Abstract
This study initially involved the isolation of a number of bifidobacteria from either the lumen or the epithelium of a porcine cecum. A total of 160 isolates were selected at random on MRS plates containing cysteine hydrochloride (0.5 g/liter) and mupirocin (50 mg/liter). All were identified as bifidobacteria based on fructose-6-phosphate phosphoketolase activity. Following genomic digestion with the restriction enzyme XbaI and pulsed-field gel electrophoresis (PFGE), the isolates produced 15 distinct macro-restriction patterns. Several of the PFGE patterns differed by only 1, 2, or 3 DNA fragments and were grouped as related patterns into seven PFGE types, termed A through G. The related patterns appeared to show genomic plasticity within the isolates arising from chromosomal mutations or possibly horizontal transfer of plasmids. The relative frequency of each PFGE type was maintained within each cecal sample, with PFGE type E representing approximately 50% of the isolates. Randomly amplified polymorphic DNA PCR, cell morphology, whole-cell protein profiling, 16S ribosomal DNA sequencing, and DNA-DNA hybridization were used to determine if the seven apparently unrelated PFGE types represented genetically distinct isolates. Four groups were identified: PFGE types A, C/D/G, B/E, and F, and these appeared to represent Bifidobacterium minimum, Bifidobacterium pseudolongum subsp. pseudolongum, and Bifidobacterium pseudolongum subsp. globosum and two new species, respectively. The data demonstrate the presence of considerable genomic diversity within a relatively simple bifidobacteria population, consisting of 15 distinct strains representing four groups, which was maintained throughout the porcine cecal contents and epithelial layer.
The genus Bifidobacterium consists of gram-positive anaerobes with a variety of rod morphologies that appear to be among the most prevalent microflora in the gastrointestinal tract (GIT) of humans and animals (5, 41). The contribution of these bacteria to good health has been recognized for quite some time and has led to the widespread exploitation of bifidobacteria as probiotics for maintaining or improving human and animal health (15, 27, 48).
At present, the genus Bifidobacterium includes 34 species (5, 16). Approximately 20 species have been identified from fecal sources, of which only 10 appear to have been recovered from GIT samples (5, 24, 29). In addition, three species, Bifidobacterium ruminatium and Bifidobacterium merycicum from bovine rumen (4) and Bifidobacterium gallinarum from chicken cecum (51), do not appear to have been recovered from fecal samples. Therefore, it seems likely that the examination of GIT samples will identify more Bifidobacterium species. Indeed, a potential new species, based on 16S ribosomal DNA (rDNA) sequence comparisons, was isolated from the porcine intestine (24), and recently, bifidobacteria were recovered from the chicken crop, a habitat not considered suitable for the growth of strict anaerobes (31).
Bifidobacteria have been cultured from the cecal contents of chickens (31, 35, 51), rats (43), rabbits (20, 35, 46), pigs (37), and humans (21). Isolates were confirmed as belonging to the genus Bifidobacterium in all but the rat and pig studies, through the detection of fructose-6-phosphate phosphoketolase (F-6-PPK) (41). However, only isolates from chicken and rabbit cecum were characterized to the species level. In the former a new species, B. gallinarum, was proposed (51), and in the latter two subspecies, Bifidobacterium pseudolongum subsp. pseudolongum and Bifidobacterium pseudolongum subsp. globosum, were identified (20, 46). Although none of the studies included an assessment of genomic diversity within the bifidobacterial population, the use of the restriction enzyme XbaI followed by pulsed-field gel electrophoresis (PFGE) was reported to be an effective method for discriminating human fecal Bifidobacterium isolates (17, 23).
In addition, a variety of molecularly based techniques have been used to establish the genetic relatedness and species identity of Bifidobacterium isolates. Randomly amplified polymorphic DNA (RAPD) PCR was used to group bifidobacteria recovered from rat intestines (13), human feces (50), and human GIT samples (22). Whole-cell protein profiling (WCPP) (3, 54), 16S rDNA sequencing (11, 16, 26), and DNA-DNA hybridization (39, 47) have been used to identify Bifidobacterium species.
Bifidobacterium strains are reported to vary in their ability to adhere to cultured epithelial cells, and this has been attributed to specific protein, polysaccharide, lipoteichoic acid, hydrophobic surface, and autoaggregation factors (2, 6, 9, 10, 19, 30). In addition, studies on the general microflora of the epithelium and contents of the porcine GIT have suggested that distinct bacterial populations exist within each region (1, 44). However, to our knowledge no study has determined the genomic diversity of bifidobacteria isolated from the contents and epithelium of the same GIT sample.
The present study had two aims: to determine the level of genomic diversity of Bifidobacterium isolates recovered from different fractions of a porcine cecum, using PFGE; and to establish genetic relatedness and species identity of isolates based on PFGE, cell morphology, RAPD PCR, WCPP, 16S rDNA sequencing, and DNA-DNA hybridization.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The Bifidobacterium strains used in the study are listed in Table 1. Cultures were grown under anaerobic conditions (anaerobic jars with Anaerocult A gas packs; Merck, Darmstadt, Germany) at 37°C in modified MRS (mMRS), comprising Lactobacilli MRS medium (Difco, Detroit, Mich.) supplemented with 0.05% (wt/vol) cysteine-hydrochloride. To facilitate the recovery of bifidobacteria from a pig cecum, 50 mg of mupirocin (Oxoid Inc., New York, N.Y.)/liter was added to mMRS from antimicrobial susceptibility test discs, as previously described (36). For WCPP (see below), selected isolates were cultured on Columbia medium and on mMRS. Lactobacillus rhamnosus ATTC 7469 was cultured on mMRS as outlined above. Cell morphology was determined by phase-contrast microscopy (cells from colonies) using a BX51 microscope and DP50 digital camera (Olympus Optical Co. Ltd., Tokyo, Japan). Cell sizes were determined from a minimum of 25 cells using the program ANALYSIS (Soft Imaging System GmbH, Münster, Germany).
TABLE 1.
Bifidobacterium strains used in this study
| Species | Collection namea | Strain name | Origin |
|---|---|---|---|
| B. thermophilum | NCIMB 702253T | J18-P2-91 biotype a | Pig feces |
| JCM 7027 | PNA-1-24 biotype a | Pig feces | |
| JCM 7028 | PN-Ro-3 biotype a | Pig feces | |
| JCM 7031 | 14-44 biotype b | Pig feces | |
| JCM 7033 | K30-P16-6 biotype b | Poultry feces | |
| JCM 7034 | I27-P23-116 biotype c | Poultry feces | |
| JCM 7035 | PN-Nu-1-6 biotype c | Poultry feces | |
| JCM 7036 | P25-111 biotype c | Poultry feces | |
| ATCC 25867 | RU445 | Cow rumen | |
| NCIMB 702554 | RU326 | Cow rumen | |
| DSMZ 20209 | C3/12 | Cow rumen | |
| B. ruminantium | DSMZ 6489T | RU687 | Cow rumen |
| LMG 18895 | RU728 | Cow rumen | |
| LMG 18896 | RU679 | Cow rumen | |
| B. pseudolongum subsp. globosum | JCM 5820T | RU224 | Cow rumen |
| JCM 7092 | RU256 | Cow rumen | |
| B. pseudolongum subsp. pseudolongum | NCIMB 702244T | PNC-2-9G biotype a | Pig feces |
| DSMZ 20094 | 28T biotype b | Chicken feces | |
| DSMZ 20095 | 29Sr-T biotype c | Chicken feces | |
| B. adolescentis | DSMZ 20087 | RU424 | Cow rumen |
| B. merycicum | LMG 11341T | RU915B | Cow rumen |
| DSMZ 6493 | RU767 | Cow rumen | |
| B. boum | LMG 10736T | RU917 | Cow rumen |
| B. suis | ATCC 27533T | SU859 | Pig feces |
| ATCC 27531 | SU868 | Pig feces | |
| ATCC 27532 | SU901 | Pig feces | |
| B. pullorum | DSMZ 20433T | P145 | Chicken feces |
| B. gallinarum | DSMZ 20670T | CH 206-5 | Chicken cecum |
| B. magnum | DSMZ 20222T | RA 3 | Rabbit feces |
| JCM 7120 | RA 206 | Rabbit feces | |
| DSMZ 20220 | RA 76 | Rabbit feces | |
| B. animalis | DSMZ 20104T | R101-8 biotype a | Rat feces |
| JCM 7117 | RA20 | Rabbit feces | |
| DSMZ 20097 | C10-45 biotype b | Calf feces | |
| DSMZ 20105 | P23 | Chicken feces | |
| B. saeculare | DSMZ 6531T | RA161 | Rabbit feces |
| DSMZ 6532 | Ra158 | Rabbit feces | |
| DSMZ 6533 | RA159 | Rabbit feces |
Culture collection. NCIMB, National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom. JCM, Japanese Collection of Microorganisms, Saitama, Japan. ATCC, American Type Culture Collection. DSMZ, German Microorganism Collection, Braunschweig, Germany. LMG, Laboratorium voor Microbiology en microbiele Genetica, Ghent, Belgium. Superscript T, type strain.
Pig cecum samples.
The cecum from a 24-week-old pig was removed at slaughter, and the contents were secured by ligature and placed on ice. Within 1 h, duplicate samples were diluted 1 in 10 with Maximum Recovery Diluent (MRD) (Oxoid Inc., New York, N.Y.) from three cecum fractions representing bacterial isolates from the cecal contents (sample I) and bacterial isolates considered to be weakly (sample II) or strongly (sample III) adherent to the cecal epithelial layer. To obtain adhering isolates, a mid-region of the cecal epithelial tissue, ∼8 cm2, was aseptically removed, washed with MRD to remove loosely adhering contents, weighed, and diluted accordingly. To obtain weakly adherent bacteria, the tissue sample was gently shaken for 5 min before serial dilution. To obtain strongly adherent bacteria, the same piece of tissue was reweighed, resuspended with fresh MRD, and vigorously shaken using a Stomacher 400 (Seward, London, United Kingdom), set at high speed, for 5 min before serial dilution (sample III). Diluents from each cecal sample were pour plated with mMRS with and without 50 mg of mupirocin/liter and incubated for 2 days at 37°C under anaerobic conditions. From each sample, 50 to 60 colonies were randomly picked, inoculated into mMRS broth, and incubated as before for 48 h, from which 1-ml samples were taken for the preparation of frozen stocks, detection of F-6-PPK activity, and PFGE preparations. Selected isolates were deposited in the Dairy Products Centre (DPC) Collection at Teagasc, Moorepark, Fermoy, County Cork, Ireland.
F-6-PPK activity.
A protocol for detecting F-6-PPK activity in the intracellular extracts from 1 ml of mMRS broth cultures using Triton X-100 detergent-mediated cell lysis was employed based on protocols previously described (7, 28). Bifidobacterium thermophilum NCIMB 702553 and Lactobacillus rhamnosus ATCC 7469 were used as positive and negative controls, respectively.
PFGE.
Preparation of high-molecular-weight DNA from mMRS broth cultures was as previously described (45) for pediococci except that 40 μg of mutanolysin/ml was added to the cell lysis buffer and the restriction enzymes XbaI and SpeI were used (New England Biolabs, Beverly, Mass.). DNA fragments were resolved using a CHEF-DR III pulsed-field system (Bio-Rad Laboratories, Richmond, Calif.) at 6 V/cm for 18 h with a 1- to 15-s linear ramp pulse time. A 0.5× Tris base-borate-EDTA buffer was maintained at 14°C during electrophoresis. Molecular size markers were included in each gel (#N0350S; New England Biolabs). Gels were stained in distilled water containing 0.5 μg of ethidium bromide/ml for 30 min and destained for 60 min in distilled water.
RAPD PCR.
RAPD PCR was performed on DNA from selected pig cecal isolates. Details on the isolation of DNA from mMRS broth cultures, random primers P1 and P2, RAPD PCR conditions, and RAPD pattern comparisons using Gelcompar software were as previously described (45).
Whole-cell protein profile.
Whole-cell protein extraction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and computer-based comparisons of the resulting protein profiles were performed by BCCM/LMG (Laboratory for Microbiology, University of Ghent, Ghent, Belgium) as previously described (32). The normalized and digitized protein patterns were numerically analyzed and clustered with the reference profiles of 33 Bifidobacterium type strains.
16S rDNA sequencing.
Two 16S rDNA primers, CO1, for the 5′ end (5′AGTTTGATCCTGGCTCAG3′), and CO2, for the 3′ end (5′TACCTTGTTACGACT3′), were used to generate an approximately 1.5-kb 16S rDNA product. The PCRs were performed in a 50-μl (each) solution containing 0.25 μM of each deoxynucleoside triphosphate (supplied as a 50× dNTP Mastermix; Bioline, USA Inc., Randolph, Mass.), 10 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 0.1% (vol/vol) Tween-20 (supplied as a 10× NH4 reaction buffer; Bioline), 5 mM MgCl2 (supplied as a 50 mM stock; Bioline), 1 μM (each) primer, 2 μl of DNA, and 1 U of BIOTAQ DNA polymerase (Bioline). The mixture was denatured at 94°C for 5 min in a DNA thermal cycler (Hybaid Ltd., Middlesex, United Kingdom) followed by 30 amplification cycles, each consisting of three 1-min stages at 94°C (denaturation), 60°C (annealing), and 72°C (extension).
PCR amplicons were separated through a 1.5% (wt/vol) agarose gel using a 1× Tris base-acetate-EDTA running buffer containing 0.5 μg of ethidium bromide/ml and gel electrophoresis conditions of 100 V for 2 h. DNA was visualized by UV trans-illumination and sized using a 100-bp ladder (Amersham-Pharmacia-Biotech, Uppsala, Sweden) as a molecular weight standard. The ∼1.5-kb fragment was extracted from the gel using a QIAquick gel extraction kit (Qiagen, Valencia, Calif.). The recovered rDNA was sequenced using two forward primers: CO1 (5′ AGT TTG ATC CTG GCT CAG 3′) and MG3f (5′ CTA CGG GAG GCA G 3′) and three reverse primers, CO2 (5′ TAC CTT GTT ACG ACT T 3′), 782R (5′ ACC AGG GTA TCT AAT CCT GT 3′), and 765R (5′ CTG TTT GCT CCC CAC GCT TTC 3′) by MWG sequencing service (Ebersberg, Germany). Overlapping contigs and a consensus sequence were established using Seqmanager (DNAstar).
Phylogenetic analysis.
The analysis of 16S rDNA sequences was as previously described (8). In brief, the 16S rDNA sequences obtained in the present study and those for each Bifidobacterium type strain (obtained from the National Center for Biotechnology Information database [www.ncbi.nlm.nih.gov]) were aligned using CLUSTAL X. Using the PHYLIP package, version 3.6 (obtained from www.evolutionary.genetics.washington.edu/phylip.html), the evolutionary distance matrices were generated with the Cantor and Juke coefficient by the DNADIST program. A phylogenetic tree was constructed according to the neighbor-joining method, using the program NEIGHBOR. The stability of the grouping was estimated by bootstrap analysis (1,000 replicons) using the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE. The phylogenetic tree was viewed using TREEVIEW software. The root of the unrooted tree based on the neighbor-joining method was estimated by using the outgroup Eubacterium combesii. Gardnerella vaginalis, Actinobaculum suis, Arcanobacterium haemolyticum, Mobiluncus curtisii, and Actinomyces bovis were also included as outgroups (16).
DNA-DNA hybridization reactions.
For selected isolates, DNA-DNA hybridization reactions to Bifidobacterium type strains were performed by BCCM/LMG (Laboratory for Microbiology, University of Ghent, Ghent, Belgium) as previously described (12). Hybridization reactions with Bifidobacterium inopinatum and Bifidobacterium denticolens were performed at 44°C; all others were performed at 47°C.
Nucleotide sequence accession numbers.
The GenBank accession numbers for PFGE types A through G are AY174103, AY174104, AY174105, AY174106, AY174107, AY174108, and AY174109, respectively.
RESULTS
The isolation of bifidobacteria from the porcine cecum.
The 38 Bifidobacterium strains representing 13 animal-derived species (Table 1) showed comparable growth on mMRS agar with and without 50 mg of mupirocin/liter. Therefore, the mupirocin medium was considered to have good elective properties. Cecal samples I, II, and III yielded 4.3 × 105 ± 0.5, 1.8 × 104 ± 0.02, and 1.9 × 103 ± 0.6 CFU/g on the mupirocin medium, respectively. On mMRS without mupirocin, the samples I, II, and III yielded 6.7 × 108 ± 1.6, 3.4 × 107 ± 0.5, and 1.9 × 106 ± 0.6 CFU/g, respectively, indicating that the bifidobacteria population in each of the three cecal samples represented ∼0.1% of the total microflora enumerated. All of the 160 selected isolates tested positive for F-6-PPK activity, indicating their bifidobacterium status (41).
PFGE strain discrimination.
Previous studies have reported that PFGE following genomic digestion with the restriction enzyme XbaI can effectively discriminate strains from a number of Bifidobacterium species (29, 38). However, no strains of animal origin were included. In the present study, the discrimination of 38 animal-derived strains, including 11 type strains (Table 1), was examined based on genomic digests with XbaI and PFGE. The 38 strains produced 35 distinct macro-restriction patterns. The PFGE patterns for three Bifidobacterium suis strains (ATCC 27531, ATCC 27532, and ATCC 27533) were identical, as were those for two Bifidobacterium magnum strains (JCM 7120 and DSMZ 20220). In each case, following genomic digestion with the restriction enzyme SpeI, identical PFGE patterns within each group of strains were again observed, suggesting that they represented only two strains. In addition, Bifidobacterium thermophilum ATCC 25867 and Bifidobacterium saeculare DSMZ 6532 differed from their respective type strains by two extra fragments (at ∼40 and 80 kb) and one extra fragment (at ∼170 kb), respectively.
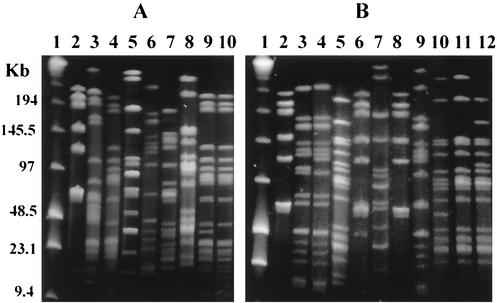
Given the effective level of strain discrimination achieved by XbaI and PFGE, 160 pig cecal isolates were examined. A total of 15 distinct macro-restriction patterns were observed (Fig. 1 and Table 2). Examples of different isolates having the same PFGE patterns are shown in Fig. 1 (see panel A, lanes 4 and 10, panel B, lanes 2 and 8, panel A, lane 5, and panel B, lane 7). In some instances, apparent differences were not recorded because they were considered to be an artifact of the weak staining associated with the low DNA content of fragments below 20 kb (see Fig. 1A, lane 8 and Fig. 1B, lane 11).
FIG. 1.
PFGE macro-restriction patterns following genomic DNA digests with the restriction enzyme XbaI. (A) Lanes 2 through 10 represent PFGE types G, F, E, D, C, B, and, PFGE subtype Ea, A, and PFGE type E, respectively. (B) Lanes 2 through 12 represent PFGE types and PFGE subtypes G, Caa, Ca, Aa, Ga, F, G, Cb, Ab, A, and Aa, respectively. Lanes 1, molecular size markers, lambda HindIII fragments with lambda concatemers.
TABLE 2.
Frequency of PFGE types and subtypes representing 160 Bifidobacterium pig cecal isolates
| PFGE typea | No. of isolates for cecal sampleb:
|
PFGE subtypes and the apparent macro-restriction patterns differences from their related PFGE typec | ||
|---|---|---|---|---|
| I | II | III | ||
| A | 12 | 8 | 13 | Aa, new XbaI site in a ∼280 kb fragment (5) |
| Ab, 25-kb deletion in a 85-kb fragment (2) | ||||
| B | 1 | 2 | 2 | None |
| C | 3 | 4 | 6 | Ca, ∼75-kb plasmid (5) |
| Caa, ∼75-kb plasmid and ∼15-kb deletion in ∼130-kb fragment (1) | ||||
| Cb, ∼300-kb plasmid (1) | ||||
| Cc, ∼5-kb deletion in ∼130-kb fragment (2) | ||||
| D | 1 | 3 | 3 | None |
| E | 25 | 25 | 22 | Ea, ∼30-kb plasmid (1) |
| F | 6 | 6 | 11 | None |
| G | 2 | 2 | 3 | Ga, ∼40-kb plasmid (1) |
| Total | 50 | 50 | 60 | (18) |
Values include subtypes.
Representing isolates recovered from the cecal contents (I) and epithelial layer, mixed lightly (II) or vigorously (III).
Number in parentheses gives the number of isolates representing the subtype.
A number of the macro-restriction patterns differed by one, two, or three fragments (Table 2), and in these cases the differences could be explained by a chromosomal mutation (49) or the presence of a putative plasmid. Grouping the related macro-restriction patterns produced seven PFGE types, termed A through G (Fig. 1A, lanes 8 through 2, respectively). Four groups, A, C, E, and G, included related PFGE subtypes (Fig. 1A, lane 9, subtype Ea; Fig. 1B, lanes 3 to 6, subtypes Caa, Ca, Aa, and Ga, respectively; lanes 9, 10, and 12, subtypes Cb, Ab, and Aa, respectively). Representative isolates from the same type were also found to have identical PFGE patterns following genomic digests with SpeI. The actual nature of the proposed chromosomal mutations, although based on established methods of interpretation, can be attributed to reciprocal rearrangements. For example, if the PFGE patterns for two strains show a two-fragment difference, the rearrangement could be interpreted as a deletion or insertion depending on which strain was the originator of the mutation. In the description of the subtypes, a convention was adopted that related patterns based on a minimum number of genomic rearrangements needed to link the subtypes. “Parental” strains were identified, and these were assigned as the PFGE type and the others as the related subtypes, using a lowercase letter. Where a succession of changes was evident, a second lowercase letter was used. For example, the three- fragment difference between type A and subtype Aa (Fig. 1B, lanes 11 and 12, respectively) could be explained by a mutation leading to a new XbaI site. The ∼280-kb fragment would then produce two smaller fragments at ∼170 and ∼110 kb, equal in size to the missing type A fragment. A single fragment of ∼85 kb in type A and an additional fragment of ∼60 kb, which comigrated with an existing fragment resulting in a fragment with a greater staining intensity, in subtype Ab, distinguished the two PFGE patterns (Fig. 1B, lanes 10 and 11, respectively). The difference could be explained by a ∼25-kb deletion in the ∼85-kb fragment. If the difference was explained by a ∼25-kb insertion in the subtype Ab fragment at ∼60 kb, a further mutation to remove an XbaI site would be required to produce subtype Aa.
The occurrence of the subtypes among the seven types was not equally distributed, with types A and C accounting for 39 and 44% of the subtypes, respectively. Both of the type A subtypes appeared to result from chromosomal mutations, unlike the majority of type C subtypes, which appeared to have putative plasmid differences (Table 2). Assuming each of the putative plasmids had a single XbaI site, the following sizes were estimated: ∼30 kb for E and Ea (Fig. 1A, lanes 10 and 9, respectively), ∼45 kb for G and Ga (Fig. 1B, lanes 8 and 6, respectively), ∼75 kb for C and Ca or Caa (Fig. 1A, lane 6, and 1B, lanes 4 and 3, respectively) and ∼300 kb for C and Cb (Fig. 1A, lane 6, and 1B, lane 9, respectively). The extreme size of the latter putative plasmid prompted analysis with a second restriction enzyme, SpeI, which also yielded a single fragment difference at ∼300 kb, suggesting that the fragment did in fact represent a single megaplasmid.
Overall, the relative distribution of the seven types was maintained within the three cecal samples analyzed (Table 2). This suggested that if bifidobacteria with specific abilities to adhere and colonize the epithelial layer do exist, these are also present at a similar relative frequency within the cecal contents, although in the latter sample, cell numbers were ∼200-fold higher. Type E was the most dominant isolate, accounting for 48, 51, and 35% of the bifidobacteria isolated from the two cecal epithelial samples and contents, respectively.
Relatedness of PFGE types.
Using five different methods of analysis, the relatedness between the seven PFGE types that appeared to represent unrelated macro-restriction patterns were determined. Results for the following sections are summarized in Table 3.
TABLE 3.
Relatedness of PFGE types A through G
| Group | Analysis
|
Species based on 16S rDNA sequence (% similarity) | ||
|---|---|---|---|---|
| Cell morphology | RAPD PCR | WCPP | ||
| 1 | A | A | A | A assigned to B. minimum (99.57) |
| 2 | C, D, G | D, G | C, D, G | G assigned to B. pseudolongum subsp. pseudolongum (100) |
| 3 | B, E | B, E | B, E | B and E related (99.72) assigned to a new species, B. aerophilum sp. nov. |
| 4 | F | F | F | F assigned to a new species, B. psychroaerophilum sp. nov. |
| 5 | C | C (99.93) and D (100) assigned to B. pseudolongum subsp. globosum | ||
(i) RAPD PCR.
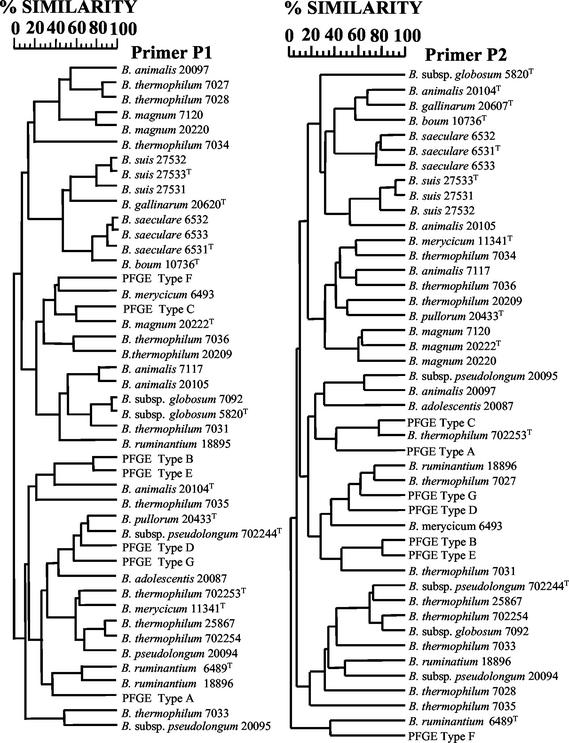
A number of studies have reported the use of RAPD PCR to cluster related bifidobacteria isolates (13, 22, 50). The dendrograms based on comparison of the RAPD PCR patterns produced by the primers P1 and P2 were considered to be similar with respect to the seven PFGE types. In both cases, the seven types showed the same interrelationship, forming five distinct clusters: types A, B/E, C, D/G, and F, respectively. Using a percentage similarity cutoff of 50%, the same clusters were maintained when RAPD PCR patterns for each culture collection strain were included (Fig. 2 and Table 3). The segregation of collection strains from the same species was considered to be poor. Therefore, RAPD PCR, with primers P1 and P2, was not considered to be a reliable tool for species identification.
FIG. 2.
Dendrograms based on computer comparisons of RAPD PCR patterns produced by random primers P1 and P2 with Bifidobacterium isolates (PFGE types A through G) recovered from a pig cecum and animal-derived culture collection strains (Table 1). The dendrograms were constructed using the Pearson product moment correlation coefficient (r) (similarity, 100 × r) and unweighted pair group algorithm method with arithmetic averages. Superscript T denotes type strain.
(ii) Cell morphology.
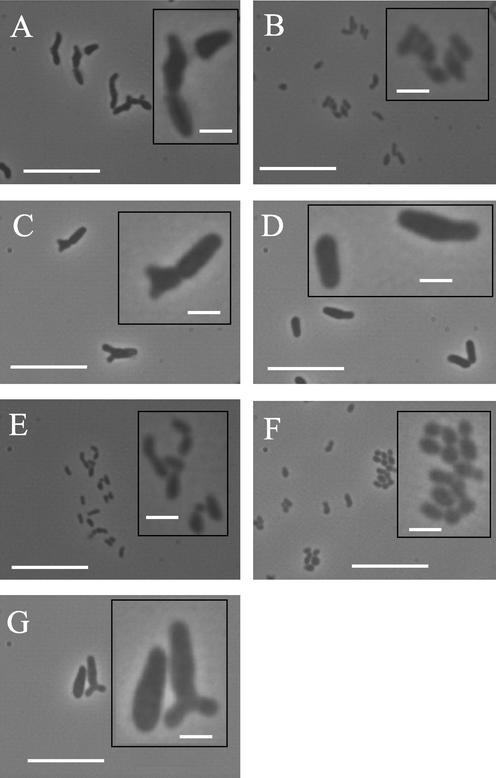
Representatives from the seven PFGE types and each subtype produced four distinct cell morphology groups (Fig. 3). Group 1 cells were irregular rods, arranged singly or in pairs, with occasional bifurcated extremities and tapered ends, ∼0.7 to 1 μm wide and ∼2 to 3 μm long, represented by type A and subtypes Aa and Ab only (Fig. 3A). Group 2 cells were also irregular rods, arranged singly or in pairs, with occasional bifurcated extremities, ∼1 to 1.2 μm wide and ∼2.5 to 5 μm long, with a tendency to form large aggregates (not shown), represented by types C, D, and G and all their related subtypes (Fig. 3C, D, and G). Group 3 cells were irregular short rods, arranged singly or in pairs, ∼0.6 to 0.9 μm wide and ∼0.8 to 1.5 μm long, represented by types B and E and subtype Ea (Fig. 3B and E). Group 4 cells were also short rods but were generally more globular than group 3 cells, arranged singly or in pairs, ∼0.7 to 1 μm wide and ∼1 to 1.5 μm long, represented by type F (Fig. 3F). Therefore, with the exception of type C, the morphology groups concurred with the genetic relatedness observed following RAPD PCR (Table 3).
FIG. 3.
Phase contrast cell morphology of Bifidobacterium isolates recovered from a pig cecum. Panels A through G represented PFGE types A through G, respectively. Four morphology groups, 1 through 4, were considered to represent type A, B and E, C, D, and G, and F, respectively (Table 3). Each subtype segregated with its related type. Scale bars in the main and inset panels are 10 and 2 μm, respectively.
(iii) WCPP.
WCPP has previously been demonstrated as a useful technique for the identification of Bifidobacterium species (3, 54). Based on WCPP after growth on Columbia and/or mMRS media, types A through G were segregated into four groups. Types C, D, and G all clustered together with a minimum similarity value of 78%, compared to 65% similarity to types A, B, E, and F. Types F and A were both distinct, with maximum similarity values of 69 and 72% towards type B, respectively. Type B clustered with type E with a similarity value of 92%. These groupings were in good agreement with those based on cell morphology (Table 3). Furthermore, based on comparisons with all 33 type strains for the genus Bifidobacterium, types C, D, and G were considered to belong to either B. pseudolongum subsp. pseudolongum or B. pseudolongum subsp. globosum (25, 41, 52). Types A, B, E, and F remained unidentified.
(iv) 16S rDNA sequencing.
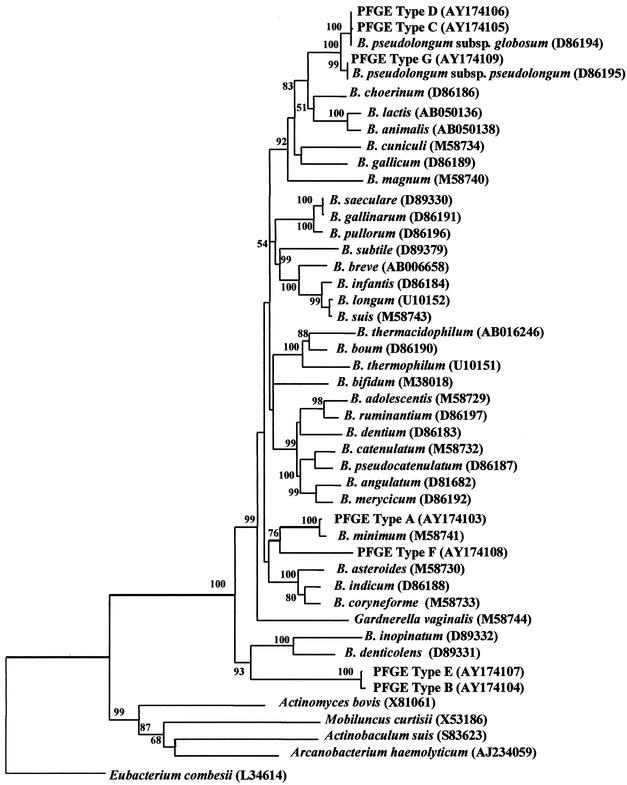
The comparison of Bifidobacterium 16S rDNA sequences has been shown to group all species within the genus and identify isolates to specific species or groups of species (11, 16, 26). In general, when 16S rDNA similarity values exceed 97%, the strains are considered to belong to the same species (47). Therefore, a tree based on comparisons between almost complete 16S rDNA sequences (>1,400 bases) for types A through G and for each Bifidobacterium type strain was constructed (Fig. 4). The seven PFGE types segregated in good agreement with the groupings based on cell morphology, WCPP, and RAPD PCR (Table 3). PFGE Type G shared 100% similarity with the B. pseudolongum subsp. pseudolongum type strain, and PFGE types C and D shared 99.93 and 100% similarity with the B. pseudolongum subsp. globosum type strain, respectively. However, in general agreement with WCPP, the assignments appeared to contradict the RAPD PCR-based data, which grouped types D and G but showed type C to be distinct. However, the 16S rDNA sequences for the two subspecies are themselves 99.14% similar, indicating the difficulty in distinguishing the two subspecies based on 16S rDNA sequence comparisons (26). Previously strains considered as intermediates between the two subspecies were proposed (52), and type D could represent an intermediate strain. Type A was identified as B. minimum since it was 99.57% similar to the species type strain. The inclusion of type F changed the position of B. minimum within the phylogenetic tree, replacing the three insect-derived Bifidobacterium species as the closest neighbor (26). However, type F was only 95.08 and 94.85% similar to type A and B. minimum, respectively, and was considered to likely represent a new Bifidobacterium species. Types B and E were 99.72% similar to each other and were considered to be highly related and probably from the same species. However, they were only 91.21 and 91.36% similar, respectively, to their nearest neighbor, Bifidobacterium denticolens, and were considered to represent a new Bifidobacterium species.
FIG. 4.
Phylogenetic tree based on 16S rDNA sequences for each Bifidobacterium species type strain and seven PFGE types, termed A through G, representing bifidobacteria isolated from a pig cecum. Database accession numbers are given in parenthesis. The tree was rooted with E. combesii and constructed using the neighbor-joining method. Bootstrap values, expressed as percentages of 1,000 replicons, are given at each branch point where above 50%.
(v) DNA-DNA similarity.
DNA-DNA hybridization is considered to be the “gold standard” for taxonomic evaluation (47). Bacteria that have DNA similarity values of 70% or above are generally considered to belong to the same species and usually have corresponding 16S rDNA similarity values of 98% or above (47). Therefore, the DNA similarity values between a number of the PFGE types and type strains of species showing a phylogenetic grouping were measured. The results were in good agreement with earlier findings (Table 4). Type A was confirmed as B. minimum, sharing 103% similarity, and the species accounted for 21% of the bifidobacteria isolated from the pig cecum. Type C and type D were 85% similar and were clearly related. However, type C and type D shared 85 and 100% similarity with the B. pseudolongum subsp. pseudolongum type strain. This finding highlights the previous contradiction between RAPD PCR and 16S rDNA sequencing (Table 3) and appears to further support the assignment of type D to an intermediary group between the two subspecies. Overall, the B. pseudolongum subsp. pseudolongum/B. pseudolongum subsp. globosum group accounted for 17% of the bifidobacteria isolated from the pig cecum. Types B and E shared 97% similarity, but both displayed low DNA similarity with B. inopinatum and B. denticolens, which were the closest neighbors within the 16S rDNA phylogenetic tree. Therefore, both were considered to represent a new species which, based on phenotypic data (unpublished data) was provisionally termed Bifidobacterium aerophilum sp. nov. The species accounted for 48% of the isolated cecal bifidobacteria. Type F displayed low similarity to the nearest 16S rDNA neighbor, B. minimum (Fig. 4), and was also considered to represent a new species which, based on phenotypic data (unpublished data), was provisionally termed Bifidobacterium psychroaerophilum sp. nov. The species accounted for 14% of the isolated cecal bifidobacteria.
TABLE 4.
DNA-DNA hybridization homologies between selected PFGE types and Bifidobacterium type strainsa
| Competitor strain | Reference strain | % DNA similarity |
|---|---|---|
| PFGE type A (DPC 5732) | PFGE type F (DPC 5740T) | 12 |
| PFGE type A (DPC 5732) | B. minimum (LMG 11592T) | 103 |
| PFGE type F (DPC 5740T) | B. minimum (LMG 11592T) | 14 |
| PFGE type D (DPC 5738) | PFGE type C (DPC 5377) | 85 |
| PFGE type C (DPC 5377) | B. pseudolongum subsp. pseudolongum (LMG 11571T) | 85 |
| PFGE type D (DPC 5738) | B. pseudolongum subsp. pseudolongum (LMG 11571T) | 100 |
| PFGE type B (DPC 5733) | PFGE type E (DPC 5739T) | 97 |
| PFGE type B (DPC 5733) | B. denticolens (LMG 18312T) | 8 |
| PFGE type E (DPC 5739T) | B. denticolens (LMG 18312T) | 7 |
| PFGE type B (DPC 5733) | B. inopinatum (LMG 18313T) | 2 |
| PFGE type E (DPC 5739T) | B. inopinatum (LMG 18313T) | 1 |
Superscript T denotes type strain.
Strain accession numbers.
Isolates representing the PFGE types A through G were deposited in the Dairy Products Collection under accession numbers DPC 5735 through 5741, respectively.
DISCUSSION
The antibiotic mupirocin was previously found to effectively select for bifidobacteria from other bacteria typically found in GIT and fecal samples (31, 34-36). The addition of mupirocin to mMRS has not been reported for the selective isolation of bifidobacteria but, along with the culture conditions used in the present study, appeared to be selective for all Bifidobacterium strains tested. This medium yielded 4.3 × 105 CFU of bifidobacteria/g for the pig cecal contents. Only a limited number of studies have reported the enumeration of bifidobacteria from cecal contents and values ranged from 3 × 103 CFU/g for rabbit (35), 5 × 106 CFU/ml for human (21), and 4 × 109 CFU/g for hen (35).
PFGE has previously been found to discriminate between Bifidobacterium strains (29, 38), a finding extended in the present study for a wide range of animal derived strains. In addition, PFGE was previously shown to be an effective technique for determining the genomic diversity of bifidobacteria isolated from human feces (17, 23). In these studies, the PFGE macro-restriction patterns for 10 Bifidobacterium isolates recovered at different times from a number of individuals were compared. Each PFGE type was considered to represent a distinct strain, and a complex population was arbitrarily assigned to subjects with four or more strains, while samples with fewer patterns were considered to be simple. Both kinds of population were observed with equal frequency. Following this convention our findings suggest that a simple bifidobacteria population, dominated by 15 Bifidobacterium strains, was maintained within a pig cecum.
A variety of studies have reported that Bifidobacterium strains vary in their ability to adhere to cultured epithelial cells (2, 6, 9, 10, 19, 30) and that the general microflora of the epithelium and contents of the porcine GIT are distinct (1, 44). However, in our study of bifidobacteria, although the total numbers of bifidobacteria associated with the cecal contents and epithelium differed considerably, distinct populations were not found. The data suggest that in vivo a relationship between “attached,” “loosely unattached,” and “unattached” cells exists. However, it is not clear if the bifidobacteria proliferate in the lumen and attach in a density-dependent manner or if proliferation occurs when they are attached to the epithelium with the lumen fraction represented an accumulation of detached bacteria.
Representatives for each PFGE type were examined by cell morphology, RAPD PCR, WCPP, 16S rDNA sequencing, and DNA-DNA hybridization to determine if a relatedness existed between the apparently unrelated macro-restriction patterns. Data from each technique were mostly complementary, and overall the seven types appeared to represent four distinct groups.
One group was identified as B. minimum, a species previously isolated only from sewage (5, 40). Another group appeared to include strains from B. pseudolongum subsp. pseudolongum and B. pseudolongum subsp. globosum (52). This study appears to be the first to report the recovery of either subspecies from a pig cecum, although they have previously been isolated from pig feces (25, 53), pork meat (14), and rabbit cecum (20, 46).
The two remaining groups were not assigned to any known Bifidobacterium species, although 16S rDNA sequencing did place them within the Bifidobacterium genus. In addition, both groups were positive for F-6-PPK activity. Based on the presented data and phenotypic analysis (unpublished data), PFGE types B, E, and subtype Ea strains were provisionally identified as a new species, termed Bifidobacterium aerophilum sp. nov. Similarly, the PFGE type F strain was provisionally identified as a new species termed Bifidobacterium psychroaerophilum sp. nov. The identification of additional species supports a recent study in which 16S rDNA sequences for 14 of 20 undefined isolates from the pig cecum were less than 95% similar to any database sequence (33).
A number of PFGE macro-restriction patterns differed by a single DNA fragment. It is possible that the extra band seen in these patterns represents an insertion, around or into a XbaI site, of a fragment with two restriction sites separated by a distance equal to the size of the insertion. However, it was considered more likely that the fragment represents the presence of a putative plasmid. In support of this, the estimated sizes for the majority of the suspected plasmids were within the range previously noted for the Bifidobacterium genus (41). However, two putative megaplasmids were also observed which have not previously been reported for the genus. Interestingly, most of the plasmids appeared to be associated with PFGE type C, which was identified as B. pseudolongum subsp. globosum, one of only four species in a survey of 25 Bifidobacterium species reported to contained plasmids (42). Work is ongoing to verify the PFGE interpretations and characterize the possible plasmids.
One interesting aspect of this study is the discovery of strains which from macro-restriction pattern analysis appear to show genomic rearrangements through reciprocal events involving either deletions or insertions. It is unclear if this genomic plasticity reflects a high rate of genomic change or a selection for very similar strains within the cecum. Interestingly, a recent study found that the commensal intestinal pathogen Bacteroides fragilis modified the structure of exopolysaccharides through chromosomal inversions within genetic promoter regions (18). The changes were considered to be part of a defense mechanism against the host immune response and allow for the long-term colonization of the GIT. It is tempting to suggest that the genomic plasticity observed in the present study may also have some bearing on long-term colonization of bifidobacteria.
In conclusion, the study presents a detailed examination of genomic diversity and relatedness of the bifidobacteria isolated from a pig cecum. The results suggest that the same relatively simple bifidobacteria population exists in the lumen and epithelium. Moreover, two potentially new species and evidence for genomic plasticity within strains were reported.
Acknowledgments
This work was supported by the Irish Government under the National Development Plan 2000-2006 and the European Union (SM&T-CT98-2235).
We are grateful to Tomotari Mitsuoka for providing B. thermophilum biotype information, Noelle Brennan for her assistance with 16S rDNA sequence comparisons and the determination of phylogenetic differences, and Shahenda Mohamed Mahmoud Al Abi for technical assistance.
REFERENCES
- 1.Allison, M. J., I. M. Robinson, J. A. Bucklin, and G. D. Booth. 1979. Comparison of bacterial populations of the pig cecum and colon based upon enumeration with specific energy sources. Appl. Environ. Microbiol. 37:1142-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1993. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl. Environ. Microbiol. 59:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biavati, B., V. Scardovi, and W. E. C. Moore. 1982. Electrophoretic patterns of proteins in the genus Bifidobacterium and proposal of four new species. Int. J. Syst. Bacteriol. 32:368-373. [Google Scholar]
- 4.Biavati, B., and P. Mattarelli. 1991. Bifidobacterium ruminatium sp. nov. and Bifidobacterium merycicum sp. nov. from the rumens of cattle. Int. J. Syst. Bacteriol. 41:163-168. [DOI] [PubMed] [Google Scholar]
- 5.Biavati, B., M. Vescovo, S. Torriani, and V. Bottazzi. 2000. Bifidobacteria: history, ecology, physiology and applications. Ann. Microbiol. 50:117-131. [Google Scholar]
- 6.Bibiloni, R., P. F. Perez, and G. L. de Antoni. 1999. Factors involved in adhesion of bifidobacterial strains to epithelial cells in culture. Anaerobe 5:483-485. [Google Scholar]
- 7.Bibiloni, R., A. G. Zavaglia, and G de Antoni. 2001. Enzyme-based most probable number for the enumeration of Bifidobacterium in dairy products. J. Food Prot. 64:2001-2006. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Microbiol. 51:843-852. [DOI] [PubMed] [Google Scholar]
- 9.Crociani, J., J.-P. Grill, M. Huppert, and J. Ballongue. 1995. Adhesion of different bifidobacteria strains to human enterocyte-like Caco-2 cells and comparison with in vivo study. Lett. Appl. Microbiol. 21:146-148. [DOI] [PubMed] [Google Scholar]
- 10.Del Re, B., A. Bussetto, G. Vignola, B. Sgorbati, and D. L. Palenzona. 1998. Autoaggregation and adhesion ability in a Bifidobacterium suis strain. Lett. Appl. Microbiol. 27:307-310. [PubMed] [Google Scholar]
- 11.Dong, X., Y. Xin, W. Jian, X. Liu, and D. Ling. 2000. Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester. Int. J. Syst. E vol. Microbiol. 50:119-125. [DOI] [PubMed] [Google Scholar]
- 12.Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Flourometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224-229. [Google Scholar]
- 13.Fanedl, L., F. V. Nekrep, and G. Avgustin. 1998. Random amplified polymorphic DNA analysis and demonstration of genetic variability among bifidobacteria isolated from rats fed with raw kidney beans. Can. J. Microbiol. 44:1094-1101. [PubMed] [Google Scholar]
- 14.Gavini, F., and H. Beerens. 1999. Origin and identification of bifidobacteria strains isolated from meat and meat products. Int. J. Food Microbiol. 46:81-85. [DOI] [PubMed] [Google Scholar]
- 15.Gomes, A. M. P., and F. X. Malcata. 1999. Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci. Technol. 10:139-157. [Google Scholar]
- 16.Hoyles, L., E. Inganas, E. Falsen, M. Drancourt, N. Weiss, A. L. McCarney, and M. D. Collins. 2002. Bifidobacterium scardovii sp. nov., from human sources. Int. J. Syst. E vol. Microbiol. 52:995-999. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human host to the predominant strain. Appl. Environ. Microbiol. 63:3394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krinos, C. M., M. J. Coyne, K. G. Weinacht, A. O. Tzaianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555-558. [DOI] [PubMed] [Google Scholar]
- 19.Lankaputhra, W. E. V., and N. P. Shah. 1998. Adherence of probiotic bacteria to human colonic cells. Biosci. Microflora 17:105-113. [Google Scholar]
- 20.Marounek, M., V. Rada, and V. Benda. 1998. Biochemical characteristics and fermentation of glucose and starch by rabbit caecal strains of Bifidobacterium globosum. Folia Microbiol. (Phara) 43:113-116. [DOI] [PubMed] [Google Scholar]
- 21.Marteau, P., P. Pochart, J. Dore, C. Bera-Maillet, A. Bernalier, and G. Corthier. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBrearty, S., P. J. Simpson, G. Fitzgerald, J. K. Collins, R. P. Ross, and C. Stanton. 2000. Probiotic bifidobacteria and their identification using molecular genetic techniques, p. 97-107. In J. Buttriss and M. Saltmarsh (ed.), Functional foods: claims and evidence. Royal Society of Chemistry, Cambridge, United Kingdom.
- 23.McCartney, A. L., W. Wenzhi, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen, L. L., and B. B. Jensen. 2001. Bifidobacteria in piglets, p. 285-288. In J. E. Lindberg and B. Ogle (ed.), Digestive physiology of pigs. CAB Int., Oxford, United Kingdom.
- 25.Mitsuoka, T. 1969. Comparative studies on bifidobacteria isolated from the alimentary tract of man and animals, including descriptions of Bifidobacterium thermophilum nov. spec. and Bifidobacterium pseudolongum nov. spec. Infektionskrankhh. Hyg. Abt. 1 Orig. Reihe A 210:52-64. [PubMed] [Google Scholar]
- 26.Miyake, T., K. Watanabe, T. Watanabe, and H. Oyaizu. 1998. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 42:661-667. [DOI] [PubMed] [Google Scholar]
- 27.Muirhead, S. 2000. Direct-fed products, p. 57-294. In S. Muirhead (ed.), 2000-2001 direct-fed microbial, enzyme and forage additive compendium, vol 5. Miller Publishing Co., ••••, Minnesota.
- 28.Orban, J. J., and J. A. Patterson. 2000. Modification of the phosphoketolase assay for rapid identification of bifidobacteria. J. Microbiol. Methods 40:221-224. [DOI] [PubMed] [Google Scholar]
- 29.O'Riordan, K., and G. F. Fitzgerald. 1997. Determination of genetic diversity within the genus Bifidobacterium and estimation of chromosomal size. FEMS Microbiol. Lett. 156:259-264. [DOI] [PubMed] [Google Scholar]
- 30.Perez, P. F., Y. Minnaard, E. A. Disalvo, and G. L. de Antoni. 1998. Surface properties of bifidobacterial strains of human origins. Appl. Environ. Microbiol. 64:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petr, J., and V. Rada. 2001. Bifidobacteria are obligate inhabitants of the crop of adult laying hens. J. Vet. Med. B. Infect. Dis. Public Health 48:227-233. [DOI] [PubMed] [Google Scholar]
- 32.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fingerprints. M. Goodfellow and A. G. O'Donnell (ed.), Chemical methods in prokaryotic systematics. J. Wiley and Sons, Chichester, United Kingdom.
- 33.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rada, V. 1997. Detection of Bifidobacterium species by enzymatic methods and antimicrobial susceptibility testing. Biotechnol. Tech. 11:909-912. [Google Scholar]
- 35.Rada, V., K. Sirotek, and J. Petr. 1999. Evaluation of selective media for bifidobacteria in poultry and rabbit caecal samples. J. Vet. Med. B 46:369-373. [DOI] [PubMed] [Google Scholar]
- 36.Rada, V., and J. Koc. 2000. The use of mupirocin for selective enumeration of bifidobacteria in fermented milk products. Milchwissenchaft 55:65-67. [Google Scholar]
- 37.Robinson, I. M., M. J. Allison, and J. A. Bucklin. 1981. Characterization of the cecal bacteria of normal pigs. Appl. Environ. Microbiol. 41:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy, D., P. Ward, and G. Champagne. 1996. Differentiation of bifidobacteria by use of pulsed-field gel electrophoresis and polymerase chain reaction. Int. J. Food Microbiol. 29:11-29. [DOI] [PubMed] [Google Scholar]
- 39.Scardovi, V., G. Zani, and L. D. Trovatelli. 1970. Deoxyribonucleic acid homology among the species of the genus Bifidobacterium isolated from animals. Arch. Mikrobiol. 72:318-325. [DOI] [PubMed] [Google Scholar]
- 40.Scardovi, V., and L. D. Trovatelli. 1974. Bifidobacterium animalis (Mitsuoka) comb. nov. and the “minimum” and “subtile” groups of new bifidobacteria found in sewage. Int. J. Syst. Bacteriol. 24:21-28. [Google Scholar]
- 41.Scardovi, V. 1986. Genus Bifidobacterium, p. 1418-1434. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, Md.
- 42.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1982. Plasmids in the genus Bifidobacterium. J. Gen. Microbiol. 128:2121-2131. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu, J., N. Tsuchihashi, K. Kudoh, M. Wada, T. Takita, and S. Innami. 2001. Dietry curdlan increases proliferation of bifidobacteria in the cecum of rats. Biosci. Biotechnol. Biochem. 65:466-469. [DOI] [PubMed] [Google Scholar]
- 44.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-179. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2002. Genomic diversity within the genus Pediococcus as revealed by randomly amplified polymorphic DNA PCR and pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 68:765-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slovakora, L., D. Duskova, and M. Marounek. 2002. Fermentation of pectin and glucose and activity of pectin-degrading enzymes in the rabbit caecal bacterium Bifidobacterium pseudolongum. Lett. Appl. Microbiol. 35:126-130. [DOI] [PubMed] [Google Scholar]
- 47.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 48.Stanton, C., G. Gardiner, H. Meehan, K. Collins, G. Fitzgerald, P. B. Lynch, and R. P. Ross. 2001. Market potential for probiotics. Am. J. Clin. Nutr. 73:476-483. [DOI] [PubMed] [Google Scholar]
- 49.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. A. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent, D., D. Roy, F. Mondou, and C. Dery. 1998. Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 43:185-193. [DOI] [PubMed] [Google Scholar]
- 51.Watabe, J., Y. Benno, and T. Mitsuoka. 1983. Bifidobacterium gallinarum sp. nov.: a new species isolated from the ceca of chickens. Int. J. Syst. Bacteriol. 33:127-132. [Google Scholar]
- 52.Yaeshima, T., T. Fujisawa, and T. Mitsuoka. 1992. Bifidobacterium globosum, subjective synonym of Bifidobacterium pseudolongum, and description of Bifidobacterium pseudolongum subsp. pseudolongum comb. nov and Bifidobacterium pseudolongum subs. globosum comb. nov. Syst. Appl. Microbiol. 15:380-385. [Google Scholar]
- 53.Zani, G., B. Biavati, F. Crociani, and D. Matteuzzi. 1974. Bifidobacteria from faeces of piglets. J. Appl. Bacteriol. 37:537-547. [Google Scholar]
- 54.Zavaglia, A. G., G. Kociubinski, P. Perez, and G. de Antoni. 1998. Isolation and characterisation of Bifidobacterium strains for probiotic formulation. J. Food Prot. 61:865-873. [DOI] [PubMed] [Google Scholar]