Abstract
The porphyrin requirements for growth recovery of Porphyromonas gingivalis in heme-depleted cultures are investigated. In addition to physiologically relevant sources of heme, growth recovery is stimulated by a number of noniron porphyrins. These data demonstrate that, as for Haemophilus influenzae, reliance on captured iron and on exogenous porphyrin is manifest as an absolute growth requirement for heme. A number of outer membrane proteins including some gingipains contain the hemoglobin receptor (HA2) domain. In cell surface extracts, polypeptides derived from HA2-containing proteins predominated in hemoglobin binding. The in vitro porphyrin-binding properties of a recombinant HA2 domain were investigated and found to be iron independent. Porphyrins that differ from protoporphyrin IX in only the vinyl aspect of the tetrapyrrole ring show comparable effects in competing with hemoglobin for HA2 and facilitate growth recovery. For some porphyrins which differ from protoporphyrin IX at both propionic acid side chains, the modification is detrimental in both these assays. Correlations of porphyrin competition and growth recovery imply that the HA2 domain acts as a high-affinity hemophore at the cell surface to capture porphyrin from hemoglobin. While some proteins involved with heme capture bind directly to the iron center, the HA2 domain of P. gingivalis recognizes heme by a mechanism that is solely porphyrin mediated.
The black-pigmented gram-negative bacterium Porphyromonas gingivalis is an important etiological agent of adult periodontal disease (reviewed in reference 23). This bacterium has been reported to absolutely require heme, usually as hemin (Hm, a chloride salt of the iron-oxidized form of heme) or hemoglobin (Hb), as a growth factor in vitro (20). While heme from Hb is thought to be the principal source of this growth factor in the environment of the gingival crevice (29), it may be supplemented by iron-porphyrins from various sequestering proteins of the host such as hemopexin, haptoglobin, and serum albumin, which are all present in the gingival crevice in significant concentrations (51). Although iron is also captured from other carrier proteins such as transferrin and lactoferrin, this additional capability under iron-limiting conditions does not appear to be correlated with P. gingivalis pathogenicity (22). Some bacteria capture heme principally as a source of essential iron by means of specialized receptors termed hemophores (19, 37). The pathogenic EB1 strain of Escherichia coli secretes a bifunctional protein which is an Hb protease and a hemophore (37). The active transport of captured heme and other iron-loaded siderophores across the outer membrane (OM) into the periplasmic space involves energy-transducing TonB proteins (28), and therefore the OM receptors are often referred to as TonB-dependent receptors.
Heme is required by P. gingivalis for a number of functions. First, cell surface heme acquisition has been postulated to be a defense mechanism against active oxygen species (49, 54). Second, heme capture is likely to be critical for energy metabolism because, while genome sequencing has identified genes (The Institute for Genomic Research's prepublished genomic sequence of P. gingivalis strain W83 [http://www.tigr.org]) coding for enzymes that utilize heme cofactors (e.g., cytochrome d ubiquinol oxidase and cytochrome c nitrite reductase), genes required for the de novo porphyrin biosynthetic pathway are absent (e.g., those encoding 5-aminolevulinic acid synthase and porphobilinogen deaminase) (40). However, a putative hemD gene (encoding uroporphyrinogen-III cosynthase) is reported in the genome, and its conservation is inconsistent with the loss of the de novo pathway (27). In iron-replete conditions the exogenous heme requirement of P. gingivalis is manifest as a protoporphyrin IX (PPIX) requirement (3, 41, 56). Of note, putative genes hemN, hemG, and hemH, encoding the last three enzymes in the heme biosynthetic pathway, are present in the P. gingivalis genome (27). The intracellular expression of HemH (porphyrin ferrochelatase) would permit exogenous PPIX, once captured and transported into the cell, to substitute as a growth factor for heme in iron-replete conditions. In addition, the presence of homologues of HemN (a coporphyrinogen oxidase) and HemG (a protoporphyrinogen oxidase) could impart an ability to generate essential porphyrins from related protoporphyrins by modification of side chains on the tetrapyrrole ring. A capability in vitro to bypass the growth requirement for heme by using other iron sources in combination with exogenous noniron porphyrins would imply that at least some of the porphyrin capture systems in P. gingivalis can recognize noniron porphyrins.
These observations are not unique to P. gingivalis, as some strains of Haemophilus spp. have an absolute growth requirement for heme (16). Haemophilus influenzae also lacks enzymes of the heme biosynthetic pathway (4) and is able to replace essential Hm with the heme precursor PPIX as a growth factor (21). A critical role for the propionic acid side chains in porphyrins has been recognized in H. influenzae because esterification of the side chains resulted in loss of growth stimulation (21). Significantly, a HemH homologue in H. influenzae reversibly catalyzes the chelation of Fe2+ and PPIX into heme (17, 31). Mutagenesis studies of H. influenzae indicate that the cytoplasmic ferrochelatase is not required for utilization of heme but is required for use of PPIX for growth (42). It has been suggested that there is an active capture and transport system in H. influenzae which is able to assimilate PPIX (39) because passive transport of PPIX and Hm through the outer membranes of gram-negative bacteria such as E. coli is insignificant (30, 32, 33, 52). Although no proteins with these functions have been identified, a number of strains of H. influenzae produce a cell surface heme-hemopexin-binding protein, HxuA (8). A second putative component of such a system is lipoprotein e (P4), which is thought to have a role in Hm and PPIX transport (39).
Heme-starved P. gingivalis cells express high- and low-affinity Hm-binding activity with dissociation constants (Kds) of ∼10−10 and ∼10−7 M, respectively (54). A number of OM proteins have been observed to bind Hm by tetramethylbenzidine (TMBZ) staining of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels (6, 26, 48). It has been proposed that some of these proteins may be, or may be derived from, the TonB-linked polypeptide IhtB (12). The affinity of Hm binding to E. coli cells expressing the TonB-dependent Hb receptor (HmuR) of P. gingivalis is low (Kd, ∼10−5 M); however, a protein cotranscribed with HmuR, HmuY (36), is also observed to bind Hm on TMBZ-stained gels (26).
In addition to TonB-dependent OM receptors, heme function in P. gingivalis requires the cell surface protein Kgp for hemolysis, Hb proteolysis (29), and efficient heme capture (7). The lysine-specific Kgp and arginine-specific RgpA cysteine proteinases (known as gingipains [10]) are multidomain proteins that contain a cysteine protease domain and additional C-terminal domains HA1 to HA4, which comprise the so called hemagglutinin domain (45). One highly conserved 15-kDa domain has been identified as an Hb receptor (referred to here as HA2) with a Kd(Hb) of ∼10−9 M and is part of three HA2-containing proteins (HA2-proteins): gingipains Kgp and RgpA and the putative hemagglutinin protein HagA (15, 34). The gingipain Kgp is an Hb protease (29), and the HA2 domain of Kgp functions to bind the substrate (15, 34). The HA2 domain is also reported to bind Hm (with an apparent Kd[Hm] of ∼10−8 M), and it has been suggested that it functions as a hemophore to capture heme (15, 36). The genes encoding these proteins, kgp, rgpA, and hagA, contribute to cell surface HA2 expression and high-affinity Hb binding (44). As PPIX competes with both Hm and Hb for HA2 binding, heme recognition by HA2 may be solely porphyrin mediated (15). Recognition of the porphyrin moiety was apparently independent of the substituents on the vinyl face of the macrocycle, i.e., the face which is not exposed at the protein surface in Hb (15).
An experimental approach used here confirms that HA2-proteins in cell surface extracts predominate in Hb binding and, by inference, in heme binding. The functions of these proteins are essential for the survival of P. gingivalis in the host organism (44). Following confirmation that recognition of heme is porphyrin mediated, the structural requirements for HA2 recognition of the porphyrin macrocycle are evaluated in vitro by using protoporphyrin and deuteroporphyrin derivatives. A wide range of structural derivatives and isomers of porphyrin were recognized by HA2. To further investigate the function of HA2 in cell surface porphyrin capture, this study reports the growth activities of P. gingivalis in media supplemented by these porphyrins. In all cases, recognition by HA2 is a requisite for growth rescue in heme-depleted cultures. While many of the porphyrins supported normal growth, variations in growth recovery rates mediated by some porphyrins are postulated to result from defects in transport and/or intracellular metabolism.
MATERIALS AND METHODS
Materials.
The translated product of a synthetic HA2 gene, derived from the sequence of RgpA, was expressed in E. coli with a C-terminal polyhistidine tag (D. B. Langley, C. A. Collyer, and N. Hunter, 2002, international patent application WO 2002061091), was functionally characterized by Hb-agarose affinity chromatography as previously described (15) and is referred to as rHA2. When analyzed under reducing conditions by SDS-PAGE, rHA2 runs anomalously (34) as a single band (estimated >98% purity) at 19 kDa (Langley et al., international patent application WO 2002061091). Nonreducing SDS-PAGE reveals a 31-kDa species confirmed by electrospray mass spectrometry to be a cystine-linked homodimer with f-methionine absent at 31,456.6 ± 1.7 Da (Langley et al., international patent application WO 2002061091). Antigingipain monoclonal antibody (MAb) 5A1 was prepared in mice and characterized as previously described (14). Murine MAb 5A1 recognizes a polypeptide epitope in HA2 (with the sequence ALNPDNYLISKDVTG) for which the only known homologues are in P. gingivalis and which are found in other domains of HA2-proteins, in HA1 domains, and within the C- and N-terminal domains of HagA (15). RgpA was purified from P. gingivalis strain ATCC 33277 as previously described (57). l-1-Chloro-3-[4-tosyl-amido]-7-amino-2-heptanone (TLCK) was supplied by Roche Diagnostics. Hm-agarose beads, Hb-agarose beads, bovine Hb, bovine Hm chloride, and PPIX were supplied by Sigma-Aldrich Company. In this study the choice of porphyrins was limited to those being stable and soluble (or sparingly soluble) in water. Deuteroporphyrin IX 2,4-bis-ethylene glycol (DBEG), deuteroporphyrin IX dihydrochloride (DPIX), deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride (DSA), deuteroporphyrin IX 2,4-disulfonic acid α,α′-dimethyl ester (DSAdiMe), deuteroporphyrin IX 2,4-bis-ethylene glycol ethylene diamine diamide (DBEG-EDD), deuteroporphyrin IX di-taurine (DPdiTA), biliverdin, and bilirubin di-taurate were supplied by Frontier Scientific Corp., and stock solutions of these porphyrins were prepared in 0.01 M NaOH.
The free acids of protoporphyrin isomers IV, VI, IX, XIII, XIV, and XII were prepared from the dimethyl ester analogues by following the method of Smith and coworkers (50). The general method is as follows. The dimethyl ester of the protoporphyrin isomer (∼50 mg) was dissolved in 25 ml of a solution of potassium hydroxide (1.0 g) in methanol (95 ml) and water (5 ml). The mixture was heated at reflux under nitrogen in the dark, and its progress was monitored by thin-layer chromatography. The solution was diluted with ethyl acetate (100 ml) and washed with hydrochloric acid (0.2 M, 50 ml) and water (50 ml). The organic phase was dried over anhydrous sodium sulfate and filtered, and the solvent was removed to yield the free acid (typically 80 to 85% yield). Gallium(III) deuteroporphyrin IX methoxide (Ga-DPIX) was also prepared from the dimethyl ester (9). Proton nuclear magnetic resonance and mass spectroscopy (not shown) confirmed the structure and purity of each porphyrin.
Competition assays.
Hb was used to coat plastic well plates in 0.1 M bicarbonate buffer (pH 9.0). Dilutions of rHA2 in 50 mM acetate buffer containing 137 mM NaCl, 0.1% Tween 20, and 10 mM NaN3 (pH 5.5) (acetate-Tween) were incubated overnight before plates were washed in phosphate-buffered saline (PBS; pH 7.4) with 0.1% Tween 20 (PBS-Tween). The primary MAb (5A1) was applied in PBS-Tween at a concentration of 0.5 μg/ml for 1.0 h at 37°C. Secondary goat anti-mouse antibodies conjugated with alkaline phosphatase (AP) (Dako Corp.) were applied at 1.1 μg/ml for 1.0 h at 37°C, and AP activity was monitored at 414 nm (A414) by hydrolysis of 4-nitrophenylphosphate (Boehringer) in 5.0 mM Tris (pH 9.5) by using a Titertek Twinreader PLUS photometer (absorbance maximum of 3.0 enzyme-linked immunosorbent assay [ELISA] units). rHA2 at concentrations which produced 50% saturation binding to an Hb-coated plate was preincubated for 1.0 h in acetate-Tween with dilutions of the porphyrins or Hb and then allowed to bind to Hb-coated plates overnight. The 50% inhibitory concentrations (IC50s) for competitive binding in solution phase assays were determined by a 6-h preincubation of rHA2 with titrations of the porphyrin followed by incubation with Hb-coated plates as previously described (15).
Growth curve assays.
P. gingivalis ATCC 33277 was used in all growth assays. Cultures were anaerobically grown at 37°C in a 5% CO2-10% H2-85% N2 atmosphere. Optical density was determined at 600 nm (A600). Growth rates of P. gingivalis were determined by addition of a 1% (vol/vol) inoculum of a late-logarithmic-phase culture (4 to 5 days; A600 = 1.0) to anaerobically equilibrated modified CDC broth (10 g of Trypticase peptone, 10 g of Trypticase soy broth, 5 g of NaCl, 10 g of yeast extract, 0.4 g of l-cysteine, 5 mg of Hm, and 2 mg of menadione per liter and 2% [vol/vol] horse serum). Culture purity was assessed by Gram staining and anaerobic subculture on modified CDC blood agar plates. A 1% inoculum was transferred to fresh CDC modified broth replete with Fe (estimated [18] concentration, ∼10 μM) but without (i) Hm and horse serum or (ii) Hm, horse serum, and menadione. Endogenous stores of heme in P. gingivalis whole cells were exhausted by serial passage of the culture at least five times into heme-depleted medium after each culture achieved stationary phase. Porphyrins to be tested were then added to the heme-depleted media with or without menadione, and the growth was monitored by measuring A600. Porphyrins were added to the Hm-depleted CDC medium at 10 μM; the control medium included Hm at the same concentration.
Extraction and biotinylation of surface proteins from P. gingivalis.
To prepare biotinylated CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}-extracted (BCE) surface proteins, P. gingivalis was grown in modified CDC broth under anaerobic conditions for 48 h. The cell pellet was washed three times in ice-cold PBS (pH 7.4) and resuspended at a concentration of approximately 25 × 106 cells/ml in sodium bicarbonate buffer (64 mM, pH 9.6). Sulfosuccinimidyl-6-(biotinamido)hexanoate-biotin was added to 0.5 mg/ml. The mixture was stirred gently at 23°C for 30 min and washed three times in ice-cold PBS (pH 7.4). The cell pellet was resuspended in 10 ml of Tris buffer with 0.25% (vol/vol) CHAPS and 2.0 mM TLCK and rotated gently overnight. The suspension was centrifuged at 3,000 × g for 15 min, and the protein concentration in the supernatant was calculated to be 2.0 mg/ml by using Coomassie Plus protein assay reagent.
Identity of Hb- and Hm-binding proteins in cell surface extract.
Hb-agarose and Hm-agarose in aliquots of 750 μl were washed twice batch-wise with 7.5 ml of PBS, pH 7.4, and twice with 7.5 ml of PBS-0.5 M NaCl and reequilibrated three times with PBS. All steps were done at 4°C unless otherwise stated. A total of 1.5 mg of BCE surface proteins in 2 ml was incubated with equilibrated beads overnight with gentle mixing. Beads were then washed five times with 10 bead volumes of PBS-0.5 M NaCl and subsequently stripped of bound proteins with 200 μl of 50 mM Tris-0.5 M NaCl-1% SDS, pH 8.0, at 37°C for 15 min. Supernatants were collected for the blotting procedure.
SDS-PAGE under reducing conditions was carried out with an optimized quantity of supernatant by using a 14% acrylamide resolving gel. Proteins were either stained with Coomassie blue or electrophoretically transferred onto a 0.2-μm nitrocellulose membrane (Bio-Rad Corp.). Membranes were blocked with 2% bovine serum albumin in PBS-10 mM NaN3 (PBS-N3) overnight at room temperature. A multichannel blotting apparatus (Milliblot-MP; Millipore Corp.) was used to probe the membrane with either AP-conjugated streptavidin or 5A1 and AP-conjugated anti-mouse immunoglobulin (Ig). Bound conjugates were detected with the AP substrate kit (Bio-Rad Corp.).
Conditions for neutralization of Hb binding.
Hb in PBS-N3 was used to coat the surfaces of the wells. Dilutions of BCE surface proteins made in PBS-Tween were incubated for 4 h at 23°C on Hb-coated plates before plates were washed in PBS-Tween. Streptavidin-AP (Strep-AP) was applied in PBS-Tween at a concentration of 0.5 μg/ml for 1 h at 37°C, and AP activity was monitored by measuring A414. The Hb binding assays were repeated with aliquots of BCE surface proteins fixed at the observed midpoint concentration but coincubated with increasing concentrations of rHA2.
It was decided to use sera with a strong neutralizing capacity for Hb binding (from a patient with chronic periodontitis) and sera with weak neutralizing capacity as a control. Venous blood was collected and frozen in aliquots at −70°C. Once the blood was thawed, NaN3 was added to a final concentration of 10 mM and the samples were kept at 4°C. The IgG fraction was isolated by protein G affinity chromatography. Protein G columns (Pharmacia Biotech) were equilibrated with 50 mM Tris-25 mM NaCl-1 mM CaCl2-10 mM NaN3 (pH 7.4) and then loaded with a 1/10 dilution of sera in the same buffer. Columns were washed with 8 column volumes of equilibration buffer, and then bound IgG was eluted with 0.1 M glycine (pH 2.7). IgG fractions were adjusted to pH 8.4 with 1/20 volume of 2.0 M Tris buffer (pH 8.4). IgG concentrations were determined by the Bradford assay.
Patient sera for the assay were selected by the relative capacity to neutralize Hb binding. Of the 38 purified IgG fractions tested, the patients with the strongest (IgG 125) and the weakest (IgG 65) neutralizing capacity were selected. By using the standard ligand binding assay described herein, BCE surface proteins at a concentration which produced 50% saturation binding to an Hb-coated plate were preincubated for 1 h in PBS-Tween with dilutions of patient IgG 125 and the control patient IgG 65 and then allowed to bind to Hb-coated plates overnight.
Inhibition of neutralization assay.
rHA2 was dialyzed overnight against 0.2 M NaHCO3 at 0.5 M NaCl (pH 9.0). N-Hydroxysuccinimide activated Sepharose 4 fast-flow beads (Pharmacia Biotech) were washed six times with ice-cold 1 mM HCl. The resin was pelleted, the rHA2 was added, and the suspension was rotated gently at 23°C overnight. The beads were washed five times with buffer to remove unbound protein. The resin was resuspended in 50 mM Tris (pH 8)- 10 mM NaN3 and rotated overnight to block unbound sites. The rHA2-bound resin was pelleted and resuspended in PBS-N3 at 700 μg of peptide/ml.
Increasing amounts of rHA2-bound resin suspension were preincubated with purified IgG fractions and rotated gently at 23°C overnight. As a resin control, beads (prepared using the same method) were preincubated with the IgG samples. The resin was pelleted, and the supernatant was added to a preblocked Hb-coated plate. BCE surface proteins at a concentration which produced 50% saturation binding to an Hb-coated plate were added, and the mixture was incubated at 23°C for 4 h and then analyzed with Strep-AP. The midpoint of the linear portion of the standard neutralization curve was calculated to the nearest twofold dilution. The midpoint was compared with the percentage of total binding observed at this dilution on the inhibition curve. The reported percentage range was representative of six separate experiments.
RESULTS
Profile of Hb- and Hm-binding proteins in detergent extracts of P. gingivalis.
To obtain qualitative information on the contribution of HA2 to the binding of Hb and Hm, surface proteins of P. gingivalis were biotinylated and extracted as BCE surface proteins. Figure 1a illustrates a typical BCE surface protein profile on SDS-PAGE gel. Proteins remaining bound to either Hb-agarose (Fig. 1b) or Hm-agarose (Fig. 1c) following a high-salt wash were stripped from beads with SDS and probed with either streptavidin to detect biotin or MAb 5A1 to detect HA2-proteins. The complexity of these profiles is compatible with autocatalytic action by the gingipains at the OM site to yield complex patterns of polypeptides derived from HA2-proteins (13). A typical MAb 5A1-reactive profile of purified HA2-protein RgpA is shown in Fig. 1d. In both affinity and detection systems there is some correspondence of the banding patterns, including common prominent peptides at positions a, c, and d; one notable exception is a 50-kDa species (position b) detected only by streptavidin. Okamoto et al. (35) suggested that the 50-kDa catalytic domain of Kgp has Hb-binding activity although this has yet to be substantiated. Perhaps the 50-kDa species was noncovalently associated with hemagglutinin domains (including HA2) and therefore copurified. The 75-kDa HmuR receptor reported by Olczak et al. (36) was not detected as a prominent band in either affinity system when probed with streptavidin. With HA2-proteins observed at positions g and h, the detection by streptavidin of other putative Hm-binding proteins at ∼30 kDa (6, 12, 26, 52) is equivocal. Detection of low-molecular-weight bands by 5A1 without a corresponding reaction with streptavidin could relate to limited access for biotinylation of OM proteins.
FIG. 1.
HA2-containing proteins extracted from the cell surface bind to Hb- and Hm-linked agarose. This series of gels demonstrates that a majority of the BCE surface proteins that bind to Hb- or Hm-linked agarose beads detected on Western blot by probing with streptavidin are also immunoreactive with 5A1. Estimates of the molecular masses of binding proteins are at the right. (a) BCE surface protein profile on SDS-PAGE gel under reducing conditions stained with Coomassie blue; (b) BCE surface proteins stripped from Hb beads with streptavidin detection of biotin (left lane) and 5A1 detection of HA2-containg proteins (right lane); (c) BCE surface proteins stripped from Hm beads with streptavidin detection of biotin (left lane) and 5A1 detection of HA2-containing proteins (right lane); (d) profile of purified RgpA probed with 5A1.
Neutralization of Hb binding.
The quantitative contribution of HA2 as a porphyrin-mediated Hb binding peptide in P. gingivalis was investigated by a competition assay and by using neutralizing antibodies. Coincubation of increasing concentrations of rHA2 with 0.5 μg of BCE surface proteins/ml (with concentration estimated from Fig. 2a) produced a progressive reduction in the binding of BCE surface proteins to Hb (data not shown). This competition assay provided no evidence for additional high-affinity receptors for Hb, nor did it assist in measuring the BCE surface protein-Hb interaction. In the second approach, we used low-titer neutralizing antibodies in sera from patients with chronic periodontitis because MAb 5A1 did not neutralize Hb binding by rHA2 and no neutralizing antibodies could be raised by immunization of New Zealand White rabbits with rHA2 (N. Hunter, unpublished data). The strategy exploited the unique sequence of HA2, which has homology only with other domains of HA2-proteins (as described in Materials and Methods). Accordingly, it was possible to selectively deplete patient sera of antibodies that recognize rHA2 (see Materials and Methods).
FIG. 2.
The importance of HA2 as a Hb-binding domain. (a) Standard curve for the binding of BCE from P. gingivalis to Hb. Dilutions of BCE were incubated on Hb-coated plates. Strep-AP was applied, and then AP activity was monitored by measuring A414. The 50% saturation point of the curve for the binding of BCE surface proteins to Hb was calculated and used as a fixed reference point for the preincubation mixture in the neutralization assay of Hb binding. (b) Curve for the binding of BCE surface proteins to Hb after neutralization of Hb binding. The patients with the strongest (IgG 125) and the weakest (IgG 65) neutralizing capacity were selected. In the standard ligand binding assay, BCE surface proteins at 0.5 μg/ml were preincubated with dilutions of patient IgG 125 and the control patient IgG 65 and then allowed to bind to Hb-coated plates overnight. 125/CE and 65/CE, positive controls where IgG 125 and 65 were used to neutralize Hb binding of the BCE surface proteins; 125/HA2 and 65/HA2, same assay after rHA2-bound resin had been used to deplete rHA2-related IgG from IgG 125 and 65, resulting in a shift in the profile of the curves. These curves are representative of six experiments.
A standard curve of the binding of BCE surface proteins to Hb is shown in Fig. 2a. This curve established that surface proteins from P. gingivalis bind Hb. The 50% saturation point of the curve for the binding of BCE surface proteins to Hb was calculated and used as a fixed reference point for the preincubation mixture in the neutralization assay of Hb binding.
The ability of human IgG fraction 125 to neutralize Hb binding is shown in Fig. 2b. In contrast, the control IgG 65 was only weakly neutralizing. This demonstrated the ability of human IgG 125 to eliminate the binding of BCE surface proteins to Hb. However, since the human IgG preparation was not specific to HA2, a further step was required to relate this neutralizing capacity specifically to HA2.
To demonstrate the importance of HA2 in Hb binding, the rHA2-bound resin was used to remove HA2-related IgG from IgG 125, resulting in a shift in the profile of the competition curve (Fig. 2b). At 10 μg of IgG fraction 125/ml there was a 50% inhibition of binding of BCE surface proteins to Hb. This was reduced to 20% inhibition at this concentration of IgG following preabsorption of the IgG fraction with immobilized rHA2. At higher concentrations of IgG fractions, nonspecific inhibition was observed. This study did not exclude a contribution by other polypeptides derived from HA2-proteins. For example the HA1 domain of RgpA is also recognized by MAb 5A1 (15). However, because MAb 5A1 does not block Hb binding (N. Hunter, unpublished data), there is no evidence to indicate that the epitope shared by HA2 and HA1 is involved in Hb or Hm binding. It has been proposed HA1 functions for hemagglutination (11). From these assays it is concluded that HA2-proteins are major contributors to cell surface Hb binding.
Recognition by HA2 for, and growth response to, noniron porphyrins.
When starved of porphyrin, P. gingivalis cells are unable to proliferate even under iron-replete culture conditions and the cells die. However, if treated soon after stationary phase, heme-starved cultures recover growth upon porphyrin supplementation. Cells grown for 48 h were transferred at a 1% (vol/vol) inoculum into heme-depleted media to exhaust internal stores of heme until growth stopped. The growth recovery of cultures was recorded after the addition of test porphyrins to the culture of growth-arrested cells (Fig. 3a). Biliverdin and bilirubin di-taurate are porphyrin oxidation products in which the tetrapyrrole macrocycle is not preserved, and both failed to stimulate growth recovery in this assay (Fig. 3b). The metal porphyrins Ga-DPIX and Hm (iron-PPIX) produced superimposable recovery profiles similar in lag phase and growth kinetics to the those produced by the nonmetal porphyrins PPIX and DPIX. These data indicate that cells capture and respond to porphyrins in a manner independent of their metal content.
FIG. 3.
Investigation of the role of iron and the tetrapyrrole macrocycle of porphyrin and porphyrin derivatives. HA2 recognition in vitro can be correlated with the capability to stimulate growth recovery in culture. (a) Selected porphyrin and porphyrin derivative structures. (b) Capacity of divergent porphyrins to support growth of P. gingivalis in iron-replete conditions. Cultures were depleted of heme stores by passage in Hm-free media (see Materials and Methods). Test porphyrins were added at 10 μM, and growth was determined under anaerobic conditions following a 1% inoculum of growth-arrested (heme-depleted) cells. Data are representative of three independent experiments. OD600, optical density at 600 nm. (c) Competition of Hb, metal, and nonmetal porphyrins with Hb-coated plates for rHA2 as measured by solid-phase ELISA (see Materials and Methods). Data are representative for triplicate determinations of three or more experiments. Estimates for IC50s were derived from curve fitting (Prism, version 3.03; Graph Pad Corp.).
A number of OM cell surface proteins of P. gingivalis are known to bind Hm. Of this group of proteins HA2, being a Hb receptor, could also have a role in facilitating heme capture from Hb. As porphyrin-induced growth recovery was not dependent on metal content, porphyrin-HA2 interactions were investigated. Hm, PPIX, DPIX, and Ga-DPIX were observed by ELISA to competitively inhibit rHA2 binding to Hb-coated plates at similar concentrations (IC50s, ∼6 to 25 μM), while Hb competition was observed over a wider range of concentrations, 0.5 to 25 μM, and is biphasic (Fig. 3c). As the Kd for the Hb tetramer-to-αβ dimer dissociation is 1 to 3 μM (2), two different Hb oligomers are present and the wide Hb concentration range for the interaction with rHA2 observed here is consistent with distinct interactions of rHA2 with the two oligomers. Biliverdin and bilirubin di-taurate did not compete in this assay (data not shown), suggesting that the tetrapyrrole ring of the porphyrin is a requirement for HA2 recognition.
Normal growth recovery requires HA2 recognition.
A number of water-soluble porphyrins with the same propionic acid face of PPIX but with modifications of the vinyl face of the macrocycle (Fig. 4a) were tested for their growth rescue and HA2-binding activities. Deuteroporphyrins were used in this study wherever possible because they lack the labile vinyl groups and are more stable in solution than protoporphyrins. For DPIX in which there is only methyl group substitution on the vinyl aspect, substitution of sulfonic acid (DSA) or ethylene glycol (DBEG) groups resulted in a prolonged lag phase and reduced rate of growth but an equal final biomasses (Fig. 4b). Supplementation of porphyrin-starved cultures with vinyl face derivatives stimulated growth recovery in the following order: Hm = PPIX = DPIX > DSA > DBEG. Porphyrins with additional derivatization of the propionic acid side chains DSAdiME and DBEG-EDD could not support growth. DPdiTA, in which the carboxylic acid groups of the propionate chains are derivatized by the amino acid taurine, supported only weak late growth. The lesser growth response to these three deuteroporphyrins could be a consequence of malfunction in capture, transport, and/or utilization. These deuteroporphyrins and the porphyrin oxidation products biliverdin and bilirubin di-taurate did not inhibit the stimulatory growth activities of DPIX (data not shown).
FIG. 4.
Solid-phase competition and growth recovery experiments showing the effects of side chain substitutions of porphyrins. Conditions were as for Fig. 2, with DPIX data reproduced for comparison. (a) Selected porphyrin structures. (b) Capacity of modified porphyrins to support growth of P. gingivalis in iron-replete conditions. Data are representative of three independent experiments. OD600, optical density at 600 nm. (c) Competition of modified porphyrins with Hb for rHA2 as measured by solid-phase ELISA (see Materials and Methods). Shown are representative data for triplicate determinations from three or more experiments. Estimates for IC50s were derived from curve fitting.
Recognition of the porphyrin derivatives by rHA2 was also investigated. DSAdiMe and DPdiTA did not compete with Hb in the competitive binding assay (data not shown). However, some deuteroporphyrins with free carboxylate side chains on the propionic acid face, such as DPIX, DSA, and DBEG, were competitive with Hb for binding to rHA2 (IC50s, 7 to 20 μM; Fig. 4c). A related compound, DBEG-EDD, with modified carboxylate side chains, unexpectedly competed effectively for rHA2 in the same concentration range but unlike DBEG did not support growth recovery.
Metabolism of protoporphyrin isomers with and without menadione supplementation.
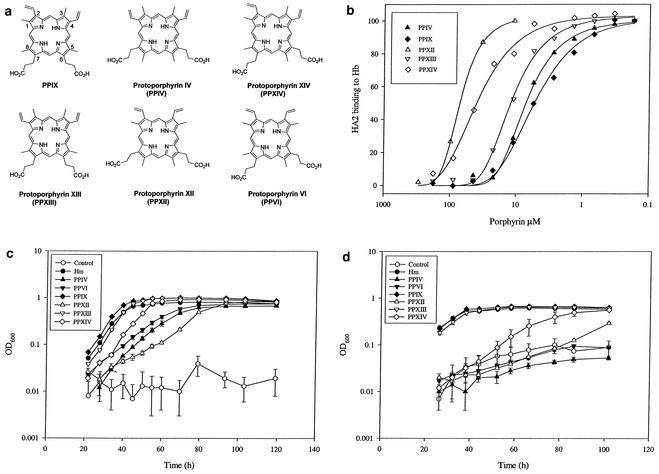
To address more precisely the effect of the disposition of the propionate side chains and vinyl groups of protoporphyrins on HA2 binding and growth-promoting activity, a series of protoporphyrin isomers was prepared (as described in Materials and Methods). The structures of the isomer pairs are shown in Fig. 5a. As the porphyrin ring is planar and the inner hydrogens on nitrogen are labile, there is pseudosymmetry in their structures (Fig. 5a, vertical twofold axes in the plane of the porphyrin rings) and each structure could be drawn flipped over.
FIG. 5.
Comparing the activities of protoporphyrin isomers. Conditions were as for Fig. 3, with Hm and PPIX data reproduced for comparison. (a) Selected porphyrin isomer structures. (b) Competition of modified porphyrins with Hb for rHA2 as measured by solid-phase ELISA (see Materials and Methods). Shown are representative data for triplicate determinations of three or more experiments. Estimates for IC50s were derived from curve fitting. (c) Capacity of modified porphyrins to support growth of P. gingivalis in iron-replete conditions. Data are representative of three independent experiments. OD600, optical density at 600 nm. (d) Capacity of protoporphyrin isomers to support growth of P. gingivalis following growth arrest by combined heme and menadione depletion. Data are representative of three independent experiments.
The competition with Hb binding determined by the solid-phase assay demonstrated that the order of strength of the interaction with HA2 is as follows: PPIX = PPIV = PPXIII (IC50 = 6 to 15 μM) > PPXIV (IC50 = 40 to 50 μM) > PPXII (estimated range for IC50, 60 to 90 μM; Fig. 5b) > PPVI (weak competition was observed [data not shown]; estimated IC50, >500 μM). Analysis of the isomer pairs provided direct evidence for the impact of substitution on the vinyl face. Thus comparison of PPIV and PPXII indicates that, while displacement of the propionate side chains from C6 and C7 to C5 and C8 is tolerated by HA2, vinyl groups at both C2 and C3 are unfavorable (Fig. 5). This effect is further demonstrated by comparison of the PPXIV-PPVI pair, where vinyl substitution at C4 on PPXIV would be favorable.
The contribution of the location of vinyl and propionate groups of the protoporphyrin isomers to the capacity to support the growth of P. gingivalis was evaluated (Fig. 5a). Each of these porphyrin isomers was able to support growth in the presence of menadione. Quinones are thought to be required as electron transport carriers for the later steps of anaerobic heme biosynthesis, and the supplementation of menadione may assist P. gingivalis in the metabolism and utilization of these protoporphyrin isomers (24). Alternative placement of the vinyl groups at positions 2 and 4 (PPIX) versus 1 and 4 (PPXIII) was well accepted by P. gingivalis (Fig. 5c). Displacement of the propionic acid side chains of PPXIII to the 5 and 8 positions to form PPIV was, however, poorly tolerated, as indicated by the extended lag phase, slower rate of growth, and reduced final biomass (Fig. 5c). Displacement of the propionic acid group at position 6 of PPIX to 5 (PPXIV) was well tolerated. Symmetrical (PPXII; Fig. 5a) or asymmetrical (PPVI; Fig. 5a) displacement of the propionic acid groups when combined with vinyl substitution at position 3 (both isomers) was unfavorable for growth support (Fig. 5c). Growth rates induced by most of these porphyrin isomers correlated with the order of the strength of the interaction with rHA2 although PPIV was an exception as it competes with Hb equally to PPIX, PPXIII, and PPXIV but induces a slower growth recovery (Fig. 5c). A closer correlation of the recovery rates with the competition order was observed when heme-depleted cultures were transferred into medium without either Hm or menadione (Fig. 5d). When cells were cultured in growth recovery media without menadione supplementation, the final biomass was reduced (Fig. 5d). Under this condition, PPIX, PPXIII, and Hm all produced superimposable growth profiles. Growth was markedly delayed in the presence of PPXIV although a similar final biomass was achieved. There was, however, minimal (PPXII) or no capacity (PPIV and PPVI) to rescue growth for the other protoporphyrin isomers. PPIV was, again, the exception, indicating that growth recovery is also dependent upon the ability of the bacteria to utilize porphyrin after its cell surface capture and transport.
DISCUSSION
The mechanism of porphyrin capture by P. gingivalis should be closely coupled to heme and Hb recognition as these are the natural sources of this essential cofactor. However, for growth heme can be replaced by PPIX, implying that at least one porphyrin acquisition pathway can function in the absence of iron in the growth factor. The HA2 domain has high affinity for both Hb and Hm. The efficiency with which such binding can be contested with a variety of nonmetal porphyrins lends weight to the postulate that this protein is intimately associated with porphyrin uptake, a process vital to the survival of the organism.
Partial neutralization of P. gingivalis Hb binding by HA2-related IgG implicates HA2-proteins as major contributors in cell surface Hb and Hm binding. This is supported by the following observations. First, spontaneous pigment-less P. gingivalis mutants are generated when the portion of the kgp gene which encodes HA2 is deleted by recombination (7). Second, Hm and Hb binding of P. gingivalis cells is decreased by 50% in Kgp truncation mutants which lack the HA2 domain (A. Sroka, M. Sztukowska, M. Bugno, J. Potempa, J. Travis, and C. Genco, Abstr. 80th Gen. Session Int. Assoc. Dent. Res., abstr. 1042, 2002). Third, a P. gingivalis kgp rgpA hagA triple deletion mutant exhibits no detectable Hb binding in a solid-phase assay (44). Last, the bacteriostatic action of lactoferrin on P. gingivalis is associated with the removal of HA2-proteins from the cell surface (43).
Structural studies of proteins that function in heme capture indicate that heme recognition is usually iron mediated, and in general these proteins are not known to capture PPIX (55). For example, the host defense protein hemopexin acquires extra cellular heme by using two histidine ligands covalently bound to the iron (38). The secreted bacterial hemophore HasA uses a histidine and tyrosine ligand pair (1). Mutagenesis studies of TonB-dependent OM receptors indicate that conserved histidine residues play a critical role in heme recognition (5). Of most relevance here, the P. gingivalis HmuR protein binds PPIX poorly (Kd > 10−5 M), implying that the active site may also contain ligands that recognize the metal ion in the porphyrin ring (36). The generality of this heme recognition mechanism in bacteria is further confirmed in a number of organisms, showing that PPIX does not inhibit heme capture and transport (28). A notable exception is the blocking of high-affinity Hm binding to P. gingivalis cells by competition with PPIX (54), indicating that in this case an alternative to the general mechanism is operative and a significant fraction of heme acquisition must be mediated by a process which can capture nonmetal porphyrins.
The recovery of growth of heme-starved P. gingivalis cells has been used here as an indicator of the ability to utilize various noniron porphyrins. Under iron-replete conditions, porphyrin-induced cell growth recovery from heme starvation appears to be independent of the metal content of the added porphyrin. However, the chemical structures of the side groups attached to the tetrapyrrole ring are critically important. Porphyrins which do not compete in vitro with Hb for rHA2, such as DPdiTA and DSAdiME, contain covalently modified propionic acid groups. As these porphyrins also fail to elicit normal growth recovery, it is likely that both the integrity of the macrocycle and the ability of its propionic acid face to be recognized by HA2 are critical factors in determining porphyrin capture and utilization by P. gingivalis. This is, however, not demonstrated by the observed competition of DBEG-EDD for Hb binding to rHA2. Perhaps this deuteroporphyrin, with ethylene diamide additions to both the carboxylic acid groups, competes with Hb for recognition by HA2 by a mechanism different from that of other porphyrins. Alternatively, such a modification might simply be tolerated in the HA2-porphyrin interaction. Regardless, the growth responses to DBEG-EDD, DPdiTA, and DSAdiME indicate that the organism cannot utilize these deuteroporphyrins.
While HA2 is thought to act only in porphyrin capture, the growth response depends on cell surface acquisition, transport, and utilization. Hence, a discrepancy between the growth recovery profiles and affinities of various porphyrins for HA2 is not surprising. However, a number of porphyrins, including DPIX, PPXIII, and PPIX, which differ only in the vinyl aspect of the tetrapyrrole ring, are comparable in the two assays. Porphyrins with less closely related structures, such as DSA, DBEG, PPVI, and PPXII, are less efficient in growth recovery and weaker in competition with Hb for HA2. These effects are more evident for PPVI and PPXII in the absence of menadione supplementation, where growth recovery is not observed.
After porphyrins are captured by HA2-proteins at the cell surface, what pathways function to transport the growth factor into the cell? A number of P. gingivalis genes, including tlr (47), hemR (25), hmuR (46), and those of the ihtABCDE locus (12), have been implicated in heme transport. Four of these genes (ihtA, tlr, hemR, and hmuR) exhibit homology to genes encoding bacterial TonB-dependent receptors; their products are thought to act as OM receptors in iron and/or heme transport, but none have been reported as high-affinity receptors. HmuR acts as a low-affinity Hm/Hb receptor (36) and inactivation of hmuR decreased the ability of P. gingivalis to use Hm or Hb as its sole iron source (46). Inactivation of tlr resulted in mutant cells which were unable to grow on low concentrations of Hm (47). The roles of HemR and IhtA in heme or iron transport in P. gingivalis have not been defined. Perhaps one of these cell surface receptors is able to transport sufficient noniron porphyrin and thereby support cell growth?
Another possible route for porphyrin import involves novel OM protein IhtB, a putative PPIX ferrochelatase, which is proposed to act in reverse and remove iron from cell surface heme for iron transport (12). However, another study proposes that IhtB is an intracellular cobalt chelatase (40). Porphyrin capture and/or transport systems can also act as portals of entry of noniron metalloporphyrins such as Ga-PPIX, which can display potent bacteriostatic activity (53). Ga-porphyrins inhibit the growth of a number of microorganisms, particularly those that express active Hm transport systems, with the toxic mechanism thought to involve incorporation of a Ga cofactor into cytochromes. The affinity of Ga-DPIX for HA2 was comparable to those of other porphyrins, but, curiously, Ga-DPIX supported the growth of P. gingivalis. One explanation is that Ga is removed from the deuteroporphyrin by a ferrochelatase.
As the structure of this protein domain is not known, the exact molecular mechanism by which HA2 functions to capture heme is yet to be elucidated. Few other bacteria, with the exception of H. influenzae, appear to capture nonmetal porphyrins, and none are known to express hemophores related to HA2. Although physiologically this mechanism functions to recognize heme, the knowledge of its porphyrin-mediated nature is potentially useful for the design of specific agents against this pathogen.
Acknowledgments
Thanks go to P. S. Clezy and C. R. Fookes for donating the dimethyl esters of the protoporphyrin isomers and also to D. Harty and L. Hunter for assistance in preparation of figures.
This work was supported by Biochemical Veterinary Research Pty Ltd. and the National Health and Medical Research Council of Australia with postgraduate scholarships to M. Paramaesvaran and K.-A. Nguyen.
REFERENCES
- 1.Arnoux, P., R. Haser, N. Izadi, A. Lecroisey, M. Delepierre, C. Wandersman, and M. Czjzek. 1999. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol. 6:516-520. [DOI] [PubMed] [Google Scholar]
- 2.Atha, D. H., and A. Riggs. 1976. Tetramer-dimer dissociation in hemoglobin and the Bohr effect. J. Biol. Chem. 251:5537-5543. [PubMed] [Google Scholar]
- 3.Barua, P. K., D. W. Dyer, and M. E. Neiders. 1990. Effect of iron limitation on Bacteroides gingivalis. Oral Microbiol. Immunol. 5:263-268. [DOI] [PubMed] [Google Scholar]
- 4.Biberstein, E. L., P. D. Mini, and M. G. Gills. 1963. Action of Haemophilus cultures on δ-aminolevulinic acid. J. Bacteriol. 86:814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracken, C. S., M. T. Baer, A. Abdur-Rashid, W. Helms, and I. Stojiljkovic. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramanti, T. E., and S. C. Holt. 1993. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J. Bacteriol. 175:7413-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, W., and H. K. Kuramitsu. 1999. Molecular mechanism for the spontaneous generation of pigmentless Porphyromonas gingivalis mutants. Infect. Immun. 67:4926-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope, L. D., S. E. Thomas, J. L. Latimer, C. A. Slaughter, U. Muller-Eberhard, and E. J. Hansen. 1994. The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13:863-873. [DOI] [PubMed] [Google Scholar]
- 9.Coutsolelos, A., and R. Guilard. 1983. Synthese et characteristiques physiochimiques de gallioporphyrines a liason σ metal-carbone. J. Organomet. Chem. 253:273-282. [Google Scholar]
- 10.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 11.Curtis, M. A., M. Ramakrishnan, and J. M. Slaney. 1993. Characterization of the trypsin-like enzymes of Porphyromonas gingivalis W83 using a radiolabelled active-site-directed inhibitor. J. Gen. Microbiol. 139:949-955. [DOI] [PubMed] [Google Scholar]
- 12.Dashper, S. G., A. Hendtlass, N. Slakeski, C. Jackson, K. J. Cross, L. Brownfield, R. Hamilton, I. Barr, and E. C. Reynolds. 2000. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 182:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeCarlo, A. A., and G. J. Harber. 1997. Hemagglutinin activity and heterogeneity of related Porphyromonas gingivalis proteinases. Oral Microbiol. Immunol. 12:47-56. [DOI] [PubMed] [Google Scholar]
- 14.DeCarlo, A. A., Jr., L. J. Windsor, M. K. Bodden, G. J. Harber, B. Birkedal-Hansen, and H. Birkedal-Hansen. 1997. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J. Dent. Res. 76:1260-1270. [DOI] [PubMed] [Google Scholar]
- 15.DeCarlo, A. A., M. Paramaesvaran, P. L. Yun, C. Collyer, and N. Hunter. 1999. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J. Bacteriol. 181:3784-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, N. M., D. D. Smith, and A. J. Wicken. 1974. Haemin and nicotinamide adenine dinucleotide requirements of Haemophilus influenzae and Haemophilus parainfluenzae. J. Med. Microbiol. 7:359-365. [DOI] [PubMed] [Google Scholar]
- 17.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, and J. M. Merrick. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 18.Genco, C. A., B. M. Odusanya, and G. Brown. 1994. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect. Immun. 62:2885-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghigo, J. M., S. Letoffe, and C. Wandersman. 1997. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbons, R. J., and J. B. Macdonald. 1960. Hemin and vitamin K compounds as required factors for the cultivation of certain strains of Bacteroides melaninogenicus. J. Bacteriol. 80:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granick, S., and H. Gilder. 1946. The porphyrin requirements of Haemophilus influenzae and some functions of the vinyl and propionic acid side chains of heme. J. Gen. Physiol. 30:1-13. [PMC free article] [PubMed] [Google Scholar]
- 22.Grenier, D., V. Goulet, and D. Mayrand. 2001. The capacity of Porphyromonas gingivalis to multiply under iron-limiting conditions correlates with its pathogenicity in an animal model. J. Dent. Res. 80:1678-1682. [DOI] [PubMed] [Google Scholar]
- 23.Haffajee, A. D., and S. S. Socransky. 2000. Microbial etiological agents of destructive periodontal diseases. Periodontology 5:78-111. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, J. M., and N. J. Jacobs. 1977. The late steps of anaerobic heme biosynthesis in E. coli: role for quinones in protoporphyrinogen oxidation. Biochem. Biophys. Res. Commun. 78:429-433. [DOI] [PubMed] [Google Scholar]
- 25.Karunakaran, T., T. Madden, and H. Kuramitsu. 1997. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J. Bacteriol. 179:1898-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S.-J., L. Chu, and S. C. Holt. 1996. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb. Pathog. 21:65-70. [DOI] [PubMed] [Google Scholar]
- 27.Kusaba, A., T. Ansai, S. Akifusa, K. Nakahigashi, S. Taketani, H. Inokuchi, and T. Takehara. 2002. Cloning and expression of a Porphyromonas gingivalis gene for protoporphyrinogen oxidase by complementation of a hemG mutant of Escherichia coli. Oral Microbiol. Immunol. 17:290-295. [DOI] [PubMed] [Google Scholar]
- 28.Lee, B. C. 1995. Quelling the red menace: haem capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, L. A., M. H. Sung, M. Gipson, K. Hartman, and D. W. Dyer. 1998. Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J. Bacteriol. 180:6043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeb, M. R. 1995. Ferrochelatase activity and protoporphyrin IX utilization in Haemophilus influenzae. J. Bacteriol. 177:3613-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McConville, M. L., and H. P. Charles. 1979. Mutants of Escherichia coli K12 permeable to haemin. J. Gen. Microbiol. 113:165-168. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto, K., K. Nishimura, T. Masuda, H. Tsuji, and H. Inokuchi. 1992. Accumulation of protoporphyrin IX in light-sensitive mutants of Escherichia coli. FEBS Lett. 310:246-248. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama, K., D. B. Ratnayake, T. Tsukuba, K. Yamamoto, and S. Fujimura. 1998. Haemoglobin receptor is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutination-encoding gene of Porphyromonas gingivalis. Mol. Microbiol. 27:51-61. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 36.Olczak, T., D. W. Dixon, and C. A. Genco. 2001. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol. 183:5599-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto, B. R., S. J. van Dooren, J. H. Nuijens, J. Luirink, and B. Oudega. 1998. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 188:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoli, M., B. F. Anderson, H. M. Baker, W. T. Morgan, A. Smith, and E. N. Baker. 1999. Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two β-propeller domains. Nat. Struct. Biol. 6:926-931. [DOI] [PubMed] [Google Scholar]
- 39.Reidl, J., and J. J. Mekalanos. 1996. Lipoprotein e (P4) is essential for hemin uptake by Haemophilus influenzae. J. Exp. Med. 183:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roper, J. M., E. Raux, A. A. Brindley, H. L. Schubert, S. E. Gharbia, H. N. Shah, and M. J. Warren. 2000. The enigma of cobalamin (vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J. Biol. Chem. 275:40316-40323. [DOI] [PubMed] [Google Scholar]
- 41.Schifferle, R. E., S. A. Shostad, M. T. Bayres-Thering, and M. E. Neiders. 1996. Effect of protoporphyrin IX limitation on Porphyromonas gingivalis. J. Endod. 22:352-355. [DOI] [PubMed] [Google Scholar]
- 42.Schlör, S., M. Herbert, M. Rodenburg, J. Blass, and J. Reidl. 2000. Characterization of ferrochelatase (hemH) mutations in Haemophilus influenzae. Infect. Immun. 68:3007-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, Y., W. Kong, and K. Nakayama. 2000. Human lactoferrin binds and removes the hemoglobin receptor protein of the periodontopathogen Porphyromonas gingivalis. J. Biol. Chem. 275:30002-30008. [DOI] [PubMed] [Google Scholar]
- 44.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 45.Shibata, Y., M. Hayakawa, H. Takiguchi, T. Shiroza, and Y. Abiko. 1999. Determination and characterization of the hemagglutinin-associated short motifs found in Porphyromonas gingivalis multiple gene products. J. Biol. Chem. 274:5012-5020. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, W., T. Olczak, and C. A. Genco. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182:5737-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slakeski, N., S. G. Dashper, P. Cook, C. Poon, C. Moore, and E. C. Reynolds. 2000. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol. Immunol. 15:388-392. [DOI] [PubMed] [Google Scholar]
- 48.Smalley, J. W., A. J. Birss, A. S. McKee, and P. D. Marsh. 1993. Haemin-binding proteins of Porphyromonas gingivalis W50 grown in a chemostat under haemin-limitation. J. Gen. Microbiol. 139:2145-2150. [DOI] [PubMed] [Google Scholar]
- 49.Smalley, J. W., A. J. Birss, and J. Silver. 2000. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the μ-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol. Lett. 183:159-164. [DOI] [PubMed] [Google Scholar]
- 50.Smith, K. M., E. M. Fujinari, K. C. Langry, D. W. Parish, and H. D. Tabba. 1983. Manipulation of vinyl groups in protoporphyrin IX: introduction of deuterium and carbon-13 labels for spectroscopic studies. J. Am. Chem. Soc. 105:6638-6646. [Google Scholar]
- 51.Sroka, A., M. Sztukowska, J. Potempa, J. Travis, and C. A. Genco. 2001. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J. Bacteriol. 183:5609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 53.Stojiljkovic, I., V. Kumar, and N. Srinivasan. 1999. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31:429-442. [DOI] [PubMed] [Google Scholar]
- 54.Tompkins, G. R., D. P. Wood, and K. R. Birchmeier. 1997. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J. Bacteriol. 179:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 56.Wyss, C. 1992. Growth of Porphyromonas gingivalis, Treponema denticola, T. pectinovorum, T. socranskii, and T. vincentii in a chemically defined medium. J. Clin. Microbiol. 30:2225-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun, P. L., A. A. DeCarlo, and N. Hunter. 1999. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect. Immun. 67:2986-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]