Abstract
Derivatives of the stationary-phase sigma factor σS of Escherichia coli lacking either of two conserved domains, the postulated N-terminal subregion 1.1 or the C-terminal region 4, were shown to be competent in vitro for transcription initiation from several σS-dependent promoters on supercoiled DNA templates. Unlike wild-type σS, however, the deletion derivatives were inactive on relaxed templates. The anomalous slow electrophoretic mobility of σS on denaturing gels was corrected by deletion of subregion 1.1, suggesting that this domain in σS may be structurally and functionally analogous to subregion 1.1 of σ70, substitutions in which have previously been shown to rectify the anomalous electrophoretic migration of σ70 (V. Gopal and D. Chatterji, Eur. J. Biochem. 244:614-618, 1997).
Enterobacterial strains, including Escherichia coli, possess two primary and several alternative σ factors that are mutually exclusive in their association with RNA polymerase core enzyme (E) to constitute different holoenzyme species, each with a different promoter specificity in the initiation of transcription (reviewed in references 15, 18, and 27). The two primary σ factors in E. coli are RpoD, or σ70, with 613 amino acid residues, which is the housekeeping or indispensable σ factor (6, 27), and RpoS, or σS, with 330 amino acid residues, which is important for gene expression under particular conditions of growth, such as the stationary phase, high osmolarity, or low temperature (17, 18, 42).
Based on sequence alignment of a large number of σ factors from both gram-positive and gram-negative bacteria, four conserved regions (numbered 1 to 4, beginning from the N-terminal end) have been delineated in σ70, and each is further divided into subregions (27). Regions 2 and 4 are the most highly conserved among the various σ factors and bind, respectively, to the −10 and −35 promoter sequence motifs (8, 30). σ70 mutants from which region 4 has been deleted are still proficient for transcription in vitro at promoters that are not dependent for their activity on a good fit to consensus at the −35 region (22, 23).
Region 1 in σ70 has further been divided into two subregions, 1.1 and 1.2, of which subregion 1.1 (more N-terminal) is highly acidic and appears to be conserved only in the housekeeping σ factors of different bacteria and to a lesser extent perhaps in σS (27, 34). Subregion 1.1 in E. coli σ70 (comprising the segment from approximately residues 30 through 100) has been ascribed various functions and activities, including those of masking the DNA-binding domain(s) in free σ (10, 11) and mediating the binding of σ with core enzyme (16, 32), thereby influencing promoter binding and the efficiency of transcription initiation by holoenzyme (4, 5, 13, 31, 46, 47). Deletion of subregion 1.1 of σ70 is associated with a dominant lethal phenotype in E. coli (10, 38). Interestingly, the σ70 polypeptide exhibits on denaturing polyacrylamide gels an anomalous slow electrophoretic mobility whose basis is not completely understood (6), but this anomaly was corrected in two different mutants with single amino acid substitutions in subregion 1.1 (13).
The second primary σ factor in E. coli σS also has the conserved regions 1 through 4 (Fig. 1). The N-terminal acidic stretch of 60 amino acids has been postulated to constitute its subregion 1.1 (27, 34), but Wilson and Dombroski (47) undertook sequence alignment of the housekeeping σ factors from nine bacteria to identify invariant and conserved residues in subregion 1.1, less than half of which are present in σS (34); furthermore, unlike the case of σ70 (10, 11, 47), σS from which this N-terminal region has been deleted appears to retain substantial functional activity in vivo (34). It has also been speculated that region 4 in σS may be dispensable for promoter recognition (19), since the recognition specificity is apparently dictated mainly by the −10 hexamer and the −13/−14 sequences at the promoter (2, 21, 41).
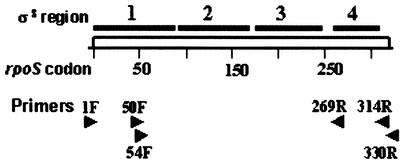
FIG. 1.
Conserved regions in σS and construction of N- and C-terminally truncated σS mutants. The rpoS coding region is shown by the open bar, and the lines above demarcate the segments encoding the conserved regions 1 through 4 in the σS polypeptide. Aligned below are arrowheads depicting the positions for annealing to the rpoS locus of forward and reverse PCR primers (suffixed F and R, respectively) employed in construction of the various His-tagged σS derivatives: σS NH, 1F and 330R; σS NHΔ2-50, 50F and 330R; σS NHΔ2-54, 54F and 330R; σS NHΔ315-330, 1F and 314R; and σS NHΔ270-330, 1F and 269R. The sequences of the primers (5′ to 3′; mismatches to rpoS sequence are shown in italics) were as follows: 1F, GGAGCCACCATATGAGTCAGAATAC; 50F, GTTATCGCAG CATATGACACAGCGTGTG; 54F, GAGCCACACAGCATATGTTGGACGC; 269R, CGTGCCAGC GCGGCCGCCTATTTGG; 314R, CCCCTGCGTTGCGGCCGCTTAGGCAA; and 330R, CTGACAGGCGGCCGCTTACTCGCGG.
In the present study, we prepared His-tagged derivatives of full-length and truncated σS polypeptides and tested their properties in vitro. Our studies provide direct evidence that holoenzyme bearing σS lacking either postulated subregion 1.1 or region 4 retains at least partial activity at σS-dependent promoters on supercoiled DNA templates. The deletion mutants were inactive on relaxed templates, whereas full-length σS under the same conditions was equally active on both supercoiled and relaxed templates. Furthermore, our data demonstrate that the anomalously slow electrophoretic mobility of σS on denaturing polyacrylamide gels (25, 42) is a property that is conferred by the N-terminal region of the polypeptide, providing additional support to the hypothesis that this region is structurally and functionally analogous to subregion 1.1 in σ70.
Evidence for expression in vivo of a partially functional σS Δ2-54 polypeptide from an rpoS allele with amber mutation in codon 33.
The ancestral E. coli K-12 strain and several of its derivatives have been shown to bear an amber mutation in codon 33 of the rpoS gene (1, 45). In the course of studies on PCR amplification and sequencing of the rpoS gene from different strains, we noted that strain W3350 and several stocks of strain W3110 (19) also carried this mutation.
The rpoS (Am) allele from W3350 was cloned downstream of the lac promoter on a pBR322-based plasmid vector, and the resulting plasmid (designated p25rpoS) was introduced into the suppressor-free null rpoS::Kan strain ZK1000 (obtained from R. Kolter). The following lines of evidence indicated that the mutant rpoS gene on the plasmid was able to direct the synthesis of a σS variant polypeptide that lacks the major part of the postulated subregion 1.1 but that still retains at least partial σS activity in vivo. (i) In Western blot experiments with anti-σS antibody, cell extracts of ZK1000/p25rpoS exhibited a cross-reacting band of apparent Mr of 34,000 (that is, smaller than the native σS band of apparent Mr of 43,000) whose intensity was approximately 20 to 30% of that for native σS from a haploid rpoS+ strain; as expected, no immunoreactive band was detected in cell extracts of either ZK1000 or W3350 (data not shown). (ii) The ZK1000/p25rpoS cell extract also possessed a moderate level of catalase HPII activity (encoded by the σS-dependent katE gene), as assayed by the method of Visick and Clarke (45); the activity was approximately 60% of that in the haploid rpoS+ strain (data not shown). (iii) Finally, N-terminal sequence analysis indicated that the protein immunoprecipitated by anti-σS antibody from the cell extract has an N-terminal Met residue followed by a sequence which is identical to that downstream of residue 54 in native σS. We refer to this protein as σS Δ2-54 and infer that it has been synthesized from the mutant gene by reinitiation of translation from codon 54 (GUG) in rpoS mRNA (eleven nucleotides upstream of which also is the sequence AGGGAG that represents a ribosome-binding-site motif).
Based on studies on an IS10 insertion mutation in rpoS, a recent report had also similarly concluded that a variant of σS whose N-terminal 50 amino acids were expected to have been substituted by six amino acids encoded by the IS10 end is partially active in vivo (34). Accordingly, in the experiments described below, we tested two N-terminally truncated σS polypeptides (Δ2-50 and Δ2-54) for their properties in vitro. We also tested two C-terminal deletions of σS (Δ315-330, with removal of residues downstream of region 4 in the segment that has been termed the C-terminal extension [33], and Δ270-330, with removal of all of region 4).
Expression and purification of His-tagged full-length or truncated σS derivatives.
Procedures for PCR and molecular cloning were as described previously (37). Appropriate forward and reverse oligonucleotide primer pairs were used along with a proofreading-proficient thermostable DNA polymerase to PCR amplify, from a chromosomal DNA preparation of wild-type E. coli, rpoS sequences that represented full-length σS or σS derivatives with N- or C-terminal truncations (Fig. 1). Each amplified rpoS sequence was flanked at its upstream and downstream ends by NdeI and NotI sites, respectively, and the products were then cloned into the NdeI-NotI sites of plasmid vector pET28a (Novagen, Madison, Wis.) so that the vector-encoded sequence provided an N-terminal fusion of 20 amino acid residues (including a six-residue His tag) to each of the full-length or truncated σS derivatives. The authenticity of each plasmid construct was confirmed by DNA sequencing (data not shown). Our decision to place the His tag at the N-terminal ends of the polypeptides was based on an earlier report that N-terminally His-tagged σS is fully functional in vitro (46).
The plasmids so constructed were introduced into one of two overexpression host strains, BL21-SI (12) or BL21(DE3) (40), and overproduction of the σS derivatives was achieved in 250-ml cultures by induction, respectively, with both isopropyl-β-d-thiogalactopyranoside and NaCl (3, 12) or with isopropyl-β-d-thiogalactopyranoside alone (40). Cell lysates were prepared after suspension in lysis buffer (50 mM Tris-Cl [pH 8], 1 mM EDTA, 100 mM NaCl) as previously described (25, 26), and the overproduced protein (present predominantly in the insoluble fraction) was purified from each of them in a single step by column chromatography through a nickel-nitrilotriacetic acid resin column (Qiagen, Chatsworth, Calif.) in the presence of 6 M guanidine hydrochloride (following the manufacturer's instructions). In each case, the His-tagged protein was eluted in a 2-ml volume with 100 mM imidazole, and the chaotrope was then removed by dialysis at 4°C against two changes of one liter each of storage buffer (10 mM Tris-Cl [pH 7.6], 200 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA, 50% glycerol). The dialysates were centrifuged at 12,000 × g for 10 min at 4°C, and the supernatants, which were judged to be substantially pure for the full-length and truncated σS polypeptides (see Fig. 2), were stored at −30°C. The His-tagged polypeptides so prepared included those of full-length σS (σS NH), N-terminal truncations of amino acid residues from 2 to 50 (σS NHΔ2-50) or from 2 to 54 (σS NHΔ2-54), and C-terminal truncations of amino acid residues from 270 to 330 (σS NHΔ270-330) or from 315 to 330 (σS NHΔ315-330).
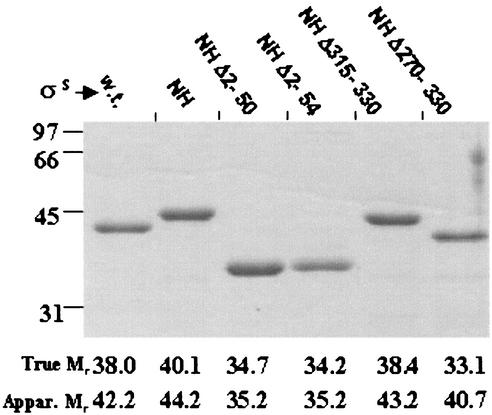
FIG. 2.
Electrophoretic migration of σS and its derivatives. The indicated σS derivatives were electrophoresed (37) on a 10% polyacrylamide gel with sodium dodecyl sulfate and visualized after staining with Coomassie blue. At left are shown the positions of migration of marker proteins of indicated Mr. For each σS derivative, the true and apparent (Appar.) Mr values, the latter as calculated from the migration rate on the gel, are given below the corresponding lane. w.t., wild-type (native) σS.
Electrophoretic mobility of σS derivatives on denaturing polyacrylamide gels.
It is known that σS, like several other σ factors (25), exhibits anomalously slow migration properties during sodium dodecyl sulfate-polyacrylamide gel electrophoresis. When the electrophoretic mobility of the various purified His-tagged σS preparations were compared by standard procedures (37) with that of native σS, it was observed that σS NH, as well as each of its C-terminal truncated derivatives σS NHΔ270-330 and σS NHΔ315-330, exhibited an anomalous mobility similar to that of native σS, whereas in the N-terminally truncated derivatives σS NHΔ2-50 and σS NHΔ2-54, the anomaly had largely been corrected (Fig. 2). The difference is particularly striking, for example, between σS NHΔ270-330 and either of the N-terminally truncated derivatives, since the former is smaller and yet migrates less rapidly than the latter. The results therefore indicate that deletion of the postulated N-terminal subregion 1.1, but not of C-terminal region 4, is associated with correction of the anomalous electrophoretic mobility of σS.
In vitro transcription with σS derivatives on supercoiled templates.
Each of the σS derivatives was reconstituted as previously described (26) with highly purified core RNA polymerase at a ratio of 4:1 or higher, and the holoenzyme preparations were used in in vitro transcription experiments. The DNA templates in the reactions were plasmids pHYD351 (36) or pHYD406, supercoiled preparations of which were obtained with the aid of a commercial plasmid preparation kit (Qiagen) from transformants of the strain DH5 (37). The two plasmids are derivatives of plasmid vector pCU22 (43) and carry the −60 to +116 and the −121 to +70 regions of the σS-dependent proU P1 (35) and glgS P2 (2) promoters, respectively, from E. coli, cloned about 50 bp upstream of the first of a tandem pair of Rho-independent transcription terminators present in the vector (that are themselves 40 bp apart).
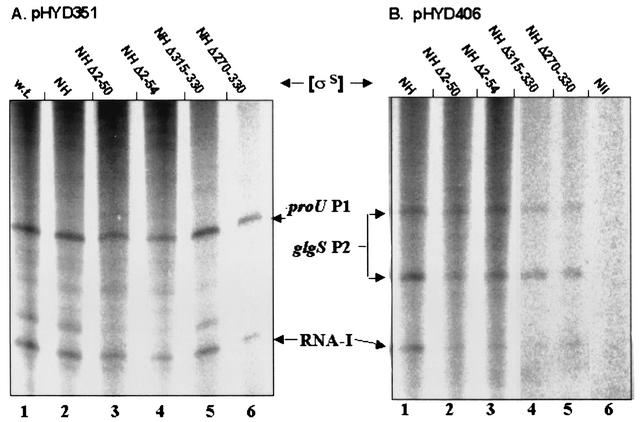
Single-round in vitro transcription reactions with [α-32P]UTP were performed, and the products were analyzed by autoradiography following electrophoresis on urea-polyacrylamide gels, as earlier described (36); the results are shown in Fig. 3. EσS NH, carrying the N-terminal His-tagged full-length σS derivative, was indistinguishable from EσS (Fig. 3A, compare lanes 1 and 2), while the negative control reaction with core enzyme E alone yielded no transcripts under these conditions for both pHYD406 (Fig. 3B, lane 6) and pHYD351 (data not shown). The two N-terminally truncated σS derivatives and the two C-terminally truncated derivatives exhibited substantial transcriptional activity at both the proU P1 and glgS P2 promoters (Fig. 3A and B, respectively). All the deletion derivatives were active also for transcription of RNA-I (whose promoter, in the pCU22 vector-derived sequence common to both plasmids, is known to be recognized by both Eσ70 and EσS [26, 35, 36]), although there was some variation in its relative band intensity between different experiments, even for the same σS mutant preparation (compare, for example, lanes 3 and 2 in panels A and B, respectively, of Fig. 3 for σS NHΔ2-50).
FIG. 3.
Single-round in vitro transcription with indicated EσS derivatives on supercoiled pHYD351 (A) and pHYD406 (B) templates, carrying the proU P1 and glgS P2 promoters, respectively. Bands corresponding to RNA-I and to the transcripts initiated from proU P1 and glgS P2 are marked. The reaction represented on lane 6 (labeled Nil) in panel B was done with core enzyme alone (without any σS).
The N- and C-terminally truncated σS derivatives were also active in vitro on the σS-dependent proU P1 promoter of Salmonella enterica serovar Typhimurium carried on a supercoiled pCU22 derivative plasmid (35; data not shown). As expected, none of the σS derivatives was active on the exclusively σ70-dependent proU P2 promoter (36; data not shown).
Ohnuma et al. (33) have previously demonstrated that holoenzyme carrying a mutant σS with deletion of the C-terminal residues from 315 to 330 is rendered salt sensitive in that it is transcriptionally inactive in the presence of 0.2 M potassium glutamate, whereas the activity of EσS itself is known to be actually stimulated under these conditions (26, 36). Using the various holoenzyme preparations of the His-tagged full-length and N- or C-terminally truncated σS derivatives on supercoiled plasmid pHYD351 (with E. coli proU P1) as template, we were able to confirm that it is only the holoenzyme reconstituted with either of the C-terminally truncated mutants (σS NHΔ315-330 or σS NHΔ270-330) whose activity is sensitive to a high potassium glutamate concentration in vitro (data not shown). Similarly, only the two C-terminal truncation mutants were consistently associated with a reduction in the background of high-molecular-weight RNA species that is a common feature of in vitro transcription with supercoiled plasmid DNA templates (35, 36, 43) (see Fig. 3A and B); a plausible explanation for this observation is given in the following section.
In vitro transcription with σS derivatives on linear templates.
Previous studies have shown both that the superhelical density of DNA in E. coli cells is less in the stationary phase than it is in the exponential phase and that σS-directed transcription in vitro from a σS-dependent promoter osmY is indeed maximal from a relaxed DNA template (26). We examined the ability of the His-tagged σS full-length and truncated derivatives to support transcription from linear DNA templates carrying the E. coli proU P1 and glgS P2 promoters. For this purpose, the template plasmids pHYD351 and pHYD406 were digested with PstI, which cuts each plasmid at a single site immediately upstream of the promoter-bearing DNA fragment cloned into the pCU22 vector.
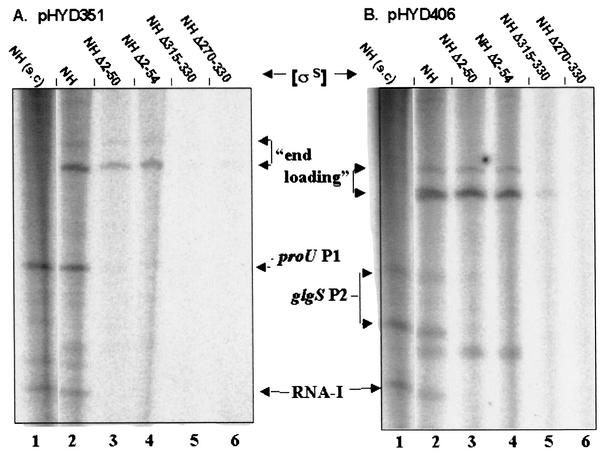
The results of these experiments are shown in Fig. 4. For either promoter tested, the full-length σS derivative σS NH was as effective for transcription from a linear template as it was from a supercoiled template (Fig. 4, compare lane 1 with lane 2 in each of the two panels). On the other hand, the N- and C-terminally truncated σS derivatives were all inactive for transcription from both proU P1 and glgS P2, as well as from the RNA-I promoter, carried on the linear templates. These data indicate that the truncations have compromised the property of supercoiling insensitivity of promoter-specific transcription that is associated with full-length σS.
FIG. 4.
Single-round in vitro transcription with indicated EσS derivatives on linear pHYD351 (A) and pHYD406 (B) templates, carrying the proU P1 and glgS P2 promoters, respectively. Lane 1 in each panel represents a control reaction in which the corresponding supercoiled (s.c.) template was employed. Bands corresponding to RNA-I and to the transcripts initiated from proU P1 and glgS P2 are marked. Bands labeled “end loading” are discussed in the text.
We also observed an interesting difference in behavior between the σS derivatives with N-terminal truncations, on the one hand, and those with C-terminal truncations, on the other. The former behaved like σS NH on either DNA template in yielding a pair of bands (designated by the term “end loading” in Fig. 4) whose sizes (approximately 230 and 270 bases) are consistent with transcription initiated from the upstream PstI-cut end (29) and proceeding through the cloned promoter fragment to the tandem terminators situated downstream. On the other hand, the C-terminal deletion derivatives did not yield such products (Fig. 4, compare lanes 1 to 3 with lanes 4 and 5 in each of the panels A and B). Therefore, the loss of postulated subregion 1.1 in σS does not apparently affect the ability of holoenzyme to load nonspecifically at a DNA nick or end and initiate transcription (29); the inability of the C-terminal deletion mutants to do so may perhaps explain the reduced background of high-molecular-weight RNA species in the cognate transcription reactions with supercoiled template preparations (Fig. 3), which are expected also to have a proportion of nicked and linear DNA molecules.
Discussion.
In this study, we have investigated the in vitro properties of mutant σS derivatives with N-terminal deletions of the postulated subregion 1.1 and with two different C-terminal deletions. In a previous study (33), the stretch of 16 amino acid residues at the extreme C terminus of σS was taken to be separate from, and downstream of, its subregion 4.2, as defined by Lonetto et al. (27). We found in this study, however, that in a variety of assays, the transcriptional activity of a mutant σS lacking its C-terminal 16 amino acid residues is indistinguishable from that with a larger deletion of all of region 4, suggesting that the extreme C terminus is also a part of region 4 in the polypeptide.
The present data provide direct and conclusive evidence that mutant σS polypeptides from which postulated subregion 1.1 or region 4 is deleted, and whose putative occurrence in some naturally occurring E. coli strains is not unlikely (1, 19, 34, 45), retain substantial promoter-specific transcription-initiation activity on supercoiled templates. In particular, the behavior of the σS mutant lacking region 4 is markedly different from that of the equivalent σ70 mutant, which is active in vitro only on the subset of promoters that possess an extended −10 recognition motif (23).
Our observations indicate that in comparison with full-length σS, both the N-terminal and the C-terminal deletion mutants are severely compromised for activity at promoters on linear or relaxed templates. Although a mechanistic explanation for this phenomenon was not obtained in this study, we suggest that holoenzymes reconstituted with the mutant σS polypeptides are unable to achieve efficient isomerization from the closed to the open complex at the various promoters and that they are able to do so only if the energy of DNA supercoiling is available for facilitating strand opening at the promoter −10 region (9). A similar phenotype of supercoiling sensitivity of transcription in vitro has been reported for mutations in subregion 2.3 of σA (the Bacillus subtilis housekeeping σ factor) and shown to be associated with a defect in melting of the DNA strands at the −10 region of the promoter (20). Since the stationary growth phase is believed to be associated with relaxation of DNA supercoiling (26), our in vitro data may also explain why it is that in vivo, even the shorter C-terminal deletion mutant of σS is inactive (33) and the subregion 1.1 deletions are only partially active (34; this study) at σS-dependent promoters.
Anomalous electrophoretic mobility is an unusual property of σ70 (6) that is also shared by a variety of other σ factors, including σS, σH, σF, and σN of E. coli (25, 42) and σA of B. subtilis (39), but whose basis is not completely understood. It had earlier been suggested (6) that an acidic region between amino acid residues 184 and 215 in σ70 may be responsible, but a mutant from which this region has been deleted retains this property (24). Gopal and coworkers have shown that substitution mutations in subregion 1.1 (13) or a mutation in subregion 2.3 (14) of σ70 abolish its aberrant migration and that the cognate holoenzymes are defective for transcription initiation on linear templates; interestingly, the residue mutated in subregion 2.3 in the latter study was the equivalent of that whose substitution in B. subtilis σA conferred supercoiling sensitivity (20).
Gopal and coworkers have therefore suggested that binding to DNA of the free σ70 subunit is inhibited by hydrophobic interaction of subregion 1.1 with the −10 promoter DNA-binding subregion 2.3 and that this interaction is also responsible for the aberrant electrophoretic mobility of σ70 (13, 14). Their suggestions are contrary to the more prevalent view (10, 11) that autoinhibition of σ70 binding to DNA involves direct interaction between subregion 1.1 and domain 4.2 that binds the −35 region of the promoter, but they are consistent with three other findings, namely, (i) that subregion 1.1 of σ70 also masks the core-binding sites in the vicinity of subregion 2.3 (16), (ii) that the only mutations or deletions in σ that affect the isomerization step (of holoenzyme bound to promoter) from the closed complex to the open complex map either to subregion 1.1 (13, 46, 47) or to subregion 2.3 (9, 14, 20), and (iii) that the property of supercoiling sensitivity in transcription initiation is shared by subregion 1.1 mutations (in σS, as shown in this study) and subregion 2.3 mutations (as shown for B. subtilis σA [20]). Furthermore, a recent study (7) has excluded the possibility of direct interaction between subregions 1.1 and 4.2 of σ70. It is noteworthy that crystals of σ70 (either as free subunit or as part of holoenzyme) have been successfully obtained only if the autoinhibitory subregion 1.1 of the polypeptide is omitted (8, 28, 31, 44), although crystals of the holoenzyme-DNA complex can include this domain (30, 31).
In this context, our finding in this study that deletions of the N-terminal region (but not of region 4) in σS abolish aberrant mobility is significant in that (i) it strengthens the case for a distinct subregion 1.1 in σS equivalent to that in σ70 and (ii) it supports an autoinhibitory role for this subregion in free σS similar to that for σ70 suggested by Gopal and coworkers (13, 14).
Acknowledgments
We thank the members of A.I.'s laboratory for providing reagents, including σS and core RNA polymerase, and those in J.G.'s laboratory for discussions and for assistance in preparation of the manuscript.
Support for this work from the India-Japan Science Co-operation Programme of the Japan Society for the Promotion of Science and the Department of Science & Technology (Government of India) is gratefully acknowledged. Research in A.I.'s laboratory is also supported by grants-in-aid from the Ministry of Education, Science and Technology of Japan, and the CREST fund from Japan Science and Technology Foundation. J.G. is Honorary Senior Fellow of the Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Atlung, T., H. V. Nielsen, and F. G. Hansen. 2002. Characterisation of the allelic variation in the rpoS gene in thirteen K12 and six other non-pathogenic Escherichia coli strains. Mol. Genet. Genom. 266:873-881. [DOI] [PubMed] [Google Scholar]
- 2.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter σS dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 3.Bhandari, P., and J. Gowrishankar. 1997. An Escherichia coli host strain useful for efficient overproduction of cloned gene products with NaCl as the inducer. J. Bacteriol. 179:4403-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers, C. W., and A. J. Dombroski. 1999. A mutation in region 1.1 of σ70 affects promoter DNA binding by Escherichia coli RNA polymerase holoenzyme. EMBO J. 18:709-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowers, C. W., A. McCracken, and A. J. Dombroski. 2000. Effects of amino acid substitutions at conserved and acidic residues within region 1.1 of Escherichia coli σ70. J. Bacteriol. 182:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, Z., R. R. Burgess, J. Lin, D. Moore, S. Holder, and C. A. Gross. 1981. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E. coli K12. Nucleic Acids Res. 9:2889-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camarero, J. A., A. Shekhtman, E. A. Campbell, M. Chlenov, T. M. Gruber, D. A. Bryant, S. A. Darst, D. Cowburn, and T. W. Muir. 2002. Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc. Natl. Acad. Sci. USA 99:8536-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 9.deHaseth, P. L., and J. D. Helmann. 1995. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol. Microbiol. 16:817-824. [DOI] [PubMed] [Google Scholar]
- 10.Dombroski, A. J., W. A. Walter, and C. A. Gross. 1993. Amino-terminal amino acids modulate σ-factor DNA-binding activity. Genes Dev. 7:2446-2455. [DOI] [PubMed] [Google Scholar]
- 11.Dombroski, A. J., W. A. Walter, M. T. Record, Jr., D. A. Siegele, and C. A. Gross. 1992. Polypeptides containing highly conserved regions of transcription initiation factor σ70 exhibit specificity of binding to promoter DNA. Cell 70:501-512. [DOI] [PubMed] [Google Scholar]
- 12.Donahue, R. A., Jr., and R. L. Bebee. 1999. BL21-SI competent cells for protein expression in E coli. Focus 21:49-51. [Google Scholar]
- 13.Gopal, V., and D. Chatterji. 1997. Mutations in the 1.1 subdomain of Escherichia coli sigma factor σ70 and disruption of its overall structure. Eur. J. Biochem. 244:613-618. [DOI] [PubMed] [Google Scholar]
- 14.Gopal, V., H.-W. Ma, M. K. Kumaran, and D. Chatterji. 1994. A point mutation at the junction of domain 2.3/2.4 of transcription factor σ70 abrogates productive transcription and restores its expected mobility on a denaturing gel. J. Mol. Biol. 242:9-22. [DOI] [PubMed] [Google Scholar]
- 15.Gross, C. A., C. Chan, A. Dombroski, T. Gruber, M. Sharp, J. Tupy, and B. Young. 1998. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 63:141-155. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, T. M., D. Markov, M. M. Sharp, B. A. Young, C. Z. Lu, H. J. Zhong, I. Artsimovitch, K. M. Geszvain, T. M. Arthur, R. R. Burgess, R. Landick, K. Severinov, and C. A. Gross. 2001. Binding of the initiation factor σ70 to core RNA polymerase is a multistep process. Mol. Cell 8:21-31. [DOI] [PubMed] [Google Scholar]
- 17.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 18.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54:499-518. [DOI] [PubMed] [Google Scholar]
- 19.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juang, Y.-L., and J. D. Helmann. 1994. A promoter melting region in the primary σ factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J. Mol. Biol. 235:1470-1488. [DOI] [PubMed] [Google Scholar]
- 21.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase Eσ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, A., B. Grimes, N. Fujita, K. Makino, R. A. Malloch, R. S. Hayward, and A. Ishihama. 1994. Role of the sigma70 subunit of Escherichia coli RNA polymerase in transcription activation. J. Mol. Biol. 235:405-413. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, A., H. S. Williamson, N. Fujita, A. Ishihama, and R. S. Hayward. 1995. A partially functional 245-amino-acid internal deletion derivative of Escherichia coli σ70. J. Bacteriol. 177:5193-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu, T. S., S. Kusano, and A. Ishihama. 1997. Promoter selectivity of Escherichia coli RNA polymerase σF holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol. 179:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusano, S., Q. Ding, N. Fujita, and A. Ishihama. 1996. Promoter selectivity of Escherichia coli RNA polymerase Eσ70 and Eσ38 holoenzymes. Effect of DNA supercoiling. J. Biol. Chem. 271:1998-2004. [DOI] [PubMed] [Google Scholar]
- 27.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malhotra, A., E. Severinova, and S. A. Darst. 1996. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell 87:127-136. [DOI] [PubMed] [Google Scholar]
- 29.Melancon, P., R. R. Burgess, and M. T. Record, Jr. 1983. Direct evidence for the preferential binding of Escherichia coli RNA polymerase holoenzyme to the ends of deoxyribonucleic acid restriction fragments. Biochemistry 22:5169-5176. [DOI] [PubMed] [Google Scholar]
- 30.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 31.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 32.Nagai, H., and N. Shimamoto. 1997. Regions of the Escherichia coli primary sigma factor σ70 that are involved in interaction with RNA polymerase core enzyme. Genes Cells 2:725-734. [DOI] [PubMed] [Google Scholar]
- 33.Ohnuma, M., N. Fujita, A. Ishihama, K. Tanaka, and H. Takahashi. 2000. A carboxy-terminal 16-amino-acid region of σ38 of Escherichia coli is important for transcription under high-salt conditions and sigma activities in vivo. J. Bacteriol. 182:4628-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajkumari, K., and J. Gowrishankar. 2002. An N-terminally truncated RpoS (σS) protein in Escherichia coli is active in vivo and exhibits normal environmental regulation even in the absence of rpoS transcriptional and translational control signals. J. Bacteriol. 184:3167-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajkumari, K., A. Ishihama, and J. Gowrishankar. 1997. Evidence for transcription attenuation rendering cryptic a σS-dependent promoter of the osmotically regulated proU operon of Salmonella typhimurium. J. Bacteriol. 179:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajkumari, K., S. Kusano, A. Ishihama, T. Mizuno, and J. Gowrishankar. 1996. Effects of H-NS and potassium glutamate on σS- and σ70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J. Bacteriol. 178:4176-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Sharp, M. M., C. L. Chan, C. Z. Lu, M. T. Marr, S. Nechaev, E. W. Merritt, K. Severinov, J. W. Roberts, and C. A. Gross. 1999. The interface of σ with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 13:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shorenstein, R. G., and R. Losick. 1973. Comparative size and properties of the sigma subunits of ribonucleic acid polymerase from Bacillus subtilis and Escherichia coli. J. Biol. Chem. 248:6170-6173. [PubMed] [Google Scholar]
- 40.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, K., S. Kusano, N. Fujita, A. Ishihama, and H. Takahashi. 1995. Promoter determinants for Escherichia coli RNA polymerase holoenzyme containing σ38 (the rpoS gene product). Nucleic Acids Res. 23:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal σ factor in Escherichia coli: the rpoS gene product, σ38, is a second principal σ factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 12:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vassylyev, D. G., S.-I. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 45.Visick, J. E., and S. Clarke. 1997. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J. Bacteriol. 179:4158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vuthoori, S., C. W. Bowers, A. McCracken, A. J. Dombroski, and D. M. Hinton. 2001. Domain 1.1 of the σ70 subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 309:561-572. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, C., and A. J. Dombroski. 1997. Region 1 of σ70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 267:60-74. [DOI] [PubMed] [Google Scholar]