level of murein hydrolase activity in KB350 suggests that the gene products are being translated.
One possibility is that the lack of complementation observed in the penicillin sensitivity assays could have been an artifact created by supplying multiple plasmid copies of cidA or cidAB, and this possibility is currently under investigation. However, these results could also reflect the notion that murein hydrolases are not the only factor that determines penicillin-induced killing. It is possible that the cidA mutation in KB350 had originally disrupted other, unknown membrane functions that are normally required for penicillin-induced killing and that these functions were not capable of being restored by the complementing plasmids. The ability of these plasmids to complement the murein hydrolase activity and not the level of tolerance to penicillin-induced killing in the cidA mutant in fact underscores the previously published observation for Streptococcus pneumoniae that penicillin-induced killing and penicillin-induced lysis are two separate and distinguishable events (33). In this previous work, genetic studies revealed the participation of two independent factors in the penicillin-induced killing of S. pneumoniae (33). The first, an amidase encoded by the lytA gene, was found to be responsible for a 1-log-unit loss of culture viability after 6 h of exposure of exponentially growing cultures of S. pneumoniae to penicillin. An additional 3 to 4 log units of killing was dependent on a second, yet-to-be-identified factor that was defective in so-called cid mutants (33). The cid mutation was able to dramatically reduce the amount of penicillin-induced killing in both wild-type and lytA mutant strains of S. pneumoniae (33), emphasizing the murein hydrolase-independent nature of this phenotype. Although the gene(s) responsible for the cid phenotype has never been identified, it was hypothesized that it encodes a protein analogous to bacteriophage-encoded holins. Similar S. aureus mutants, which are tolerant to the killing effects of penicillin, have been previously reported (52) and also generated by our laboratory (unpublished results).
In the present study, the cidAB operon was shown to enhance penicillin-induced killing of S. aureus, as mutation of cidA resulted in a decreased susceptibility to penicillin-induced killing. It is envisioned that the effect that cidAB expression has on penicillin-induced killing could occur via a mechanism directly involving the cidAB and lrgAB gene products (1), similar to that previously proposed by Moreillon et al. (33). In the presence of penicillin, the CidA and/or CidB proteins are envisioned to form membrane lesions that cause the membrane potential to collapse, effectively allowing the cells to “bleed” to death (1). This lethality would still occur in the presence of an intact cell wall and, thus, would explain the observations that penicillin-induced killing precedes lysis and still occurs in the absence of murein hydrolase activity (33). As a putative antiholin, LrgA and/or LrgB would be expected to inhibit the formation of the CidA/B holin-like complexes within the membrane, reducing penicillin-induced lethality as described previously (19). A more detailed understanding of how the cidAB and lrgAB gene products function should provide important insight into the regulation of murein hydrolase activity and into the mechanism of penicillin-induced killing.
Acknowledgments
This work was funded by NIH grant no. R29-AI38901, NIH-NRRI grant no. P20RR15587, and NSF-Idaho EPSCoR grant no. EPS-9720634.
We thank the Staphylococcus aureus Genome Sequencing Project (University of Oklahoma Health Sciences Center) and Chia-Yen Lee for providing plasmid pCL52.2.
REFERENCES
- 1.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 2.Brunskill, E. W., and K. W. Bayles. 1996. Identification and characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calamita, H. G., and R. J. Doyle. 2002. Regulation of autolysins in teichuronic acid-containing Bacillus subtilis cells. Mol. Microbiol. 44:601-606. [DOI] [PubMed] [Google Scholar]
- 5.Calamita, H. G., W. D. Ehringer, A. L. Koch, and R. J. Doyle. 2001. Evidence that the cell wall of Bacillus subtilis is protonated during respiration. Proc. Natl. Acad. Sci. USA 98:15260-15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, A. J. 1993. Extent of peptidoglycan O-acetylation in the tribe Proteae. J. Bacteriol. 175:4550-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the Dense Alignment Surface Method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 9.Dyer, D. W., and J. J. Iandolo. 1983. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 46:283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fein, J. E. 1979. Possible involvement of bacterial autolytic enzymes in flagellar morphogenesis. J. Bacteriol. 137:933-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, S. J. 1992. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 174:464-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gally, D., and A. R. Archibald. 1993. Cell wall assembly in Staphylococcus aureus: proposed absence of secondary crosslinking reactions. J. Gen. Microbiol. 139:1907-1913. [DOI] [PubMed] [Google Scholar]
- 15.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1277. [DOI] [PubMed] [Google Scholar]
- 16.Garrett, J. M., and R. Young. 1982. Lethal action of bacteriophage λ S gene. J. Virol. 44:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghuysen, J. M., J. Lamotte-Brasseur, B. Joris, and G. D. Shockman. 1994. Binding site-shaped repeated sequences of bacterial wall peptidoglycan hydrolases. FEBS Lett. 342:23-28. [DOI] [PubMed] [Google Scholar]
- 18.Goerke, C., S. Campana, M. G. Bayer, G. Döring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 98:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Holtje, J. V., and E. I. Tuomanen. 1991. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J. Gen. Microbiol. 137:441-454. [DOI] [PubMed] [Google Scholar]
- 24.Horne, D., and A. Tomasz. 1985. Competence-specific autolysis in Streptococcus sanguis. J. Gen. Microbiol. 131:533-541. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 26.Jolliffe, L. K., R. J. Doyle, and U. N. Streips. 1981. The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25:753-763. [DOI] [PubMed] [Google Scholar]
- 27.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 28.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda, A., Y. Asami, and J. Sekiguchi. 1993. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J. Bacteriol. 175:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda, A., and J. Sekiguchi. 1993. High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation. J. Bacteriol. 175:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquez, L. M., J. D. Helmann, E. Ferrari, H. M. Parker, G. W. Ordal, and M. J. Chamberlin. 1990. Studies of σD-dependent functions in Bacillus subtilis. J. Bacteriol. 172:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreillon, P., Z. Markiewicz, S. Nachman, and A. Tomasz. 1990. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob. Agents Chemother. 34:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novak, R., E. Charpentier, J. S. Braun, E. Park, S. Murti, E. Tuomanen, and R. Masure. 2000. Extracellular targeting of choline-binding proteins in Streptococcus pneumoniae by a zinc metalloprotease. Mol. Microbiol. 36:366-376. [DOI] [PubMed] [Google Scholar]
- 35.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins, H. R., S. J. Thorpe, and C. A. Brown. 1980. Tunicamycin and wall-polymer synthesis in Micrococcus luteus and Neisseria gonorrhoeae. Biochem. Soc. Trans. 8:163-164. [DOI] [PubMed] [Google Scholar]
- 39.Ramadurai, L., and R. K. Jayaswal. 1997. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J. Bacteriol. 179:3625-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rashid, M. H., A. Kuroda, and J. Sekiguchi. 1993. Bacillus subtilis mutant deficient in the major autolytic amidase and glucosaminidase is impaired in motility. FEMS Microbiol. Lett. 112:135-140. [DOI] [PubMed] [Google Scholar]
- 41.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romeis, T., U. Kohlrausch, K. Burgdorf, and J. V. Holtje. 1991. Murein chemistry of cell division in Escherichia coli. Res. Microbiol. 142:325-332. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Laboratory Press, Cold Spring Harbor, N.Y.
- 44.São-José, C., R. Parreira, G. Vieira, and M. A. Santos. 2000. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 182:5823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-630. [DOI] [PubMed] [Google Scholar]
- 47.Sekiguchi, J., B. Ezaki, K. Kodama, and T. Akamatsu. 1988. Molecular cloning of a gene affecting the autolysin level and flagellation in Bacillus subtilis. J. Gen. Microbiol. 134:1611-1621. [DOI] [PubMed] [Google Scholar]
- 48.Shafer, W. M., and J. J. Iandolo. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect. Immun. 25:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shockman, G. D., and J.-V. Holtje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 50.Smith, D. L., C. Y. Chang, and R. Y. Young. 1998. The lambda holin accumulates beyond the lethal triggering concentration under hyperexpression conditions. Gene Expr. 7:39-52. [PMC free article] [PubMed] [Google Scholar]
- 51.Tokunaga, T., M. H. Rashid, A. Kuroda, and J. Sekiguchi. 1994. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon of Bacillus subtilis. J. Bacteriol. 176:5177-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomasz, A., and M. L. de Vegvar. 1986. Construction of a penicillin-tolerant laboratory mutant of Staphylococcus aureus. Eur. J. Clin. Microbiol. 5:710-713. [DOI] [PubMed] [Google Scholar]
- 53.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 54.Ward, J. B., and R. Williamson. 1985. Bacterial autolysins: specificity and function, p. 159-166. In C. Nombela (ed.), Microbial cell wall synthesis and autolysis. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 55.Wong, W., A. N. Chatterjee, and F. E. Young. 1978. Regulation of bacterial cell walls: correlation between autolytic activity and cell wall turnover in Staphylococcus aureus. J. Bacteriol. 134:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, R., and U. Blasi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191-205. [DOI] [PubMed] [Google Scholar]