Abstract
Cholesterol 7α-hydroxylase (CYP7A1) of the bile acid biosynthesis pathway is suppressed by bile acids and inflammatory cytokines. Bile acids are known to induce inflammatory cytokines to activate the MAPK/JNK signaling pathway that inhibits CYP7A1 gene transcription. c-Jun has been postulated to mediate bile acid inhibition of CYP7A1. However the c-Jun target involved in regulation of CYP7A1 is not known. Human primary hepatocytes and HepG2 cells were used as models to study chenodeoxycholic acid (CDCA) and interleukin-1β (IL-1β) regulation of human CYP7A1 gene expression by real time PCR, reporter assays, co-immunoprecipitation (Co-IP) and chromatin immunocipitation assay (ChIP) assays. IL-1β and CDCA reduced CYP7A1 but induced c-Jun mRNA expression in human primary hepatocytes. IL-1β inhibited human CYP7A1 reporter activity via the HNF4α binding site. A JNK-specific inhibitor blocked the inhibitory effect of IL-1β on HNF4α expression and CYP7A1 reporter activity. c-Jun inhibited HNF4α and PPARγ co-activator-1α (PGC-1α) co-activation of CYP7A1 reporter activity while a dominant negative c-Jun did not. Co-IP and ChIP assays revealed that IL-1β and CDCA reduced HNF4α bound to the CYP7A1 chromatin and c-Jun interacted with HNF4α and blocked HNF4α recruitment of PGC-1α to the CYP7A1 chromatin. In conclusion, IL-1β and CDCA inhibit HNF4α but induce c-Jun, which in turn blocks HNF4α recruitment of PGC-1α to the CYP7A1 chromatin and results in inhibition of CYP7A1 gene transcription. The JNK/c-Jun signaling pathway inhibits bile acid synthesis and protects hepatocytes against the toxic effect of inflammatory agents.
Keywords: Bile acid biosynthesis, nuclear receptors, HNF4α cholestasis, inflammation
Introduction
Bile acid synthesis from cholesterol is the predominant pathway for eliminating excess cholesterol in the body. Bile acids are physiological detergents that facilitate the absorption, transport and distribution of dietary lipids, lipid-soluble vitamins and steroids. Bile acids are reabsorbed in the ileum and transported via the enterohepatic circulation to the liver, where they inhibit bile acid synthesis (1). The bile acid feedback regulation is primarily achieved through the transcriptional regulation of CYP7A1, the rate-limiting enzyme in the classic bile acid biosynthetic pathway. Studies in mice demonstrate that bile acid-activated farnesoid X receptor (FXR, NR1H4) induces the expression of a negative nuclear receptor, small heterodimer partner (SHP, NR0B2), which interacts with liver-related homologue (LRH, NR5A2, or called α-fetoprotein transcription factor FTF in human liver) and down regulates CYP7A1 gene transcription (2, 3). However bile acid feeding to Shp knockout mice reduces CYP7A1 mRNA levels similar to the wild-type mice (4, 5) suggesting the existence of mechanisms independent of the FXR/SHP/FTF pathway. The alternative mechanisms include the PKC/JNK pathway (6), the cytokine/JNK pathway (7), the FXR/FGF (fibroblast growth factor)/FGFR4 (FGF receptor 4) pathway (8, 9) and the pregnane X receptor (PXR, NR1I2)-mediated pathway (10).
It has been reported that bile acids induce the synthesis and excretion of inflammatory cytokines such as tumor necrosis factor α (TNFα) and IL-1β in hepatic macrophages (Kupffer cells) and concomitantly repress CYP7A1 expression in mouse hepatocytes and macrophage cell lines (11). Inhibition of cytokine expression also prevents bile acid inhibition of CYP7A1 expression suggesting that cytokines inhibit CYP7A1 expression in hepatocytes. In acute phase response, inflammatory cytokines inhibit CYP7A1 mRNA and activity (12). It has been suggested that TNFα and CDCA inhibit HNF4α trans-activating activity and CYP7A1 gene transcription via the MAPK signaling pathway (7). Cytokines have also been shown to inhibit other bile acid biosynthetic genes such as sterol 12α-hydroxylase (CYP8B1) (13) and sterol 27-hydroxylase (CYP27A) (14). In hepatic inflammatory conditions, bile acids and cytokines induce intrahepatic cholestasis (15), hypercholesterolemia and dyslipidemia (16-18). Several studies have shown that both cytokines (19, 20) and bile acids (cholic acid and deoxycholic acid) (20, 21) can activate the c-Jun NH2-terminal kinase (JNK)/c-Jun pathway and inhibit the CYP7A1 gene. It has been suggested that c-Jun either forms a repressive complex with an unknown factor or activates SHP to inhibit the CYP7A1 gene (20, 22, 23). However, the events downstream of JNK/c-Jun have not been established.
In this study, we showed that CDCA and IL-1β stimulated c-Jun mRNA levels in primary human hepatocytes and that c-Jun interacted with HNF4α both in vivo and in vitro and disrupted HNF4α recruitment of PGC-1α to the CYP7A1 chromatin resulting in the suppression of the human CYP7A1 gene transcription. Our studies defined a novel mechanism for bile acid and cytokine inhibition of the human CYP7A1 gene and protection against the toxicity of bile acids during cholestasis and acute phase response.
Materials and Methods
Cell culture. The human hepatoblastoma cells (HepG2, HB8065) were obtained from the American Type Culture Collection (Manassas, VA). The cells were grown in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and F-12 (50:50), (Life Technologies, Inc.) supplemented with 100 units/ml penicillin G/streptomycin sulfate (Celox Corp., Hopkins, MN) and 10% (v/v) heat-inactivated fetal calf serum (Irvine Scientific, Santa Ana, CA). Primary human hepatocytes (HH1115, 22 yr male; HH1117, 68 yr female; HH1119, 29 yr female; HH1122, 46 yr female and HH1209, 30 yr female) were obtained through the Liver Tissue Procurement and Distribution System (LTPADS) of NIH (S. Strom, University of Pittsburgh, Pittsburgh, PA). Cells were maintained in HMM modified Williams E medium (Clonetics, US) supplemented with 10-7 M of insulin and dexamethasone, and used within 24 h after receiving.
Plasmids. Human CYP7A1/Luc reporter plasmid (ph-298 to +24) and mutant plasmids were constructed as described previously (24). PXR (mPXR-CYP7A1-Luc), FTF (mFTF-CYP7A1-Luc), or HNF4α (mHNF4α-CYP7A1-Luc) binding site mutations were introduced into ph-298/Luc plasmid. The expression plasmids were obtained as follows-HNF4α (pCMX-HNF4α): originally from Dr. William Chin (Lilly Research Laboratories, Indianapolis, IN), subcloned to pcDNA3; PGC-1α (pcDNA3-HA-PGC-1α): from Dr. A. Kralli (The Scripp Research Institute, La Jolla, CA); c-Jun: originally from Dr. Inder Verma (The Salk Institute, San Diego, CA), subcloned into pcDNA3 and dominant negative c-Jun (DN c-Jun, pcDNA3.1HisTAM67): from Dr. Nancy Colburn (NCI, Frederick, MD).
RNA Isolation and Quantitative Real-Time PCR. Primary hepatocytes were obtained in 6-well plates. Cells were treated with 10 ng/ml IL-1β (PeproTech, Rocky Hill, NJ) or 50 μM CDCA for a period of time indicated. RNA was extracted from cells using Tri-Reagent (Sigma). Quantitative real time PCR assays were performed according to the PCR Taqman Universal Master Mix 2X protocol (Applied Biosystems, Foster City, CA). Amplification of ubiquitin C (UBC) was used in the same reactions as an internal reference gene. Quantitative PCR analysis was conducted on the ABI 7500 Sequence Detection System. Relative mRNA expression was quantified using the comparative Ct (ΔΔCt) method according to the ABI manual. Assay-on-Demand PCR primers and Taqman MGB probe mix used are: human CYP7A1 (Cat. # Hs00167982_m1); human HNF4α(Cat. #: Hs00230853_m1); human cJun (Cat. #: Hs00277190_s1); human SHP (Cat. #: Hs00222677_m1); human CYP8B1 (Cat. #: Hs00244754_s1) and UBC (Cat. # Hs00824723_m1) (Applied Biosystems, Foster City, CA).
Immunoblot Analysis. Primary human hepatocytes were maintained in 6-well plates and used the next day. Cells were treated with IL-1β or CDCA (Sigma) as indicated with or without pre-incubation with 50 μM of the JNK specific inhibitor SP600125 for one hr (Calbiochem, San Diego, CA). Cell lysates were isolated for immunoblot analysis using antibody against HNF4α, FTF or actin (Santa Cruz Biotech, Santa Cruz, CA).
Transient Transfection Assay. HepG2 cells were grown to ∼80% confluence in 24-well tissue culture plates. Human CYP7A1/Luciferase reporter plasmids (ph-298, 1 μg) and its mutants were transfected into HepG2 cells in transient transfection assay as described previously (13). Cells were treated with IL-1β at concentrations and incubation periods as indicated. In some experiments, cells were incubated with 50 μM SP600125 for 1 hr before IL-1β was applied. Cells were harvested for assay of luciferase reporter activity using Luciferase Assay System (Promega). Luciferase activity was determined using a Lumat LB 9501 luminometer (Berthold Systems, Inc., Pittsburgh, PA), and normalized by dividing the relative light units (RLU) by β-galactosidase activity. Each assay was done in triplicate and individual experiments were repeated at least three times. Data are plotted as means ± standard deviation.
Co-Immunoprecipitation Assay (Co-IP). HepG2 cells were cultured in T150 flasks. The cells were treated with IL-1β (5 ng/ml) for the specified time periods and harvested for immunoprecipitation assay as described previously (10). Rabbit anti-HNF4α antibody (20 μg, Santa Cruz Biotechnology, CA) was used for co-immunoprecipitation assay. The immunoprecipitants (IP) were analyzed by immunoblot analysis using goat anti-HNF4α, anti-c-Jun or anti-phospho-c-Jun antibodies (Santa Cruz). Equal amount of the “Input” set aside from IP was also run on an SDS-polyacrylamide gel and immunoblotted with anti-Actin antibody (Santa Cruz).
Chromatin Immunoprecipitation Assay (ChIP). HepG2 cells were either grown in 100 mm culture dishes to 80% confluence and then treated with 10 ng/ml of IL-1β or 50 μM CDCA for 20 h, or grown to 50% confluence for transfection of a c-Jun expression plasmid or a HA-tagged PGC-1α plasmid using Lipofectamine 2000 (Invitrogen). Cell extracts were used for ChIP assays using a ChIP Assay kit (Upstate Cell Signaling Solutions, Lake Placid, NY) according to manufacturer’s protocol and as described previously (10). An antibody against HNF4α, c-Jun, or HA-tag (Santa Cruz) was used to immunoprecipitate DNA-protein complexes and a 391 bp DNA fragment (-432 to -41) was PCR amplified for 40 cycles and analyzed on a 1.5 % agarose gel as described previously (10).
Statistical analyses: Statistical analyses of treated vs. untreated controls were performed using Student’s t-test. A p<0.05 is considered statistically significant.
Results
Effect of IL-1β and CDCA on mRNA expression in primary human hepatocytes.
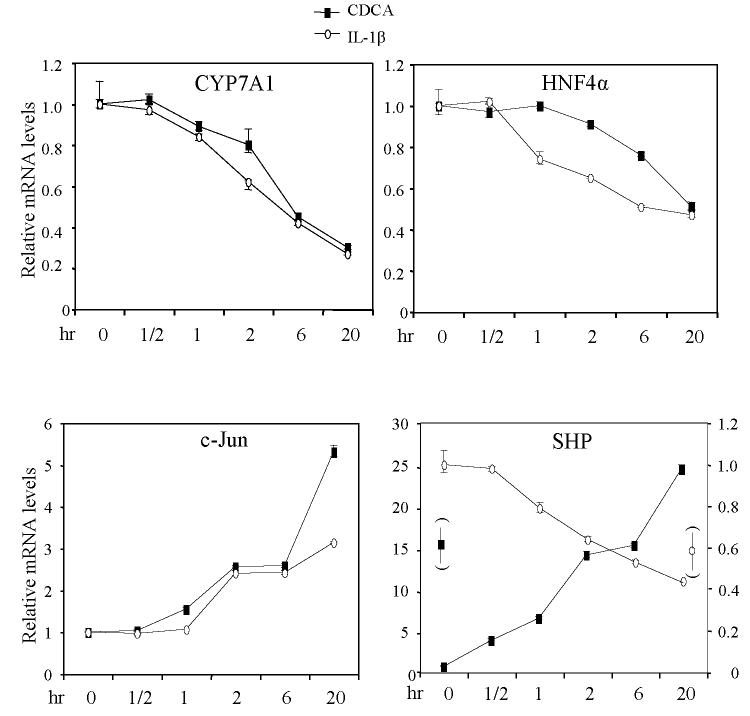
Previously we reported that IL-1β reduced CYP8B1 and CYP7A1 mRNA expression levels in primary human hepatocytes in a dose-dependent manner (13). Here we have studied the time course of IL-1β (10 ng/ml) effect on CYP7A1 expression levels in human primary hepatocytes by quantitative real time PCR assay. The effect of IL-1β on c-Jun, HNF4α and SHP mRNA were also assayed. As shown in Fig.1, IL-1β time-dependently reduced CYP7A1, HNF4α, and SHP mRNA expression, but induced c-Jun mRNA expression levels in primary human hepatocytes. On the other hand, a FXR ligand CDCA (50 μM) time-dependently reduced CYP7A1 and HNF4α, but induced SHP and c-Jun mRNA expression levels in primary human hepatocytes. The opposite effect of CDCA on CYP7A1 and SHP mRNA expression is expected since SHP is a negative regulator induced by bile acids to inhibit CYP7A1 expression (3). In contrast, IL-1β inhibited both SHP and CYP7A1 mRNA expression suggesting that IL-1β inhibition of CYP7A1 is independent of SHP. The parallel inhibition of HNF4α and CYP7A1 mRNA expression by both IL-1β and CDCA suggests that the IL-1β signaling and SHP pathway inhibit HNF4α and results in inhibiting CYP7A1. IL-1β and CDCA induce c-Jun mRNA expression and inhibit CYP7A1 suggests a hypothesis that both CDCA and IL-1β signaling converge to induce c-Jun which inhibits CYP7A1 expression.
Figure 1.
Effect of IL-1β and CDCA on mRNA expression in human primary hepatocytes. Human primary hepatocytes were treated with IL-1β (10 ng/ml) or CDCA (50 μM) for time periods indicated. Quantitative Real time PCR was performed as described in Materials and Methods. The relative mRNA levels of CYP7A1, HNF4α, SHP and c-Jun were calculated with respect to the UBC mRNA. The mRNA expression at 0 hr (un-treated) was set as 1. The error bars represent the standard deviation of from three different experiments. N=3.
Effect of IL-1β and CDCA on HNF4α protein expression in human primary hepatocytes.
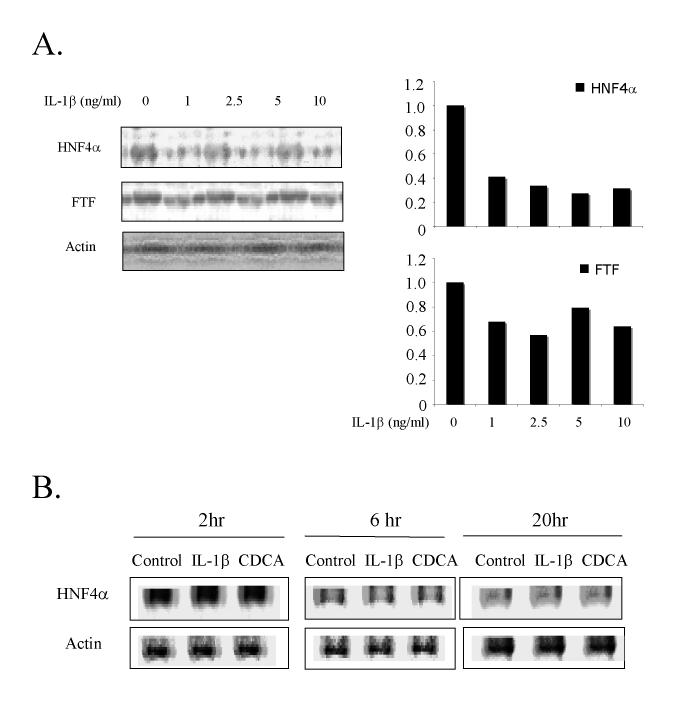
Previously we reported that IL-1β reduced HNF4α protein expression in HepG2 cells (13). We studied the effect of IL-1β on HNF4α protein expression in primary human hepatocytes using immunoblot analysis. Fig. 2A shows that HNF4α protein levels were strongly reduced in primary human hepatocytes after a 20 hr incubation with IL-1β at concentrations as low as 1 ng/ml. In contrast, the expression of FTF that binds to an overlapping site with HNF4α on human CYP7A1 promoter was reduced by IL-1β to a much less extent. A quantitative measurement of the band intensity shows that IL-1β reduced HNF4α and FTF protein by about 70% and 30%, respectively (Fig.2A, right panel). The time courses of the inhibitory effect of IL-1β (5 ng/ml) and CDCA (25μM) on HNF4α protein expression in primary human hepatocytes were similar (Fig. 2B). Thus IL-1β and CDCA inhibited HNF4α protein and mRNA expression levels in parallel (13, 25).
Figure 2.
Effect of IL-1β and CDCA on HNF4α protein expression in human primary hepatocytes. A. Human primary hepatocytes were treated with increasing concentrations of IL-1β for 20 hrs. Left panel: western blotting of HNF4α and FTF. Right panel: quantitative analysis of the western blotting on the left panel. HNF4α and FTF protein levels were normalized to actin levels. Controls are set as “1”. B. Human primary hepatocytes were treated with either IL-1β (5 ng/ml) or CDCA (25 μM) for 2, 6 and 20 hrs, respectively. HNF4α and FTF protein were detected by western blotting analysis. Actin was blotted as an internal standard to normalize the total protein amount in each sample.
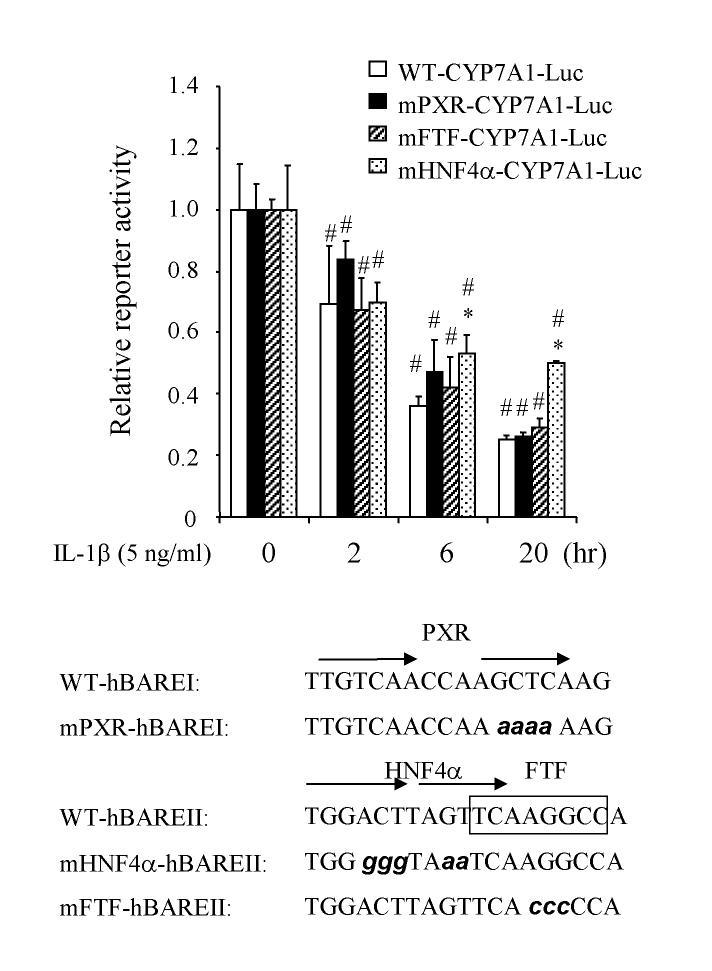
Effect of IL-1β on human CYP7A1 gene transcription. We then studied the effect of IL-1β on CYP7A1 gene transcription by transient transfection assay of a human CYP7A1-Luciferase reporter (ph-298/Luc) in HepG2 cells. We performed the IL-1β dose response and time course of inhibition of CYP7A1 reporter activity. IL-1β (5 ng/ml) significantly suppressed human CYP7A1 reporter activity as early as 2 hr, inhibited more than 60 % activity at 6 hrs and reached a maximal inhibition of ∼70% at 20 hrs (Fig 3, open bar). We have reported previously that nuclear receptors HNF4α, FTF and PXR bind human CYP7A1 promoter and regulate its gene transcription (10, 25, 26). To test if any of these nuclear receptors is involved in the IL-1β suppression of human CYP7A1, mutations were introduced into each of these nuclear receptor binding sites in the human CYP7A1/luciferase reporter construct (ph-298/Luc). Previously electrophoretic mobility shift assays have shown that the same mutations abolish their respective nuclear receptor binding (10). The mutant reporters were transiently transfected into HepG2 cells and the effect of IL-1β on these reporter activities were tested. It should be noted that the relative reporter activity of wild type and mutant reporters were drastically reduced by IL-1β in a time-dependent manner, apparently because IL-1β reduces endogenous HNF4α in HepG2 cells (Fig. 3). Mutations of the HNF4α binding site significantly prevented the IL-1β suppression of CYP7A1 reporter at 6 hr and 20 hr. However, neither the PXR binding site nor the FTF binding site mutation affected the IL-1β suppression of human CYP7A1. The decrease of reporter activities of wild type and mutant reporters is likely due to IL-1β inhibition of endogenous HNF4α and induction of cJun as shown in Fig 1. These results suggest that HNF4α may play a specific role in mediating the IL-1β suppression of human CYP7A1 gene transcription.
Figure 3.
Effect of IL-1β on human CYP7A1 reporter activity. Wild type human CYP7A1 luciferase reporter (WT-CYP7A1-Luc) and mutant reporters with PXR, FTF or HNF4α binding site mutated (mPXR-CYP7A1-Luc, mFTF-CYP7A1-Luc, mHNF4α-CYP7A1-Luc) were transfected into HepG2 cells and assayed for luciferase reporter activity. Cells were treated with IL-1β (5 ng/ml) for the time indicated. The relative luciferase activities of reporter plasmid assays without IL-1β treatment were set as 1. The basal reporter activity (RLU/β-gal) for WTCYP7A1-Luc, mPXR-CYP7A1-Luc, mFTF-CYP7A1-Luc and mHNF4α-CYP7A1-Luc are 8 χ 106, 6.7 χ 105, 3 χ 105, and 5 χ 104, respectively. The error bars represent the standard deviation from the mean of triplicate assays of an individual experiment. N=3. Sequences of mutations introduced into human CYP7A1 promoter/luciferase construct ph-298/luc are shown at the bottom. *, statistically significant (P<0.05): mutant vs. wild type at same time point; #, statistically significant (P<0.05): treatment vs. control of 0 h.
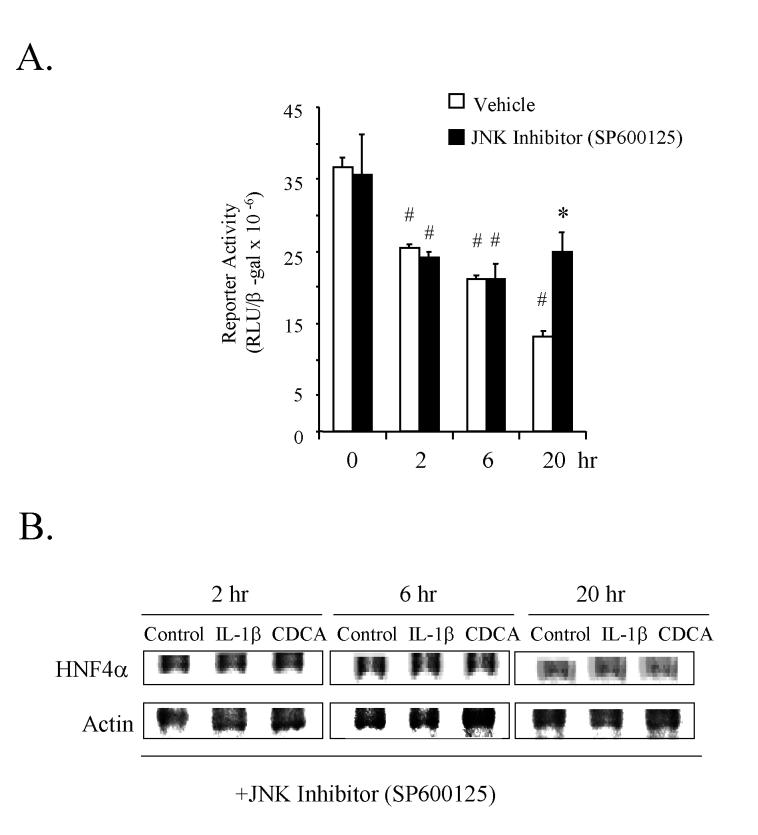
JNK-specific inhibitor prevents the IL-1β and CDCA suppression of the human CYP7A1 gene transcription and HNF4α expression. IL-1β binds to its cell surface receptor and activates the MAPK signaling pathways including JNK, ERK and p38MAPK pathways (27). Several studies have indicated that the JNK pathway activated by cytokines during acute phase response suppresses CYP7A1 gene transcription (7, 20). We demonstrated previously that JNK inhibitor SP600125, but not ERK and p38MAPK inhibitors blocked IL-1β inhibition of HNF4α expression (13). To test the role of JNK in the IL-1β suppression of the human CYP7A1 gene, we pretreated HepG2 cells with a JNK-specific inhibitor (SP600125, 50 μM) 1hr prior to IL-1β (5 ng/ml) treatment in a transient transfection assay. As shown in Fig. 4A, SP600125 treatment prevented IL-1β inhibition of CYP7A1 reporter activity indicating that JNK played a major role in mediating the IL-1β suppression of human CYP7A1. SP600125 completely blocked IL-1β and partially blocked CDCA inhibition of HNF4α protein expression in human primary hepatocytes (Fig. 4B), when compared to non-JNK inhibitor treated controls (Fig.2B), which were performed at the same time using the same primary hepatocyte preparation. These results suggest that the JNK pathway mediates IL-1β and CDCA down regulation of the CYP7A1 gene by inhibiting HNF4α expression.
Figure 4.
Effect of a JNK inhibitor on IL-1β suppression of human CYP7A1 reporter activity and HNF4α protein expression in human primary hepatocytes. A. Human CYP7A1 promoter/luciferase construct ph-298/luc (1μg) was transfected into HepG2 cells. Cells were pretreated with a JNK-specific inhibitor SP600125 (50 μM) for 1 hr before IL-1β (5 ng/ml) was added and cells were incubated for the time indicated. The error bars represent the standard deviation from the mean of triplicate assays of an individual experiment. N=3. *, Statistically significant (P<0.05): JNK inhibitor treatment vs. vehicle at 20h; #, Statistically significant (P<0.05): treatment vs. control of 0 h. B. Same human primary hepatocytes as in Fig. 2B were also pretreated with 50 μM SP600125 for 1 hr before being incubated with IL-1β (5 ng/ml) or CDCA (25μM) for the time indicated. Immunoblotting analysis was performed as in Fig. 2 in parallel.
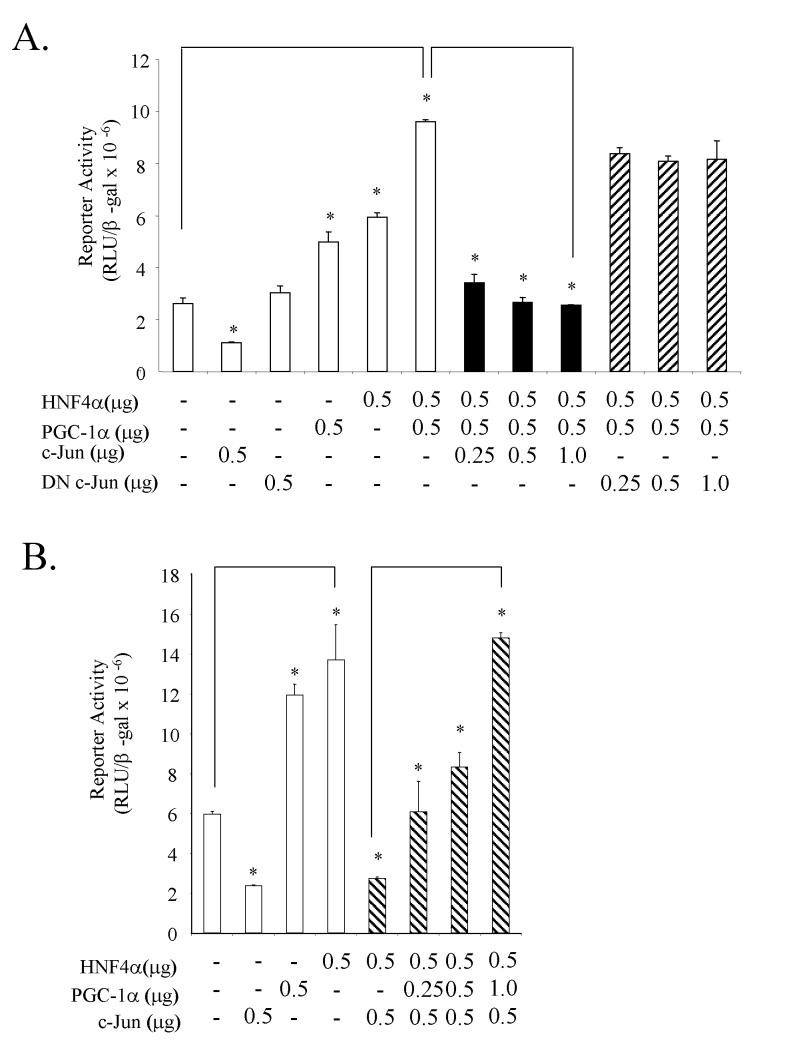
Effect of c-Jun on CYP7A1 gene transcription. The marked up regulation of c-Jun mRNA levels seen on both IL-1β and CDCA treatments led us to investigate if this factor played a role in the inhibition of CYP7A1 gene transcription. In transient transfection assays using the human CYP7A1/luciferase reporter (ph-298/Luc) in HepG2 cells, over expression of c-Jun led to 80% suppression of the CYP7A1 reporter activity (Fig. 5A), while over expression of a dominant negative c-Jun (DN c-Jun) protein showed no effect on the CYP7A1 reporter activity. HNF4α and PGC-1α synergistically stimulate CYP7A1 reporter activity. Transfection of c-Jun markedly inhibited CYP7A1 reporter activity activated by HNF4α and PGC-1α in a dose-dependent manner, while DN c-Jun had no effect on HNF4α and PGC-1α activation of CYP7A1. Fig. 5B shows that c-Jun inhibition of HNF4α stimulation of CYP7A1 reporter activity could be reversed by PGC-1α in a dose-dependent manner. These results suggest that c-Jun may prevent the HNF4α and PGC-1α co-activation of the CYP7A1 and result in inhibition of CYP7A1 gene transcription.
Figure 5.
Effect of cJun on PGC-1α and HNF4α co-activation of human CYP7A1 promoter activity- Human CYP7A1 promoter/luciferase construct (ph-298/luc, 1μg) was transfected into HepG2 cells. The expression plasmids for cJun, dncJun, PGC-1α and HNF4α were cotransfected in the concentrations indicated in A and B. The error bars represent the standard deviation from the mean of triplicate assays of an individual experiment. N=3. *, statistically significant (P<0.05).
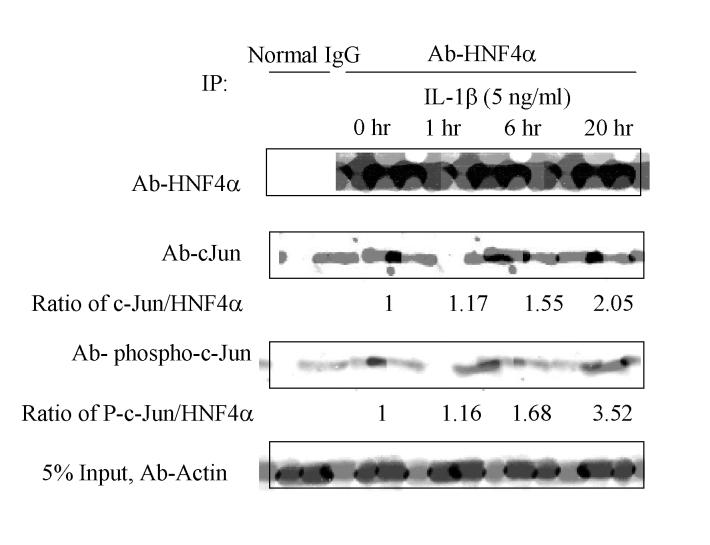
HNF4α interacts with c-Jun. Since c-Jun inhibits HNF4α stimulation of the CYP7A1 gene, we postulated that these two factors might interact with each other. We thus performed co-IP assays using HepG2 cells treated with IL-1β (5 ng/ml) and immunoprecipitated the cell extracts with anti-HNF4α antibody. An antibody against HNF4α, c-Jun or phospho-c-Jun was then used to detect HNF4α, c-Jun or phospho-c-Jun in the IP complexes. As expected HNF4α protein levels were decreased by IL-1β (5 ng/ml) in a time-dependent manner (Fig. 6). Antibodies against c-Jun and phospho-c-Jun detected c-Jun and phospho-c-Jun in IP complexes, indicating that both c-Jun and phospho-c-Jun interact with HNF4α. Semi-quantitative analysis of the ratio of c-Jun/HNF4α levels after normalizing with the actin levels showed that IL-1β treatment increased the amount of c-Jun (2-fold) associated with HNF4α, even though the HNF4α levels were decreased by IL-1β. Phosphorylation state of c-Jun associated with HNF4α is also increased by IL-1β. This is expected since IL-1β is known to activate JNK to phosphorylate cJun.
Figure 6.
Co-immunoprecipitation assay of HNF4α and c-Jun interaction-HepG2 cell extracts treated with IL-1β (5 ng/ml) for the time indicated were immunoprecipitated with rabbit anti-HNF4α antibody as described in Materials and Methods. Immunobolot analysis was performed with goat anti-HNF4α, anti-c-Jun and anti-phospho c-Jun antibodies. Five percent of the cell lysate was immunoblotted with anti-actin antibody as internal control. Rabbit non-immune IgG was used as a negative control. The ratios of c-Jun to HNF4α and phospho-c-Jun to HNF4α are indicated below the panels.
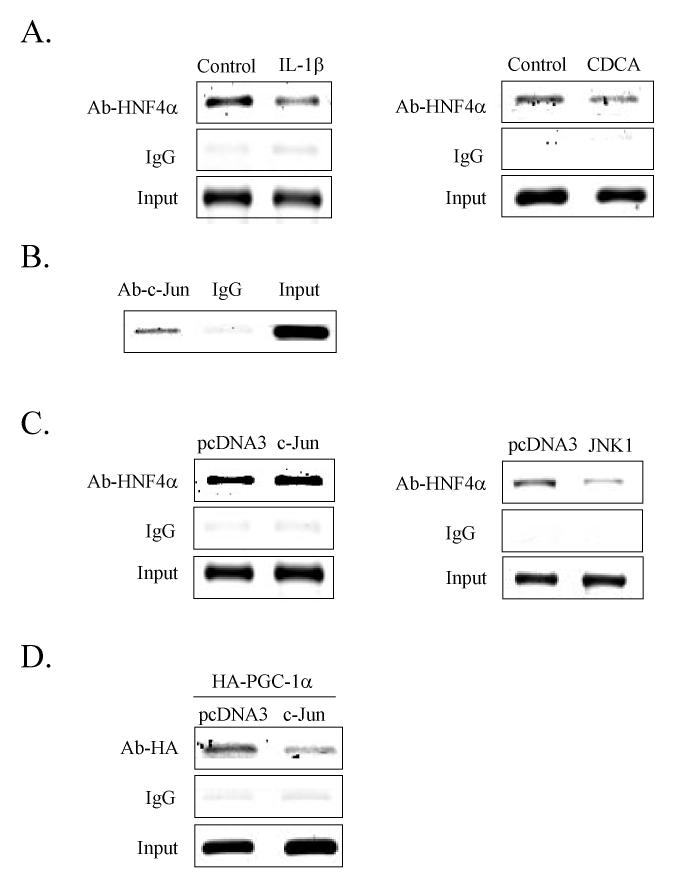
Effect of c-Jun on the recruitment of HNF4α and PGC-1α to the CYP7A1 chromatin. ChIP assay was used to study the effect of IL-1β and CDCA on HNF4α recruitment of PGC-1α to the CYP7A1 chromatin. This cell-based assay detects transcription factors bound to chromatin either directly to DNA or indirectly via interaction with other DNA-bound transcription factors. An antibody against HNF4α was used to immunoprecipitate HepG2 extracts for PCR amplification of a 391 fragment containing the HNF4α binding site in the human CYP7A1 promoter. As shown in Fig. 7A, IL-1β (10 ng/ml) or CDCA (50 μM) treatment decreased the amount of HNF4α protein associated with the CYP7A1 chromatin. These results are likely to be due to both IL-1β mediated inhibition of HNF4α protein levels and phosphorylation of HNF4α, which reduces its DNA binding activity (13). To further test if IL-1β induced c-Jun may interfere with HNF4α DNA binding activity and thus contribute to the decreased HNF4α amount in CYP7A1 chromatin, we first used a c-Jun antibody for ChIP assay to detect c-Jun in the CYP7A1 chromatin. Fig.7B shows that that c-Jun is associated with the CYP7A1 chromatin that contains an HNF4α binding site in BARE-II region. The association of c-Jun to the CYP7A1 chromatin is likely via an interaction with HNF4α because c-Jun does not bind to this DNA fragment (data not shown).
Figure 7.
Chromatin immunoprecipitation assay of HNF4α, PGC-1α, and c-Jun binding to the CYP7A1 chromatin. A. Anti-HNF4α antibody was used to precipitate chromatin from HepG2 cells treated with IL-1β (10 ng/ml) or CDCA (50 μM). B. Anti-c-Jun antibody was used to immunoprecipitate chromatin. C. HepG2 cells were transfected with cJun or JNK1 expression plasmid or pcDNA3 empty vector, and anti-HNF4α antibody was used to immunoprecipitate chromatin. D. HepG2 cells were transfected with HA-PGC-1α, and co-transfected with a c-Jun expression plasmid or pcDNA3 empty vector. A 391 bp fragment containing the BAREI and BAREII regions of the CYP7A1 promoter was PCR amplified and analyzed on a 1.5% agarose gel.
We then over expressed c-Jun or JNK1 in HepG2 cells and performed ChIP assays to determine if c-Jun or JNK1 affects HNF4α binding to CYP7A1 chromatin. Interestingly, c-Jun did not affect HNF4α binding to the CYP7A1 chromatin (left panel, Fig. 7C). On the other hand, JNK1 reduced HNF4α binding to chromatin likely because JNK1 phosphorylated HNF4α to reduce its DNA binding activity (right panel, Fig. 7C). Since PGC-1α interacts with HNF4α to stimulate human CYP7A1, we tested if c-Jun may inhibit CYP7A1 by blocking PGC-1α interaction with HNF4α. Because HepG2 cells express very low level of PGC-1α, we over-expressed HA-tagged PGC-1α in HepG2 cells. The HA-PGC-1α expressing cells were also transfected with a c-Jun expression plasmid and immunoprecipitated with an anti-HA antibody for ChIP assay. Fig. 7D shows that PGC-1α was associated with the CYP7A1 chromatin. Since PGC-1α is a co-activator that does not bind to DNA, HNF4α bound to the CYP7A1 chromatin must have recruited PGC-1α to the CYP7A1 chromatin (10). Over-expression of c-Jun markedly reduced HNF4α recruitment of PGC-1α to the chromatin. Consistent with the results shown in Fig. 5, these data indicate that c-Jun may block HNF4α recruitment of PGC-1α to the CYP7A1 chromatin, and result in the inhibition of CYP7A1 gene transcription.
Discussion
In this study we demonstrated that HNF4α is the down stream target of the JNK/c-Jun pathway that mediates cytokine inhibition of CYP7A1. We have shown that both IL-1β and CDCA inhibit CYP7A1 and HNF4α mRNA, but induce c-Jun mRNA expression in primary human hepatocytes. However, CDCA induces whereas IL-1β inhibits SHP mRNA expression. Therefore, IL-1β inhibition of CYP7A1 is independent of the FXR/SHP pathway. It appears that the JNK/c-Jun pathway is activated by bile acids, cytokines, protein kinase C, and maybe also FGF19, which are all converged to c-Jun to inhibit the CYP7A1 gene.
In this study, we provide several evidences that demonstrate the interaction of c-Jun with HNF4α. c-Jun has been suggested to interact with many nuclear receptors as either a co-activator or a co-repressor in a cell- and gene-specific manner. It has been reported that c-Jun inhibits the nuclear receptor transactivating activity by blocking nuclear receptor interactions with co-activators or general transcription factors (28, 29). Our data suggest that c-Jun does not block HNF4α binding to the CYP7A1 chromatin. Instead, cJun may interfere with PGC-1α interaction with HNF4α and results in inhibiting the CYP7A1 gene. Currently, it is not clear if phosphorylation state of c-Jun affects its interaction with HNF4α and its inhibition on CYP7A1. Our ChIP assay shows that overexpression of JNK1 reduces HNF4α binding to the CYP7A1 chromatin suggesting that JNK1 phosphorylates cJun and/or HNF4α and blocks HNF4α binding to chromatin. However, overexpression of c-Jun is sufficient to inhibit CYP7A1 reporter activity. This and our previous studies demonstrate that bile acids and cytokines inhibit HNF4α trans-activation of the CYP7A1 gene by inhibiting HNF4α gene transcription and its mRNA and protein expression levels (13, 25). In addition, JNK directly phosphorylates HNF4α (13). It has been reported that protein kinase A, extra cellular regulated kinase (ERK) and AMP-activated kinase (AMPK) phosphorylated HNF4α (30-32) reduce DNA-binding and trans-activation activity of HNF4α (33).
Blocking JNK signaling by JNK inhibitor or mutations of HNF4α binding site only partially blocked IL-1β inhibition of CYP7A1. These results indicate that additional mechanism may exist to mediate IL-1β inhibition of CYP7A1, and factors other than HNF4α may serve as downstram target in IL-1β inhibition of CYP7A1. This is expected since IL-1β signaling is known to crosstalk with other signaling pathways and CYP7A1 is inhibited by multiple intracellular signals. In this study, we also show that FTF protein levels are reduced by IL-1β, however, in contrast to rat CYP7A1, FTF is a repressor of human CYP7A1 and may not play a significant role in mediating CYP7A1 inhibition. Future studies may identify other downstream factors that mediate IL-1β inhibition of human CYP7A1.
It seems intriguing that cytokines inhibit SHP but stimulate c-Jun, whereas CDCA induces both SHP and c-Jun expression. It is possible that during inflammation cytokines may suppress SHP as an adaptive response to prevent SHP from inhibiting nuclear receptor regulated gene transcription. On the other hand, cytokines activate the JNK/c-Jun pathway, which may be the predominant pathway for suppressing CYP7A1 and protecting against bile acid toxicity during cholestasis. The FXR/SHP pathway requires protein synthesis and may be the major pathway for bile acid feedback regulation of bile acid synthesis under normal physiological conditions.
It is known that during inflammation, serum triglycerides and cholesterol levels increase (34). The hypercholesterolemia during inflammation is caused by activation of the positive acute response gene, HMG-CoA reductase, which is the rate-limiting enzyme in de novo cholesterol synthesis (35). The decrease in CYP7A1 gene transcription in response to cytokines would also lead to hypercholesterolemia. During bile duct ligation and cytokine-induced cholestasis, sinusoidal sodium taurocholate co-transport peptide (Ntcp or Slc10a1), apical sodium-dependent bile acid transporter (Asbt or Slc10a2) and canalicular multi-drug resistant protein 2 (mrp2 or Abcc2) are inhibited and increased intrahepatic bile acid concentrations (15, 36, 37). On the other hand, sinusoidal mrp3 is induced as an adaptive response to cholestasis to excrete bile acids (38). In such conditions, the inhibition of bile acid synthesis may serve as an additional protective mechanism for the liver against accumulation of toxic bile acids in hepatocytes.
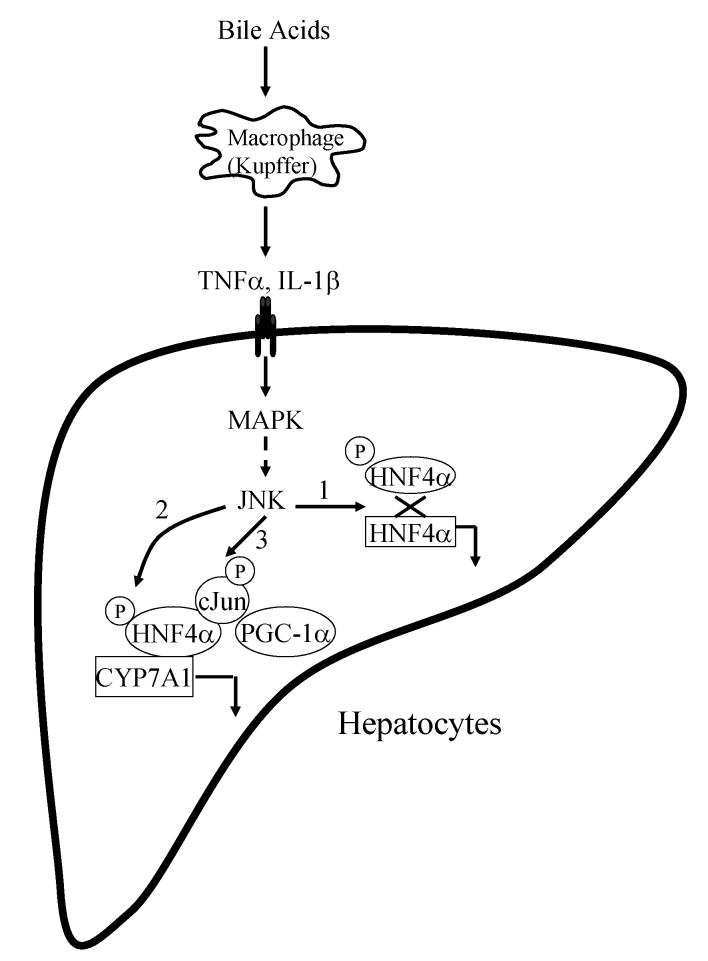
In summary, our study suggests that cytokines inhibit CYP7A1 gene transcription, in a SHP independent manner, mainly via the JNK pathway, leading to a decrease in HNF4α expression (Fig. 8). This study provides the first evidence that c-Jun expression is induced by cytokines and the c-Jun can interact with HNF4α. The interaction of c-Jun with HNF4α interferes with the PGC-1α recruitment by HNF4α thereby leading to suppression of CYP7A1 gene transcription (Fig. 8).
Figure 8.
Mechanisms of bile acid and cytokine inhibition of CYP7A1 gene transcription. Bile acids induce IL-1β synthesis and release from Kupffer cells. These cytokines activate JNK pathway in hepatocytes. The JNK pathway inhibits CYP7A1 via three possible mechanisms. 1) JNK inhibits HNF4α mRNA and protein expressions. 2) JNK phosphorylates HNF4α and reduces its DNA binding and trans-activation activity. 3) JNK induces c-Jun, which interacts with HNF4α and blocks HNF4α interaction with PGC-1α and results in inhibition of CYP7A1.
Acknowledgment:
This study was supported by NIH grants DK 44442 and DK58379.
Footnotes
- ChIP
- chromatin immunoprecipitation assay
- CDCA
- chenodeoxycholic acid
- Co-IP
- co-immunoprecipitation
- CYP7A1
- cholesterol 7α-hydroxylase
- FGF
- fibroblast growth factor
- FTF
- α-fetoprotein transcription factor
- FXR
- farnesoid X receptor
- HNF4α
- hepatocyte nuclear factor α
- IL-1β
- interleukin-1β
- JNK
- c-Jun N-terminal kinase
- PGC-1α
- peroxisome proliferator activated receptor γ co-activator 1α
- SHP
- small heterodimer partner
- TNFα
- tumore necrosis factor α
References:
- 1.Chiang JY. Regulation of bile acid synthesis. Front Biosci. 1998;3:D176–193. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 3.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 4.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 6.Stravitz RT, Rao YP, Vlahcevic ZR, Gurley EC, Jarvis WD, Hylemon PB. Hepatocellular protein kinase C activation by bile acids: implications for regulation of cholesterol 7 alpha-hydroxylase. Am J Physiol. 1996;271:G293–303. doi: 10.1152/ajpgi.1996.271.2.G293. [DOI] [PubMed] [Google Scholar]
- 7.De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- 8.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 11.Miyake JH, Wang SL, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha-hydroxylase. J Biol Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- 12.Feingold KR, Spady DK, Pollock AS, Moser AH, Grunfeld C. Endotoxin, TNF, and IL-1 decrease cholesterol 7 alpha-hydroxylase mRNA levels and activity. J Lipid Res. 1996;37:223–228. [PubMed] [Google Scholar]
- 13.Jahan A, Chiang JY. Cytokine regulation of human sterol 12alpha-hydroxylase (CYP8B1) gene. Am J Physiol Gastrointest Liver Physiol. 2005;288:G685–695. doi: 10.1152/ajpgi.00207.2004. [DOI] [PubMed] [Google Scholar]
- 14.Memon RA, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. In vivo and in vitro regulation of sterol 27-hydroxylase in the liver during the acute phase response. potential role of hepatocyte nuclear factor-1. J Biol Chem. 2001;276:30118–30126. doi: 10.1074/jbc.M102516200. [DOI] [PubMed] [Google Scholar]
- 15.Denson LA, Auld KL, Schiek DS, McClure MH, Mangelsdorf DJ, Karpen SJ. Interleukin-1beta suppresses retinoid transactivation of two hepatic transporter genes involved in bile formation. J Biol Chem. 2000;275:8835–8843. doi: 10.1074/jbc.275.12.8835. [DOI] [PubMed] [Google Scholar]
- 16.Feingold KR, Pollock AS, Moser AH, Shigenaga JK, Grunfeld C. Discordant regulation of proteins of cholesterol metabolism during the acute phase response. J. Lipid Res. 1995;36:1474–1482. [PubMed] [Google Scholar]
- 17.Hardardottir I, Moser AH, Memon R, Grunfeld C, Feingold K. Effects of TNF, IL-1, and the combination of both cytokines on cholesterol metabolism in Syrian hamsters. Lymphokine and Cytoline Research. 1994;13:161–166. [PubMed] [Google Scholar]
- 18.Grunfeld C, Pang M, Doerrler W, Shigenaga JK, Jensen P, Feingold KR. Lipids, lipoproteins, triglyceride clearance, and cytokines in human immunodeficiency virus infection and the acquired immunodeficiency syndrome. J Clin Endocrinol Metab. 1992;74:1045–1052. doi: 10.1210/jcem.74.5.1373735. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Zimmerman TL, Thevananther S, Lee HY, Kurie JM, Karpen SJ. Interleukin-1 beta-mediated suppression of RXR:RAR transactivation of the Ntcp promoter is JNK-dependent. J Biol Chem. 2002;277:31416–31422. doi: 10.1074/jbc.M204818200. [DOI] [PubMed] [Google Scholar]
- 20.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi H, Grambihler A, Canbay A, Bronk SF, Gores GJ. Bile acids up-regulate death receptor 5/TRAIL-receptor 2 expression via a c-Jun N-terminal kinase-dependent pathway involving Sp1. J Biol Chem. 2004;279:51–60. doi: 10.1074/jbc.M309476200. [DOI] [PubMed] [Google Scholar]
- 22.Stravitz RT, Vlahcevic ZR, Hylemon PB. Signal transduction pathways regulating cholesterol 7α-hydroxylase transcription by bile acids. Kluwer Acad. Pub.; Dordrecht, Boston, London: 1999. pp. 39–50. [Google Scholar]
- 23.Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J Biol Chem. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- 24.Wang DP, Stroup D, Marrapodi M, Crestani M, Galli G, Chiang JYL. Transcriptional regulation of the human cholesterol 7α-hydroxylase gene (CYP7A) in HepG2 cells. J. Lipid Res. 1996;37:1831–1841. [PubMed] [Google Scholar]
- 25.Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): Roles of hepatocyte nuclear factor 4α (HNF4α) in mediating bile acid repression. J Biol Chem. 2001;276:41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- 26.Stroup D, Chiang JYL. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7α-hydroxylase gene (CYP7A) J. Lipid Res. 2000;41:1–11. [PubMed] [Google Scholar]
- 27.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 28.Pfahl M. Nuclear receptor/AP-1 interaction. Endocr Rev. 1993;14:651–658. doi: 10.1210/edrv-14-5-651. [DOI] [PubMed] [Google Scholar]
- 29.Li LA, Chiang EF, Chen JC, Hsu NC, Chen YJ, Chung BC. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol Endocrinol. 1999;13:1588–1598. doi: 10.1210/mend.13.9.0349. [DOI] [PubMed] [Google Scholar]
- 30.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol. Cell. Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy S, Yang W, Taylor DG, Shen X, Oxender D, Kust G, Leff T. Mitogen-activated protein kinase regulates transcription of the ApoCIII gene. Involvement of the orphan nuclear receptor HNF4. J Biol Chem. 1999;274:33050–33056. doi: 10.1074/jbc.274.46.33050. [DOI] [PubMed] [Google Scholar]
- 32.Hong YH, Varanasi US, Yang W, Leff T. AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J Biol Chem. 2003 doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
- 33.Ktistaki E, Ktistaki T, Papadogeorgaki E, Talianidis I. Recruitment of hepatocyte nuclear factor 4 into specific intranuclear compartiments depends on tyrosine phosphorylation that affects its DNA-binding and transactivation potential. Proc. Natl. Acad. Sci. USA. 1995;92:9876–9880. doi: 10.1073/pnas.92.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardardottir I, Grunfeld C, Feingold KR. Effects of endotoxin and cytokines on lipid metabolism. Curr Opin Lipidol. 1994;5:207–215. doi: 10.1097/00041433-199405030-00008. [DOI] [PubMed] [Google Scholar]
- 35.Feingold KR, Hardardottir I, Memon R, Krul EJ, Moser AH, Taylor JM, Grunfeld C. Effect of endotoxin on cholesterol biosynthesis and distribution in serum lipoproteins in Syrian hamsters. J Lipid Res. 1993;34:2147–2158. [PubMed] [Google Scholar]
- 36.Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. Tumor necrosis factor alpha-dependent up-regulation of Lrh-1 and Mrp3(Abcc3) reduces liver injury in obstructive cholestasis. J Biol Chem. 2003;278:36688–36698. doi: 10.1074/jbc.M304011200. [DOI] [PubMed] [Google Scholar]
- 37.Denson LA, Bohan A, Held MA, Boyer JL. Organ-specific alterations in RAR alpha:RXR alpha abundance regulate rat Mrp2 (Abcc2) expression in obstructive cholestasis. Gastroenterology. 2002;123:599–607. doi: 10.1053/gast.2002.34758. [DOI] [PubMed] [Google Scholar]
- 38.Chen F, Ma L, Sartor RB, Li F, Xiong H, Sun AQ, Shneider B. Inflammatory-mediated repression of the rat ileal sodium-dependent bile acid transporter by c-fos nuclear translocation. Gastroenterology. 2002;123:2005–2016. doi: 10.1053/gast.2002.37055. [DOI] [PubMed] [Google Scholar]