Abstract
Transferrin-binding protein B (TbpB) is one component of a bipartite receptor in several gram-negative bacterial species that binds host transferrin and mediates the uptake of iron for growth. Transferrin and TbpB are both bilobed proteins, and the interaction between these proteins seems to involve similar lobe-lobe interactions. Synthetic overlapping peptide libraries representing the N lobe of TbpB from Moraxella catarrhalis were prepared and probed with labeled human transferrin. Transferrin-binding peptides were localized to six different regions of the TbpB N lobe, and reciprocal experiments identified six different regions of the C lobe of transferrin that bound TbpB. Truncations of the N lobe of TbpB that sequentially removed each transferrin-binding determinant were used to probe an overlapping peptide library of the C lobe of human transferrin. The removal of each TbpB N-lobe transferrin-binding determinant resulted in a loss of reactivity with peptides from the synthetic peptide library representing the C lobe of transferrin. Thus, individual peptide-peptide interactions between ligand and receptor were identified. A structural model of human transferrin was used to map surface regions capable of binding to TbpB.
Iron is an essential element for many biological processes and is not readily available to pathogenic bacteria in the environment of the host (31). Pathogenic bacteria of the families Neisseriaceae and Pasteurellaceae acquire iron by a receptor-mediated interaction at the bacterial cell surface with the host iron-binding proteins, transferrin (Tf) and lactoferrin (Lf). Recent studies have demonstrated that these surface receptors are essential for survival in vivo in humans (11), indicating that these proteins are potential targets for the development of vaccines and therapeutic agents.
Tf and Lf proteins comprise a family of related, bilobed, monomeric glycoproteins that sequester iron to the extent that levels of free iron are insufficient to support growth. The main function of Tf is the transport of Fe3+ through the body to cells requiring iron. The iron is transported to the cytosol by receptor-mediated endocytosis (20). Lf is mainly found on mucosal surfaces, where it is thought to play a role in scavenging iron (1).
The two lobes of Tf and Lf are almost identical in overall structure. Each lobe in turn is made up of two equally sized domains that are connected by a central hinge region. A cleft located between the domains in each lobe serves as the binding site for the metal ion and a carbonate anion (5, 17). Each lobe of Tf is able to bind one molecule of Fe3+, with a binding constant of about 1020. Due to the high binding affinity and a normal state of 30% iron saturation, it is a very effective iron-scavenging protein in serum. The spare iron-binding capacity is crucial to sequester any free iron that may be released from lysed cells or freed as a result of infection (31).
With one exception (28), the Tf receptor in members of the families Neisseriaceae and Pasteurellaceae consists of two iron-repressible outer membrane proteins, Tf-binding proteins A and B (TbpA and TbpB). TbpA is an integral outer membrane protein that is thought to serve as a channel for the transport of iron across the outer membrane in a TonB-dependent fashion (15). TbpA is required for the utilization of iron from Tf, but either of the binding proteins (TbpA or TbpB) can bind iron-loaded Tf (4, 29). TbpA is relatively well conserved in amino acid sequence, at least within a bacterial species (9, 27).
TbpA is a homologue of the transmembrane siderophore receptors FhuA and FepA, for which the structures have been determined (6, 12, 24). The structures consist of 22 antiparallel β strands that form a barrel or pore through the outer membrane; the amino terminus of the protein forms a plug in the center of the barrel. These structures have been used as a basis for the development of a topology model for TbpA (26). A pentameric sequence called a TonB box is present in the plug region of FhuA, FepA, and TbpA (6, 12, 19). This sequence is thought to be the point of protein interaction for periplasmic protein TonB, which transduces energy from the inner to the outer membrane, thus allowing molecules like iron to move against their concentration gradient into the cell (19, 22).
TbpB is a peripheral outer membrane lipoprotein anchored to the membrane by N-terminally linked fatty acids (15) and is largely exposed to the extracellular environment. Alignments of the predicted amino acid sequences of TbpB proteins from different organisms show several regions of homology between the N- and C-terminal halves of the proteins (35). This homology is present in spite of the variable sequences and sizes of the TbpB proteins, with molecular masses ranging from about 65 kDa to more than 85 kDa (10, 21, 37). The homology between the protein halves suggests that TbpB may have a bilobed structure analogous to that of Tf, a notion which is supported by the ability to produce separate recombinant lobes capable of binding Tf (32, 35). Overlapping peptide libraries of the N lobe and the C lobe of human Tf (hTf) have shown that sequentially homologous peptides in each of the hTf libraries bind both the N- and the C-terminal lobes of TbpB proteins from the human pathogens Moraxella catarrhalis and Neisseria meningitidis, confirming that they are functional homologues (35).
The Tf-TbpB interaction involves two lobe-lobe interactions that appear to be similar (35). Although the precise nature of the lobe-lobe interactions in the receptor-ligand complex has not been experimentally established, the evidence suggests that the TbpB N lobe may bind to the hTf C lobe and, thus, the TbpB C lobe would be expected to bind to the hTf N lobe. A recombinant TbpB N lobe from the human pathogens M. catarrhalis and N. meningitidis preferentially binds to chimeric proteins containing the C lobe and not the N lobe of hTf (35). Unfortunately, the limited binding activity of the TbpB C-lobe preparations precluded determining which hTf lobe was preferentially bound. Since the C lobe of Tf is preferentially recognized by TbpA (2, 3, 42), it likely is proximal to the outer membrane and would be positioned appropriately to interact with the membrane-anchored N lobe of TbpB.
The similarities of the lobe-lobe interactions combined with the complexity of the overall interaction between the intact proteins potentially complicate studies aimed at further delineation of the interaction. Thus, we decided to focus on individual lobe-lobe interactions to simplify the analysis. In this study, the interaction between the TbpB N lobe from the human pathogen M. catarrhalis and the hTf C lobe is investigated.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Protein expression was carried out in Escherichia coli strain ER2507 (801-J) or ER2508 (801-K) from New England BioLabs. These strains contain a deletion in the malE gene and thus do not produce native maltose-binding protein (MBP). Strain ER2508 (801-K) also contains a Lon protease mutation and was used for expressing the truncations encoded by segment 1 (no binding regions) and by segments 1 and 2 (binding region 1 only), due to reduced degradation of the truncated proteins during storage. All cloning and expression steps were carried out with the pMal-c2 plasmid (New England BioLabs), which provides in-frame fusions with the malE gene. E. coli cells containing the plasmids were maintained on Luria-Bertani medium (Gibco BRL) containing 100 μg of ampicillin (Sigma) ml−1. The expression of MBP or the MBP fusion proteins was induced by the addition of 200 μM isopropyl-β-d-thiogalactoside (IPTG) to the medium and incubation for 3 h at 37°C before harvest.
Construction of overlapping peptide libraries.
Generation of the overlapping peptide libraries representing hTf or the TbpB N-lobe regions from different bacteria was carried out by the SPOTSCAN method (Genosys Biotechnologies) (13). Essentially, the library consisted of 15-amino-acid linear peptides that were immobilized on cellulose membranes by 9-fluorenylmethoxy carbonyl amino acid-based synthesis.
One membrane was used to generate a library of the C lobe of hTf consisting of 96 15-amino-acid peptides with an 11-amino-acid overlap. The overlapping hTf peptide represented amino acids 285 to 679 of the intact hTf protein (GenBank accession number NP_001054). The first peptide library representing the TbpB N lobe from M. catarrhalis 4223 (GenBank accession number AAC34277) consisted of 32 15-amino-acid peptides with a 4-amino-acid overlap and included N-lobe amino acids 12 to 421. The amino acid numbers refer to the amino acids in the predicted mature, processed protein (i.e., the N-terminal cysteine is amino acid 1). The second peptide library representing M. catarrhalis 4223 TbpB N-lobe peptides included amino acids 27 to 421 but with an 11-amino-acid overlap.
Production of proteins.
TbpB truncations were generated by PCR-based cloning into the pMal expression system (New England BioLabs) as described previously (33). Essentially, amplified tbpB gene segments were subcloned into the pMal-c2 expression vector to generate in-frame fusions with the malE gene. Intact M. catarrhalis 4223 TbpB consisted of amino acids 3 to 680, where 1 represents the N-terminal cysteine of the mature processed protein. Amino acids 1 and 2 from the mature protein were removed to ease cloning and subsequent expression. The N-terminal half of this protein consisted of amino acids 3 to 421. The truncated TbpB proteins were expressed as MBP fusions in E. coli ER2507 or ER2508 (for segment 1 or segments 1 and 2, respectively) and purified by amylose affinity chromatography.
Two different methods for labeling proteins for library screening were used. Iron-loaded hTf (Sigma) was labeled by chemical cross-linking (by using 3-maleimidobenzoic acid-N-hydroxysuccinimide ester) to a beta-galactosidase (β-Gal) reporter enzyme as described elsewhere (18). This construct was used as a probe for colorimetric detection on the composite peptide library. The second method involved conjugating the truncations of the N lobe of TbpB from M. catarrhalis 4223 to horseradish peroxidase (HRP). HRP-conjugated proteins can be detected by chemiluminescence, providing a more sensitive screen. The truncations were conjugated to HRP by using a Linx HRP rapid protein conjugation kit (Invitrogen). The labeled fusion proteins were then used as probes for chemiluminescence detection.
In addition, HRP-conjugated hTf (Jackson ImmunoResearch) was used to probe the M. catarrhalis TbpB N-lobe peptide library.
Screening of peptide libraries.
Binding experiments with hTf and the first peptide library were carried out by the colorimetric method (Genosys Biotechnologies). Essentially, hTf in blocking solution consisting of 10% blocking buffer (Genosys Biotechnologies), 50 mg of sucrose/ml, and T-TBS (0.137 M NaCl, 2.68 mM KCl, 50.4 mM Tris base, 0.05% Tween 20 [pH 8.0]) was incubated with the membrane containing the peptide library for 4 h at room temperature. The membrane was then washed sequentially in T-TBS and phosphate-buffered saline before detection of bound beta-galactosidase by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Positive binding was determined by the development of a blue spot.
The membrane was then regenerated by the following sequence of washes for 10 min each at room temperature on a rocking table: three changes of 20 ml of double-distilled H2O (ddH2O); three changes of 20 ml of N,N-dimethylformamide (Sigma); two changes of ddH2O; three changes of regeneration buffer A (8.0 M urea, 1% sodium dodecyl sulfate [SDS], 0.1% 2-mercaptoethanol); and three changes of regeneration buffer B (50% ethanol and 10% acetic acid in ddH2O). Finally, the membrane was washed with two changes of 20 ml of methanol, allowed to air dry, and stored at −20°C. The same membrane could be reused up to five times if the regeneration protocol was followed carefully.
For increased sensitivity, the TbpB N-lobe truncations from M. catarrhalis were conjugated to HRP for the production and detection of a chemiluminescent product. The membrane was rinsed with methanol and washed with three changes (5 min each) of TBS (0.137 M NaCl, 2.68 mM KCl, 50.4 mM Tris base [pH 8.0]) on a rocking platform. The membrane was blocked by rocking at room temperature in blocking solution consisting of 20 ml of 0.5% skim milk powder (Bio-Rad) in TBS for 2.5 h. The membrane was washed three times with T-TBS before being incubated with one of the HRP-conjugated truncations diluted 1:1,000 in skim milk blocking solution for 4 h at room temperature. The membrane was then washed again three times with T-TBS, drained, and incubated with 10 ml of Lumiglo mixture (Kirkegaard & Perry Laboratories) for 5 min. It was then exposed to X-Omat Blue XB-1 X-ray film (Kodak) for at least 30 s before development. Positive peptides showed up as dark spots on a light background.
The membrane was regenerated and stored for reuse as described above.
RESULTS
Identification of hTf-binding peptides in the TbpB N lobe of M. catarrhalis.
A prior study demonstrated that at least six regions on each hTf lobe bound to TbpB proteins from N. meningitidis and M. catarrhalis (34). This finding infers that each lobe of TbpB should possess six complementary binding regions. Thus, a strategy for demonstrating these binding determinants was adopted and included (i) identification of hTf-binding peptides by probing peptide libraries from the TbpB N lobe and (ii) generating TbpB N-lobe truncations lacking the identified peptides to demonstrate loss of binding to peptides in the hTf library. M. catarrhalis was selected for our experiments since recombinant TbpB proteins and TbpB subfragments (lobes) from this species were the most stable and retained the strongest binding activity (data not shown).
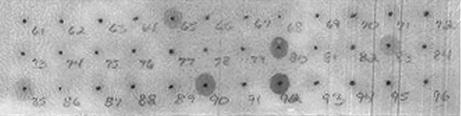
Initially, an overlapping peptide library representing the TbpB N lobe of M. catarrhalis was prepared and probed for binding to labeled hTf. This library comprised 32 15-mer peptides with a 4-amino-acid overlap representing amino acids 12 to 421 of mature M. catarrhalis TbpB. The library was screened for binding to β-Gal-conjugated hTf (β-Gal-hTf), and a number of peptides positive for binding were detected with various intensities of colored products (Fig. 1). Three strongly positive peptides (80, 90, and 92), two moderately positive peptides (65 and 83), and one weakly positive peptide (85) were identified and represented the six binding peptides (Table 1) for our truncation analysis (see below). As a negative control, the library was screened with β-Gal alone to demonstrate that the reactivity of the peptides recognized by labeled hTf (Fig. 1) was due to the binding of hTf.
FIG. 1.
Overlapping peptide library of the TbpB N lobe from M. catarrhalis. A library consisting of 32 15-amino-acid overlapping peptides representing amino acids 12 to 421 of the TbpB N lobe of M. catarrhalis was synthesized. The library was probed with β-Gal-hTf and developed as described in Materials and Methods.
TABLE 1.
hTf binding by peptides from the M. catarrhalis TbpB N lobea
| Binding peptide region | Peptide library (spot no.) | Amino acid sequence | Overlap sequence |
|---|---|---|---|
| 1 | 1 (65) | MGYGMALSKINLHNR | MGYGMALSKINLHN |
| 2 | AMGYGMALSKINLHN | ||
| 2 | 1 (80) | GPVGGVFYNGTTTAK | GPVGGVFYNGTT |
| 2 | WNLGPVGGVFYNGTT | ||
| 3 | 1 (83) | FMTDVANRRNRFSEV | FMTDVA |
| AVKYKGHWDFMTDVA | |||
| 2 | KGHWDFMTDVANRRN | ||
| DFMTDVANRRNRFSE | |||
| 4 | 1 (85) | AGWYYGASSKDEYNR | AGWYYG |
| 2 | FSEVKENSQAGWYYG | ||
| 5 | 1 (90) | FSNLQDRHKGNVTKT | FSNL |
| NFKEKKLTGKLFSNL | |||
| 2 | KKLTGKLFSNLQDRH | ||
| GKLFSNLQDRHKGNV | |||
| 6 | 1 (92) | DANIHGNRFRGSATA | DANIHGNR |
| 2 | KTERYDIDANIHGNR | ||
| YDIDANIHGNRFRGS |
A preliminary overlapping peptide library representing amino acids 12 to 421 of the N lobe of TbpB from M. catarrhalis was prepared. The library consisted of 32 overlapping 15-amino-acid peptides with a 4-amino-acid overlap (peptide library 1). The library was probed with β-Gal-hTf, and six positive peptides were identified, representing six peptide-binding regions. A second peptide library consisting of 96 15-amino-acid peptides with an 11-amino-acid overlap, representing the same region, was prepared (peptide library 2). The second library was probed with HRP-conjugated hTf to detect binding peptides. Spots refer to numbers on Fig. 1.
To confirm and refine the identification of the binding regions, a second overlapping TbpB N-lobe peptide library was prepared and probed with HRP-conjugated hTf. The use of an HRP conjugate allowed more sensitive detection of a chemiluminescent product. The second library consisted of 96 15-amino-acid peptides with an 11-amino-acid overlap representing amino acids 23 to 421 of mature M. catarrhalis TbpB. The greater degree of overlap and the use of the more sensitive chemiluminescence detection method increased the chance of detecting binding peptides. Thus, a total of 11 binding peptides were detected in this peptide library, compared to the 6 binding peptides that were identified in the first library (Table 1). None of these peptides was detected when the library was screened with HRP alone. This second library revealed the same six binding regions as the first library but included overlapping peptides for several regions.
The availability of overlapping positive peptides allowed further definition of the binding determinants, since the common amino acid sequence could be implicated as the recognition sequence (Table 1). Thus, binding region 6, represented by two overlapping peptides in the second library, shared a hendecapeptide (YDIDANIHGNR) and, by inclusion of the peptide recognized in the first library, an octapeptide (DANIHGNR) could be implicated as the binding region. Similarly, binding region 3, which is represented by three adjacent overlapping peptides in the second library, tentatively identified a heptapeptide (DFMTDVA) sequence which overlapped a hexapeptide sequence (FMTDVA) identified in the first library.
Three of the binding regions (binding regions 1, 2, and 4; Table 1) were represented by a single peptide from both libraries. Since adjacent peptides in the second library contain 11 of 15 amino acids in common, it is unlikely that the lack of binding to the neighboring peptides is solely due to the absence of the amino acids directly involved in the binding interaction. Rather, we suggest that conformational or structural features of the peptides were also important for detectable binding. For two of the regions (binding regions 1 and 2; Table 1), binding peptides from the composite library effectively provided adjacent peptides with a greater degree of overlap (14 of 15 for segment 1 and 12 of 15 for segment 2) and thus provided a minor refinement in localizing the binding determinants. The binding peptide from the first peptide library for binding region 4 provided less overlap (6 of 15) yet retained binding activity, for unknown reasons.
Production of truncations in the TbpB N lobe of M. catarrhalis.
The binding peptides listed in Table 1 were identified by binding to labeled hTf and, by inference, would be expected to interact with one of the TbpB-binding peptides identified in the hTf peptide libraries (35). To provide direct evidence for this interaction, one strategy was to eliminate a putative binding determinant from TbpB and to determine whether it resulted in a loss of reactivity with one of the TbpB-binding peptides from hTf. The approach that we adopted was to generate a series of truncations of the TbpB N lobe that sequentially eliminated the segments of TbpB containing hTf-binding determinants and to use the truncated proteins to probe the hTf peptide libraries. Although producing site-directed mutants of the TbpB N lobe was another alternative, it would have required preparation of a large number of mutants due to the lengths of the identified peptide regions.
To plan the truncation experiments, we divided the N lobe of M. catarrhalis into seven segments with the identified binding peptides at their junctions (Fig. 2). Segment 1 includes the amino acid sequence up to but not including binding region 1, and segment 2 includes binding region 1 and the amino acid sequence up to but not including binding region 2. Continuing with this convention, segment 7 includes binding region 6 and the remaining amino acid sequence to the end of the N lobe of TbpB.
FIG. 2.
Segments of the M. catarrhalis TbpB N lobe used in binding and truncation analyses. (A) The amino acids representing the mature N lobe of TbpB (amino acids [aa] 1 to 421) are broken down into seven segments. Bold text represents binding peptides from peptide library 1. Underlined text represents the common overlap of binding peptides from both peptide libraries 1 and 2. Italic type represents the regions of identity recognized previously (35). (B) Linear representation of the amino acids shown in panel A. The areas marked by black bars represent the regions that bind hTf. The areas marked by white bars represent the amino acids not recognized by labeled hTf. The lines under the drawing represent each segment as described in the text. S1, segment 1; S2, segment 2; and so forth.
To generate the TbpB N-lobe truncations, PCR mutagenesis was used to introduce a stop codon and an appropriate restriction site at the junction of the segments illustrated in Fig. 2. Using a plasmid expressing the functional TbpB N lobe as a template, PCR amplifications were performed. The PCR products were subcloned into the pMal-c2 vector, and the sequence was confirmed. After expression, the resulting MBP fusion proteins were isolated by amylose affinity chromatography. Initial experiments showed that the smaller expressed truncations that contained binding regions 1 and 2 were degraded more quickly than the other truncations or the intact proteins during storage. In order to make more stable preparations for these truncations, a protease-deficient expression strain was used.
As illustrated in Fig. 3, relatively pure and stable preparations of all of the truncations were isolated by amylose affinity chromatography. Each protein was electroblotted onto nitrocellulose and probed with anti-M. catarrhalis TbpA or TbpB antibodies. All the bands observed in the Coomassie blue gel reacted with the antibodies (data not shown), indicating that they were breakdown products of the fusion protein and not impurities. Solid-phase binding assays with HRP-conjugated hTf to probe immobilized fusion proteins showed an increasing reduction or loss of detectable binding activity with successive truncations. The fusion proteins containing segments 1 to 4 or less (Fig. 2) did not yield detectable binding activity in solid-phase binding assays with the colorimetric substrate chloronaphthol (data not shown). In addition, when the truncations were probed with labeled hTf after SDS-polyacrylamide gel electrophoresis and electroblotting, even the larger truncated proteins were not detected, similar to what was observed for TbpB from N. meningitidis (40).
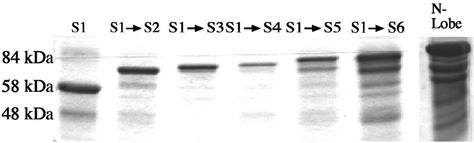
FIG. 3.
Affinity-purified M. catarrhalis TbpB N-lobe truncations. Approximately 5 μg of each purified protein was analyzed by SDS-polyacrylamide gel electrophoresis (10% acrylamide) after boiling for 5 min in sample buffer containing dithiothreitol. The proteins were detected by Coomassie blue staining. The numbers on the left indicate approximate molecular masses. The notation above each lane indicates the protein content in the respective lane: S1, segment 1 (49.8 kDa); S1 → S2, segments 1 and 2 (66.8 kDa); S1 → S3, segments 1, 2, and 3 (70.0 kDa); S1 → S4, segments 1, 2, 3, and 4 (72.4 kDa); S1 → S5, segments 1, 2, 3, 4, and 5 (78.0 kDa); S1 → S6, segments 1, 2, 3, 4, 5, and 6 (80.4 kDa); N-Lobe, entire N lobe of TbpB (segments 1 to 7) (88.2 kDa). All proteins contain both the TbpB segment and the 39.6-kDa MBP fusion partner.
Identification of hTf-binding peptides with TbpB N-lobe truncations.
The lack of binding to hTf by some of the TbpB truncations in the screening solid-phase binding assay could have been attributed to many factors, including the sensitivity of the colorimetric detection or constraints imposed by immobilization of the protein on the cellulose. Thus, we decided to include them for probing the overlapping peptide library of hTf, since the format of this assay may permit the detection of binding activity for these truncations.
To provide a potentially more sensitive means of detection that would not compromise the life span of the peptide libraries, HRP conjugates of the fusion proteins and a chemiluminescence system for detecting binding activity were used. A 15-residue overlapping peptide library representing the C-terminal half of hTf with an 11-amino-acid overlap was prepared. This library is essentially identical to the library used in a prior study (35), and the intact N lobe of M. catarrhalis TbpB recognized an almost identical set of peptides (segments 1 to 7; Table 2).
TABLE 2.
Identification of TbpB-binding peptides in the hTf C lobe with labeled TbpB N-lobe truncationsa
| Binding peptideb | Binding by truncations including the following segment(s)c:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Sequence | Domain (color) | 1-7 (intact) | 1-6 | 1-5 | 1-4 | 1-3 | 1 and 2 | 1 |
| 12 | KKSASDLTWDNLKGK | 2 (Green) | + | + | + | + | − | − | − |
| 13 | NIPMGLLYNKINHCR | 2 (Blue) | + | + | + | − | − | − | − |
| 14 | LEMGSGLNLCEPNNK | 2 (Red) | + | + | + | + | + | − | − |
| 15 | GYYGYTGAFRCLVEK | 2 (Red) | + | + | + | + | + | − | − |
| 16 | CHLARAPNHAVVTRK | 2 (Yellow) | + | + | − | − | − | − | − |
| 18 | GSNVTDCSGNFCLFR | 1 (Brown) | + | + | + | + | + | + | − |
| 19 | TDCSGNFCLFRSETK | 1 (Brown) | + | + | + | + | + | + | − |
| 20 | KYLGEEYVKAVGNLR | Tail (Cyan) | + | − | − | − | − | − | − |
Fusion proteins containing the indicated segments of the M. catarrhalis TbpB N lobe were conjugated to HRP and used to probe an overlapping peptide library representing the C lobe of hTf. The sequences of positive binding peptides and their locations are indicated.
Peptide number and color refer to the numbering and color scheme used in a prior illustration of the binding regions (35).
Segments are as detailed in Fig. 2. +, binding; −, no binding.
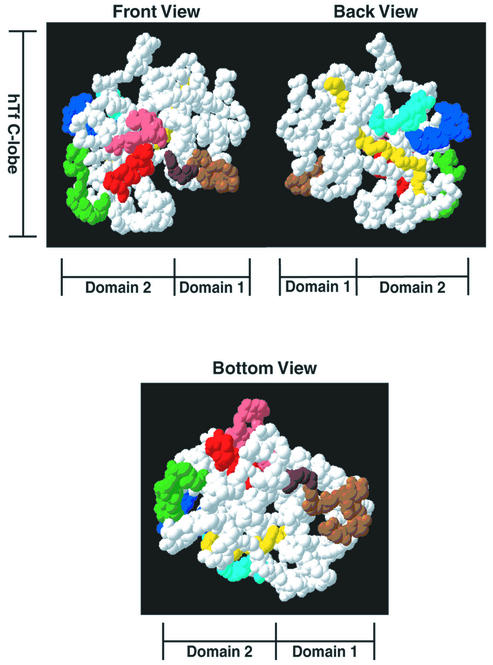
The truncation lacking segment 7 bound all of the peptides except the one localized to the C-terminal tail region of hTf (20; cyan), suggesting that segment 7 was responsible for binding to the C-terminal tail region (segments 1 to 6; Table 2). The truncation lacking both segment 6 and segment 7 (segments 1 to 5; Table 2) lost reactivity to an additional peptide localized to domain 2 (16; yellow). Although this peptide is quite distant from peptide 20 in the linear amino acid sequence, it is immediately adjacent on the surface of hTf (Fig. 4) (35). Since the binding peptides from segments 6 and 7 are fairly close in the linear amino acid sequence of TbpB (Fig. 2), the implication that they bind adjacent regions on the hTf surface seems reasonable.
FIG. 4.
Space-filling model of the hTf C lobe showing binding peptides of the M. catarrhalis TbpB N-terminal half. The images represent three views of the C lobe of hTf. The diagrams at the top illustrate the “front” and“back” of the protein. In this view, the N lobe (not shown) would be above the C lobe; both domains of the C lobe are easily seen. The diagram at the bottom illustrates the “bottom” of the protein. The amino acids colored cyan, yellow, blue, green, red, and brown represent peptides that bound to a probe made up of the N-terminal half of TbpB from M. catarrhalis as well as the truncations shown in Table 2. The model was created as described previously (35). The model was then manipulated by using Swiss-PDB Viewer (version 3.7 b2) (http://www.expasy.ch/spdbv/) (16).
The results obtained with the subsequent three truncations followed a pattern similar to that observed with the first two truncations; the removal of an additional segment of TbpB resulted in the loss of binding to a peptide that maps to an adjacent position on the hTf surface. Thus, the truncation containing segments 1 to 4 lost reactivity to peptide 13 (blue), the truncation containing segments 1 to 3 lost reactivity to peptide 12 (green), and the truncation containing segments 1 and 2 lost reactivity to peptides 14 and 15 (red). The binding peptides from these segments of TbpB are fairly close in the linear amino acid sequence and map to adjacent regions on the surface of domain 2 of the hTf C lobe.
The final truncation (segment 1; Table 2) removed a segment containing over 140 amino acids between the binding peptides identified in the TbpB library (Fig. 2). This truncation resulted in the loss of binding to two peptides that map to the surface of domain 1 (peptides 18 and 19; brown), relatively close to the prior peptides (14 and 15; red) on the surface of domain 2 in the iron-loaded form of hTf. However, these two regions would be substantially further apart in the apoprotein, in which there is considerable separation of the two domains.
DISCUSSION
There is considerable cumulative evidence supporting the use of TbpB as a vaccine antigen against organisms such as the human pathogens N. meningitidis (23), M. catarrhalis (27), and Haemophilus influenzae (41), the bovine pathogen Mannheimia (Pasteurella) haemolytica (30), and the porcine pathogen Actinobacillus pleuropneumoniae (7). As with many surface antigens, there can be considerable antigenic variation in TbpB proteins, a fact which complicates the process of vaccine development and could ultimately limit the life span of any products that are developed (36). Evidence supporting the concept of a conserved Tf-TbpB interaction suggests that it may be possible to target conserved functional domains (35). Further studies delineating the details of this interaction are required to explore this possibility and to provide additional insights into the iron removal process.
Since a previous study (35) had tentatively identified six or more TbpB-binding peptides on each lobe of hTf, we attempted to identify the complementary hTf-binding regions on TbpB proteins from M. catarrhalis. Our results with overlapping peptide libraries representing the N lobe of TbpB from M. catarrhalis implicated six distinct hTf-binding regions (Fig. 1 and Table 1). The subsequent analysis with truncated proteins (Table 2) provided independent confirmation that the defined segments (Fig. 2) contained binding determinants and thus supported the identification of these six hTf-binding regions in the M. catarrhalis library.
Prior studies with overlapping peptides libraries of TbpB proteins from A. pleuropneumoniae (38) and N. meningitidis (33) led to the identification of a smaller number of Tf-binding peptides. Three Tf-binding peptides were identified in the A. pleuropneumoniae TbpB N lobe, and seven peptides were identified as positive in a library representing the intact TbpB from N. meningitidis, two from the N lobe and five from the C lobe. Since recombinant TbpB (or its individual lobes) from N. meningitidis recognized the same set of hTf peptides as TbpB from M. catarrhalis (35), it follows that the N lobe of TbpB from N. meningitidis must also have six or more hTf-binding regions. A variety of factors might have contributed to the identification of a smaller number of binding peptides, such as the sensitivity of the detection method or the specific amino acid sequence of the peptide not favoring the formation of a required binding conformation. However, it is certainly possible that the expectation of a relatively small number of positive binding peptides would lead an investigator to assume that weakly binding peptides (Fig. 1) are due to nonspecific interactions and thus to underestimate the number of positive binding peptides. The complementary data that we provide in this study and the results of the truncation analysis provide strong evidence that all six binding peptides are authentic and compensate for any deficiencies in the intensities of the detection methods.
One limitation of synthetic peptide libraries is the lack of specific criteria for deciding whether a detected binding activity is genuine or is due to nonspecific interactions. This factor may be partially responsible for their primary use in identifying peptide epitopes recognized by antibodies (8, 14), since a small number of peptides representing a single epitope are usually identified and the antibody-binding pocket may provide the opportunity for an induced fit to contribute to the strength of the binding interaction. In this study, we demonstrated the utility of overlapping synthetic peptide libraries in characterizing the extensive set of peptide-peptide interactions between Tf and the bacterial binding protein TbpB. Our results may also suggest that, in spite of the potential limitations, this approach could be used more extensively for the study of protein-protein interactions. It is noteworthy to mention that due to the number of sites involved in the interaction (34, 35), alternative approaches, such as the use of chimeric proteins or site-directed mutagenesis, would not have been able to yield such detailed information.
One strategy for facilitating the identification of binding peptides was the synthesis of relatively long peptides (15 amino acids), based on the rationale that they might be able to more readily provide secondary structures required for effective binding. The fact that several of the binding regions were identified by a single peptide (segments 1, 2, and 4; Table 1) despite considerable overlap (11 of 15) indicates that the presence of a specific amino acid sequence is not sufficient to mediate binding and that other factors, such as the ability to attain an appropriate secondary structure, may be important (25, 39). Similarly, some of the stronger binding peptides detected in the hTf library (peptides 13, 15, and 20; Table 2) formed an alpha helix in the intact hTF protein (data not shown), and the peptides were long enough to encompass the entire alpha-helical region (35). The adjacent peptides in the library that showed little or no binding did not include the complete helix. However, the use of longer peptides cannot overcome a primary limitation of peptide libraries, the inability to represent conformational epitopes comprising amino acids from distant positions along the polypetide sequence. Thus, we must consider the possibility that the TbpB-Tf interactions could be much more extensive than our results suggest.
A drawback of using longer peptides is that they do not provide a very precise localization of the binding determinant. A smaller binding determinant can be inferred when several adjacent peptides are positive for binding (segments 3, 5, and 6; Table 1) but not when adjacent peptides are negative (segments 1, 2, and 4). The use of random or systematic changes to each Tf-binding peptide in the TbpB N lobe to further delineate the binding interaction is possible, but without structural information as a guide, the number of possible combinations is large. Further insights into the binding interaction will be facilitated when structural information becomes available for TbpB or, preferably, for a Tf-TbpB complex.
Our results illustrate that most of the binding determinants cluster in a region near the C-terminal end of the N lobe of TbpB, similar to what was observed for the regions of sequence identity found by Retzer et al. (35). These previously identified regions are shown in italic type in Fig. 2. However, it is apparent that, with some exceptions, the binding peptides and the regions of identity are distinct from one another. This fact indicates that the regions of identity are not directly responsible for the binding interactions and likely play an alternate role, possibly structural.
The structure of hTf (Fig. 4) provides some insights into the overall structural features of TbpB when considered in conjunction with the results from the analysis with the TbpB N-lobe truncations (Table 2). The TbpB-binding peptides on hTf that are recognized by binding regions 3, 4, and 5 (on segments 4, 5, and 6) of TbpB (Table 2) are approximately 10 to 15 Å apart on the surface of hTf (green, blue, and yellow) (Fig. 4). The hTf-binding determinants on these segments are separated by approximately 20 amino acids in the linear amino acid sequence of TbpB (Fig. 2). Although this fact does not implicate a specific secondary structure in this region of the protein, the spatial constraints enable us to exclude some structural models.
The TbpB-binding peptide on hTf that is recognized by binding sites on segment 3 (red) of TbpB (Table 2) is approximately 22 Å away from the segment 2 (brown) TbpB-binding peptide on the surface of hTf (Fig. 4). However, the red binding region on hTf is part of domain 2 and the brown binding region is part of domain 1, and the separation of these two regions would be substantially increased in the apoprotein form of hTf. In this context, it is interesting that there are 135 amino acids in the linear sequence of the N lobe of TbpB that separate these two binding segments (Fig. 2). It is tempting to speculate that this “extra” length attributed to TbpB may allow the two domains to separate when the TbpA-TbpB complex removes the iron from hTf.
This study details the interaction between the M. catarrhalis TbpB N lobe and the C lobe of hTf. In a prior study, the TbpB N lobe and the TbpB C lobe from N. meningitidis recognized an identical set of peptides in an overlapping peptide library representing hTf (35), suggesting that the lobe-lobe interactions were equivalent. Thus, it is anticipated that there are at least six hTf-binding determinants in each lobe of TbpB from N. meningitidis and, by inference, in each TbpB lobe from M. catarrhalis. Experimental evidence supporting the peptide-peptide interactions could presumably be obtained by implementing similar studies with the N. meningitidis TbpB lobes and with the M. catarrhalis TbpB C lobe. However, M. catarrhalis TbpB and TbpB N lobe were specifically selected for their stability and strength of binding activity, and it is uncertain whether weaker binding activity, particularly of the TbpB C lobe (35), would compromise the success of these studies.
In the current study, we used overlapping peptide libraries to identify peptide-peptide interactions between a bacterial receptor and its ligand, hTf. Refinements of this approach could lead to a more detailed understanding of the specific amino acids involved in the molecular interactions between receptor and ligand. However, a full appreciation of the structural aspects of these interactions awaits results from biophysical approaches, such as X-ray crystallography, in which the structure of a ligand-receptor complex would be particularly informative.
Acknowledgments
This work was supported by grant MT 10350 from the Canadian Institutes for Health Research (CIHR).
REFERENCES
- 1.Abdallah, F. B., and J. M. E. H. Chahine. 2000. Transferrins: iron release from lactoferrin. J. Mol. Biol. 303:255-266. [DOI] [PubMed] [Google Scholar]
- 2.Alcantara, J., and A. B. Schryvers. 1996. Transferrin binding protein two interacts with both the N-lobe and C-lobe of ovotransferrin. Microb. Pathog. 20:73-85. [DOI] [PubMed] [Google Scholar]
- 3.Alcantara, J., R.-H. Yu, and A. B. Schryvers. 1993. The region of human transferrin involved in binding to bacterial transferrin receptors is localized in the C-lobe. Mol. Microbiol. 8:1135-1143. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, J. A., P. F. Sparling, and C. N. Cornelissen. 1994. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol. 176:3162-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, E. N., and P. F. Lindley. 1992. New perspectives on the structure and function of transferrins. J. Inorg. Biochem. 47:147-160. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, M. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 7.Bunka, S., C. Christensen, A. A. Potter, P. J. Willson, and G.-F. Gerlach. 1995. Cloning and characterization of a protective outer membrane lipoprotein of Actinobacillus pleuropneumoniae serotype 5. Infect. Immun. 63:2797-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choulier, L., D. Laune, G. Orfanoudakis, H. Wlad, J. Janson, C. Granier, and D. Altschuh. 2001. Delineation of a linear epitope by multiple peptide synthesis and phage display. J. Immunol. Methods 249:253-264. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen, C. N., J. E. Anderson, I. C. Boulton, and P. F. Sparling. 2000. Antigenic and sequence diversity in gonococcal transferrin binding protein A. Infect. Immun. 68:4725-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelissen, C. N., J. E. Anderson, and P. F. Sparling. 1997. Characterization of the diversity and the transferrin-binding domain of gonococcal transferrin-binding protein 2. Infect. Immun. 65:822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelissen, C. N., M. Kelley, M. M. Hobbs, J. E. Anderson, J. G. Cannon, M. S. Cohen, and P. F. Sparling. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 27:611-616. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 13.Frank, R. 1992. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217-9232. [Google Scholar]
- 14.Geginat, G., S. Schenk, N. Skoberne, W. Goebel, and H. Hoff. 2001. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 166:1877-1884. [DOI] [PubMed] [Google Scholar]
- 15.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends. Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 16.Guex, N., and M. C. Peitsch. 1997. Swiss-Model and the Swiss-PDB Viewer: an environment for comparative protein modelling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 17.Guo, M., H. Sun, H. J. McArdle, L. Gambling, and P. J. Sadler. 2000. Ti (IV) uptake and release by human serum transferrin and recognition of Ti (IV)-transferrin by cancer cells: understanding the mechanism of action of the anticancer drug titanocene dichloride. Biochemistry 39:10023-10033. [DOI] [PubMed] [Google Scholar]
- 18.Harlow, E., and W. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Killmann, H., M. Braun, C. Herrmann, and V. Braun. 2001. FhuA barrel-cork hybrids are active transporters and receptors. J. Bacteriol. 183:3476-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurokawa, H., B. Mikami, and M. Hirose. 1995. Crystal structure of diferric ovotransferrin at 2.4 A resolution. J. Mol. Biol. 254:196-207. [DOI] [PubMed] [Google Scholar]
- 21.Legrain, M., E. Jacobs, S. W. Irwin, A. B. Schryvers, and M. J. Quentin-Millet. 1993. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin binding proteins Tbp1 and Tbp2. Gene 130:73-80. [DOI] [PubMed] [Google Scholar]
- 22.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol. Microbiol. 24:271-283. [DOI] [PubMed] [Google Scholar]
- 23.Lissolo, L., G. Maitre-Wilmotte, P. Dumas, M. Mignon, B. Danve, and M.-J. Quentin-Millet. 1995. Evaluation of transferrin-binding protein 2 within the transferrin-binding protein complex as a potential antigen for future meningococcal vaccines. Infect. Immun. 63:884-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structure of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 25.Mademidis, A., H. Killmann, W. Kraas, I. Flechsler, G. Jung, and V. Braun. 1997. ATP-dependent ferric hydroxamate transport system in Escherichia coli: periplasmic FhuD interacts with a periplasmic and with a transmembrane/cytoplasmic region of the integral membrane protein FhuB, as revealed by competitive peptide mapping. Mol. Microbiol. 26:1109-1123. [DOI] [PubMed] [Google Scholar]
- 26.Masri, H. P., and C. N. Cornelissen. 2002. Specific ligand binding attributable to individual epitopes of gonococcal transferrin binding protein A. Infect. Immun. 70:732-740. [DOI] [PMC free article] [PubMed]
- 27.Myers, L. E., Y.-P. Yang, R.-P. Du, Q. Wang, R. E. Harkness, A. B. Schryvers, M. H. Klein, and S. M. Loosmore. 1998. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect. Immun. 66:4183-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogunnariwo, J. A., and A. B. Schryvers. 2001. Characterization of a novel transferrin receptor in bovine strains of Pasteurella multocida. J. Bacteriol. 183:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pintor, M., J. A. Gómez, L. Ferrón, C. M. Ferreirós, and M. T. Criado. 1998. Analysis of TbpA and TbpB functionality in defective mutants of Neisseria meningitidis. J. Med. Microbiol. 47:757-760. [DOI] [PubMed] [Google Scholar]
- 30.Potter, A. A., A. B. Schryvers, J. A. Ogunnariwo, W. A. Hutchins, R. Y. Lo, and T. Watts. 1999. Protective capacity of Pasteurella haemolytica transferrin-binding proteins TbpA and TbpB in cattle. Microb. Pathog. 27:197-206. [DOI] [PubMed] [Google Scholar]
- 31.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 32.Renauld-Mongénie, G., M. Latour, D. Poncet, S. Naville, and M. J. Quentin-Millet. 1998. Both the full-length and the N-terminal domain of the meningococcal transferrin-binding protein B discriminate between human iron-loaded and apo-transferrin. FEMS Microbiol. Lett. 169:171-177. [DOI] [PubMed] [Google Scholar]
- 33.Renauld-Mongénie, G., D. Poncet, L. Von Olleschik-Elbheim, T. Cournez, M. Mignon, M. A. Schmidt, and M. J. Quentin-Millet. 1997. Identification of human transferrin-binding sites within meningococcal transferrin-binding protein B. J. Bacteriol. 179:6400-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Retzer, M. D., A. Kabani, L. Button, R.-H. Yu, and A. B. Schryvers. 1996. Production and characterization of chimeric transferrins for the determination of the binding domains for bacterial transferrin receptors. J. Biol. Chem. 271:1166-1173. [DOI] [PubMed] [Google Scholar]
- 35.Retzer, M. D., R.-H. Yu, and A. B. Schryvers. 1999. Identification of sequences in human transferrin that bind to the bacterial receptor protein, transferrin-binding protein B. Mol. Microbiol. 32:111-121. [DOI] [PubMed] [Google Scholar]
- 36.Rokbi, B., M. Mignon, D. A. Caugant, and M. J. Quentin-Millet. 1997. Heterogeneity of tbpB, the transferrin-binding protein B gene, among serogroup B from Neisseria meningitidis strains of the ET-5 complex. Clin. Diagn. Lab. Immunol. 4:522-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schryvers, A. B., and B. C. Lee. 1989. Comparative analysis of the transferrin and lactoferrin binding proteins in the family Neisseriaceae. Can. J. Microbiol. 35:409-415. [DOI] [PubMed] [Google Scholar]
- 38.Strutzberg, K., L. Von Olleschik, B. Franz, C. Pyne, M. A. Schmidt, and G. F. Gerlach. 1995. Mapping of functional regions on the transferrin-binding protein (TfbA) of Actinobacillus pleuropneumoniae. Infect. Immun. 63:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenidis, K., M. Waldner, J. Bernhagen, W. Fischle, M. Bergmann, M. Weber, M.-L. Merkle, W. Voelter, H. Brunner, and A. Kapurniotu. 2000. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol. 295:1055-1071. [DOI] [PubMed] [Google Scholar]
- 40.Vonder Haar, R. A., M. Legrain, H. V. J. Kolbe, and E. Jacobs. 1994. Characterization of a highly structured domain in Tbp2 from Neisseria meningitidis involved in binding to human transferrin. J. Bacteriol. 176:6207-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webb, D. C., and A. W. Cripps. 1999. Immunization with recombinant transferrin binding protein B enhances clearance of nontypeable Haemophilus influenzae from the rat lung. Infect. Immun. 67:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, R.-H., and A. B. Schryvers. 1994. Transferrin receptors on ruminant pathogens vary in their interaction with the C-lobe and N-lobe of ruminant transferrins. Can. J. Microbiol. 40:532-540. [DOI] [PubMed] [Google Scholar]