Abstract
The endmost chromosome I ORF is silenced by a natural telomere position effect. YAR073W/IMD1 was found to be transcribed at much higher levels in sir3 mutants and when its adjacent telomere was removed from it. These results suggest that telomeres play a role in silencing actual genes.
IN the yeast Saccharomyces cerevisiae subtelomeric sequences are distinctly different from the rest of the chromosomal DNA because they are mostly repetitive and contain a relatively low density of ORFs. Subtelomeric sequences compose ∼7% (∼25 kb × 32 ends/12,000 kb) of the genome and are largely made up of sequences termed W′, Y′, or X. The number, distribution, and arrangement of these sequences vary on each chromosome and in different strains. W′ and Y′ sequences contain some short ORFs but they are likely to be pseudogenes. Other subtelomeric ORFs belong to repetitive gene families and appear to be nonessential for routine growth in the laboratory (Pryde and Louis 1997). Subtelomeric sequences from S. cerevisiae have been proposed to play a role in preventing nonfunctional meiotic crossovers near chromosome ends (Barton et al. 2003). Otherwise, their precise function is not known. Subtelomeric DNA from S. cerevisiae has been compared to the telomeric heterochromatin of higher organisms, which also contains repetitive sequences and has few active genes.
Most subtelomeric genes or ORFs either are not expressed at all or are expressed at relatively low levels during routine growth in the laboratory. A few such as those from the MAL, SUC, and PHO gene families are abundantly expressed but only under certain conditions (Pryde and Louis 1997). When these genes are repressed, the amount of RNA produced from any individual gene is probably quite low. It is possible that low transcript levels from subtelomeric genes are in some part due to the well-characterized telomere position effect (TPE) that was first revealed by an epigenetic repression of genes inserted within or near telomeres (Gottschling et al. 1990; Pirrotta and Gross 2005). About 60 genes show increased transcript levels in the presence of mutations that alleviate TPEs such as sir2, sir3, and rap1 and many of these genes are relatively close to a telomere (Wyrick et al. 1999; Vega-Palas et al. 2000; Halme et al. 2004). Nevertheless, a true TPE dependent on actual telomere proximity has never been demonstrated for a naturally occurring yeast gene other than a small effect on HMR, the silenced copy of the mating-type locus on the right arm of chromosome III (Thompson et al. 1994).
The three endmost ORFs on the right arm of chromosome I are paralogous to three ORFs near the right end of chromosome VIII and have almost identical DNA sequences. The most prominent differences between the two gene sets are the chromosome VIII sequences are ∼6 kb farther from the telomere than the chromosome I sequences and the chromosome VIII sequences are transcribed while the chromosome I genes appear silent. These observations led to the suggestion that the chromosome I genes could be repressed by a TPE (Barton et al. 1997). The endmost chromosome I ORFs are YAR073W/IMD1, encoding an IMP dehydrogenase ortholog (Hyle et al. 2003); YAR071W/PHO11, encoding an inducible acid phosphatase (Kaback et al. 1989; Shnyreva et al. 1996); and YAR068W, encoding a nonessential putative membrane protein of unknown function that when overexpressed produces a drug-resistance phenotype (Barton et al. 1997; Launhardt et al. 1998). In this report we demonstrate that the endmost chromosome I ORF, YAR073W/IMD1, is indeed silenced by its telomere, suggesting that telomeres play a role in silencing actual genes.
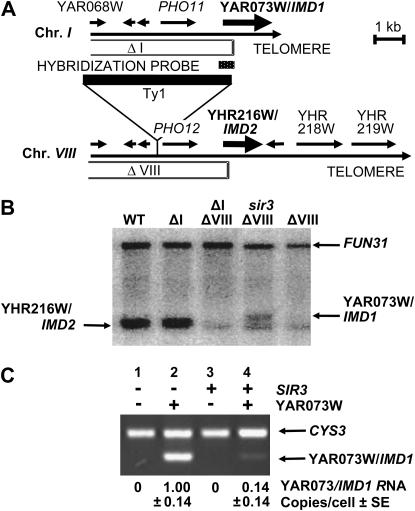
To determine whether the three endmost chromosome I ORFs were subject to a TPE, their expression was examined in sir3 mutant and wild-type control strains that contained a deletion of the paralogous chromosome VIII genes (Table 1; Barton et al. 1997). Experiments using blot hybridization revealed a YAR073W/IMD1 RNA species only in sir3 mutant strains. Quantitation of the autoradiogram using FUN31 RNA as an internal standard (Barton and Kaback 1994) indicated that this RNA was present at a rate of approximately one copy per cell. Interestingly, this RNA was ∼50 nucleotides larger than the paralogous YHR216 transcript (Figure 1B). A low level of the smaller RNA species was still present in the chromosome VIII deletions. This previously observed species (Barton et al. 1997) does not appear to be affected by sir3 and is probably transcribed from the additional orthologous copies of this ORF found on chromosomes XII and XIII. A quantitative RT–PCR assay specific for YAR073W/IMD1 (Barton et al. 1993) was used to show that sir3 mutant strains contained approximately sevenfold more YAR073W/IMD1 RNA than the wild-type controls (Figure 1C). The controls also show that this transcript could not be detected when chromosome I sequences were deleted. Similar assays were carried out for YAR071/PHO11 or YAR068W (data not shown) using the same chromosome VIII deletion strains but there was no evidence for any SIR3-dependent effects. However, results for YAR071/PHO11 were inconclusive since multiple remaining acid phosphatase genes could have obscured any effect. Therefore, at present there is evidence that only the endmost ORF, YAR073W/IMD1, is repressed by SIR3.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| D139-11B | MATα ade1 his3-11,15 leu2-3,112 trp1 can1 | J. Szostak |
| DK330-2B | MATahis3-11,15 leu2-3,112 ura3-1 can1 | Congenic to D139-11B |
| DK392 | MATa/MATα ADE1/ade1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 TRP1/trp1 URA3/ura3-1 can1/can1 | DK330-2B × D139-11B (Barton and Kaback 1994) |
| DK420 | MATa/MATα ADE1/ade1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 TRP1/trp1 URA3/ura3-1 can1/can1 ΔI(LEU2)/WT | DK392 with ΔI |
| DK420-1D | MATaade1 his3-11,15 leu2-3,112 ΔI(LEU2) | |
| DK421 | MATa/MATα ADE1/ade1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 TRP1/trp1 URA3/ura3-1 can1/can1 ΔI(LEU2)/WT ΔVIII(HIS3)/WT | DK420 with ΔVIII |
| DK421-1A | MATα his3-11,15 leu2-3,112 trp1 ura3-1 can1 ΔVIII(HIS3) | |
| DK421-1Asir3 | MATα his3-11,15 leu2-3,112 trp1 ura3-1 can1 ΔVIII(HIS3)sir3∷TRP1 | |
| DK421-2D | MATaade1 his3-11,15 leu2-3,112 can1 ΔI(LEU2) ΔVIII(HIS3) | |
| DK423 | MATa/MATα ade1/ADE1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 TRP1/trp1 ura3-1/ura3-1 can1/can1 WT/ΔVIII(HIS3) | DK392-1D × DK421-1A |
| DK424 | MATa/MATα ade1/ADE1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1/TRP1 ura3-1/ura3-1 can1/can1 ΔVIII(HIS3)/ΔVIII(HIS3) | DK423-1A × DK423-2B |
| DK425-14A | MATα his3-11,15 leu2-3,112 ade1 can1 sir3∷TRP1 ΔI(LEU2) ΔVIII(HIS3) | Haploid spore from DK421-2D × DK421-1Asir3 |
S. cerevisiae strains were constructed, grown, sporulated, and transformed with DNA using standard procedures (Burke et al. 2000). All recombinant DNA procedures were carried out using Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) and plasmid pBluescriptII (Stratagene, San Diego) using standard procedures (Sambrook et al. 1989). All gene replacements and insertions were verified by either Southern blot hybridization (Southern 1975) or PCR (Mullis and Faloona 1987). Construction of Δ2LEU2, herein labeled ΔI for the chromosome I right-end deletion, and Δ4HIS3, herein labeled ΔVIII for the chromosome VIII right-end deletion, were previously described (Barton et al. 1997). These constructs were introduced sequentially into strain DK392, yielding strains DK420 and DK421, respectively, which were used to breed strains with appropriate genotypes. SIR3 was deleted by one-step gene replacement (Rothstein 1983) using plasmid pKL12 provided by R. Sternglanz (Stone et al. 1991) as described.
Figure 1.
YAR073W/IMD1 is repressed by SIR3. (A) Physical maps of the right end of chromosomes I and VIII. Arrows indicate open reading frames. Extent of deletion constructs that eliminate cross-hybridizing genes are noted (ΔI and ΔVIII). (B) YAR073W/IMD1 expression analyzed by blot hybridization of poly(A+) RNA extracted from cells carrying noted deletions and sir3. WT, wild-type strain D139-11B; ΔI, chromosome I deletion strain DK420-1D; ΔI ΔVIII, chromosomes I and VIII double-deletion strain DK421-2D; sir3ΔVIII, sir3∷trp1 chromosome VIII deletion strain DK421-1Asir3 strain; ΔVIII, chromosome VIII deletion strain DK421-1A. The ∼5-kb FUN31 transcript, present at approximately one copy per cell (Barton et al. 1997), was used as an abundance standard to quantitate YAR073W/IMD1 RNA. (C) RT–PCR analysis of YAR073W/IMD1 RNA in haploid strains deleted for the end of chromosome VIII. Relevant genotypes are shown above each lane. Lanes: 1, DK425-14A; 2, DK421-1Asir3; 3, DK421-2D; 4, DK421-1A. YAR073W/IMD1 transcript abundance (RNA copies per cell) was determined by RT–PCR using simultaneously amplified CYS3 RNA as a loading control (Barton et al. 1993). Nucleic acid isolation and manipulation were as described (Sambrook et al. 1989). Transcript analysis using blot hybridization was carried out as described (Barton and Kaback 1994). The FUN31 probe was a cloned 1.4-kb HindIII fragment (SGD chromosome I coordinates 122,392–122,832 and the YAR073W/IMD1 probe was a 0.4-kb KpnI–BglII fragment from λ4599 (Riles et al. 1993) [Saccharomyces Genome Database (SGD, http://www.yeastgenome.org) chromosome I coordinates 227,378–227,767]. RT–PCR was performed using the Superscript first strand synthesis system (Life Technologies, Rockville, MD) using poly(A+) RNA and AmpliTaq Gold (Roche Molecular Systems, Branchberg, NJ) according to the manufacturer's directions. Oligonucleotide primers were 5′-AGACTACAAGACCGCACTAGAT and 5′-TGGCACCCACCAACTTTGCAT for YAR073W/IMD1 and 5′-GGAAAACACCAAATTGGTCTG and 5′-CAAAAACACCAGAGGCCTCTC for CYS3. To provide specificity, the forward YAR073W/IMD1 primer sequence was absent from chromosome VIII due to the ΔVIII deletion and the reverse primer was from a part of the YAR073W ORF with the least homology to the orthologs on chromosomes XII and XIII. Reactions were run in a GeneAmp PCR System 2400 (Perkin-Elmer, Norwalk, CT) utilizing 24 cycles of 94° for 30 sec, 55° for 30 sec, and 72° for 1 min. Quantitation of blots and gels was carried out using a phosphorimager and Image Quant Software (Molecular Dynamics, Sunnyvale, CA) and serial dilutions to ensure that band intensities were in the linear range of the photo scanner. Four measurements were carried out on each band and the average ± the standard error is shown. CYS3 RNA showed virtually constant levels relative to template RNA added in all strains examined and was used as a loading control (Barton et al. 1993). RNA copies per cell are calculated from the amount of the YAR073W/IMD1 RNA level measured in the sir3 strain by blot hybridization (one copy/cell).
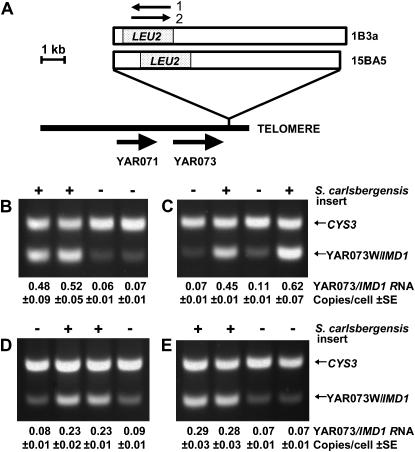
To examine whether YAR073W/IMD1 was controlled by a bona fide TPE, two different homeologous fragments from the middle of Saccharomyces carlsbergensis chromosome III, 7.0 and 8.4 kb in length, were inserted between YAR073W/IMD1 and its proximal telomere and transcript levels were determined by quantitative RT–PCR as above. LEU2, the selectable marker was introduced in both orientations in each insert to better ensure that its presence would not affect YAR073W/IMD1 expression and all strains lacked the paralogous chromosome VIII genes. The results showed that, when YAR073W/IMD1 was separated from its telomere, it produced three- to ninefold more RNA (Figure 2), an effect that was similar in magnitude to that observed in sir3 mutants. Quantitation of the results indicated that cells containing the S. carlsbergensis insert contained ∼0.3–0.6 copies of the YAR073W/IMD1 RNA/cell or slightly less than was observed in sir3 cells. However, the controls that did not contain inserts also were slightly lower than in the previous experiment. Consistent with these observations was a report that YAR073W/IMD1 RNA could only be detected when this gene was contained on a plasmid (Hyle et al. 2003). Thus, telomere proximity appears to be involved in the silencing of YAR073W/IMD1.
Figure 2.
Removal of the telomere from YAR073/IMD1 relieves repression. (A) Structure of the right end of chromosome I showing the location of the S. carlsbergensis DNA inserts 1B3a and 15BA5 that moved YAR073W/IMD1 from its proximal telomere. Arrows above indicate the LEU2 transcript direction for each numbered construct. (B–E) Analysis of YAR073W/IMD1 expression in haploid spores from strain DK424 transformants. Relative transcript abundance was determined by RT–PCR using CYS3 RNA as a loading control. B–E each show four spores from a single ascus from each heterozygous S. carlsbergensis DNA insert; two spores contain the insert (+) and two spores lack it (−). (B) Insert 1B3a-1. (C) Insert 1B3a-2. (D) Insert 15BA5-1. (E) Insert 15BA5-2. 1 and 2 indicate LEU2 orientation. Two BamHI fragments from chromosome III of S. carlsbergensis, 1B3a (7.3 kb) and 15BA5 (8.3 kb), provided by C. Newlon (Yang et al. 1999) were inserted adjacent to the telomere at position 229,850 (coordinates from SGD, http://www.yeastgenome.org) by one-step gene replacement (Rothstein 1983) using the chromosome I insert from plasmid pEL56H2 provided by Ed Louis as the target and a PCR-amplified S. cerevisiae (Mullis and Faloona 1987) 1.6-kb LEU2 gene with primer-encoded BglII ends as the selectable marker inserted at BglII sites within each S. carlsbergensis sequence. Fragment 15BA5 had a 1.6-kb BglII fragment deleted where LEU2 was inserted. In spite of repeated attempts and for unknown reasons, one-step gene replacements involving sequences near telomeres were unsuccessful using haploids. Accordingly, all constructs were introduced successfully in diploids and haploid segregants containing the inserted DNA obtained by sporulation and tetrad dissection. Quantitation of the PCR products was carried out as described in Figure 1. “Copies/cell RNA” was estimated from the CYS3 RNA normalized relative intensity of the PCR products using RT–PCR-amplified RNA from strain DK421-1Asir3 as an abundance standard that was run in parallel using the same gel (not shown).
YAR068W and YAR071/PHO11 were also examined but produced the same low level of transcript with and without the S. carlsbergensis insert (data not shown). Thus, the TPE appeared specific for YAR073W/IMD1.
On the basis of its apparent lack of expression and a sequence that either truncates or uses frameshifting to change the reading frame of the last 20% of the protein, it has been suggested that YAR073W/IMD1 is a pseudogene and cannot encode functional IMP dehydrogenase (Hyle et al. 2003). A functional IMD gene makes cells resistant to the antifungal mycophenolic acid, providing a simple assay that was carried out on strains where an IMD1 transcript could be detected (Hyle et al. 2003). We found that sir3 and SIR3+ control strains deleted for the end of chromosome VIII (ΔVIII), where the functional IMD2 gene is located, were equally sensitive to mycophenolic acid. Furthermore, all S. carlsbergensis insertion strains that removed YAR073W/IMD1 from its proximal telomere also were equally sensitive. Control strains containing the end of chromosome VIII were fully resistant as previously described (Hyle et al. 2003; data not shown). These results are consistent with previous studies that demonstrate that YAR073W/IMD1 does not encode this enzyme (Hyle et al. 2003). Nevertheless, it is possible that this sequence has a different but unknown function, especially in light of the finding that both the mRNA and its frameshifted open coding region are longer than that found for the active IMD2 gene.
Genome-array expression studies with different SIR mutants and histone depletion revealed that telomere-proximal genes were more likely than telomere-distal genes to be affected (Wyrick et al. 1999). In these studies, Wyrick et al. reported only a twofold sir3-dependent effect on YAR073W/IMD1, which was not considered significant. This discrepancy is likely due to either strain variations or uncertainties in measuring relatively low levels of RNA using arrays.
While it has been suggested that TPEs play a role in gene expression, it is also likely that many telomere-proximal genes are unaffected due to insulator elements that prevent the effects from spreading (Louis 1995). Nevertheless, HMR showed a modest telomere proximity effect and now YAR073W/IMD1 has been shown to have bona fide TPE. As there are now ∼60 telomere-proximal genes that are affected by various SIR gene mutants, it is likely that some of these genes are directly regulated by their nearby telomeres.
The function of subtelomeric DNA is still not entirely understood. In addition to its suggested structural role preventing nonfunctional crossing over during meiosis, these sequences could also provide an area of the genome where rarely needed genes and gene progenitors are efficiently prevented from being expressed, especially if their expression could be deleterious.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM053924) and the National Science Foundation (MCB0136278).
References
- Barton, A. B., and D. B. Kaback, 1994. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the genes in the FUN38–MAK16–SPO7 region. J. Bacteriol. 176: 1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, A. B., D. B. Kaback, M. W. Clark, T. Keng, B. F. Ouellette et al., 1993. Physical localization of yeast CYS3, a gene whose product resembles the rat gamma-cystathionase and Escherichia coli cystathionine gamma-synthase enzymes. Yeast 9: 363–369. [DOI] [PubMed] [Google Scholar]
- Barton, A. B., H. Bussey, R. K. Storms and D. B. Kaback, 1997. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: characterization of the 54 kb right terminal CDC15–FLO1–PHO11 region. Yeast 13: 1251–1263. [DOI] [PubMed] [Google Scholar]
- Barton, A. B., Y. Su, J. Lamb, D. Barber and D. B. Kaback, 2003. A function for subtelomeric DNA in Saccharomyces cerevisiae. Genetics 165: 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Halme, A., S. Bumgarner, C. Styles and G. R. Fink, 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Hyle, J. W., R. J. Shaw and D. Reines, 2003. Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J. Biol. Chem. 278: 28470–28478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback, D. B., H. Y. Steensma and P. de Jonge, 1989. Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86: 3694–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launhardt, H., A. Hinnen and T. Munder, 1998. Drug-induced phenotypes provide a tool for the functional analysis of yeast genes. Yeast 14: 935–942. [DOI] [PubMed] [Google Scholar]
- Louis, E. J., 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573. [DOI] [PubMed] [Google Scholar]
- Mullis, K. B., and F. A. Faloona, 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155: 335–350. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V., and D. S. Gross, 2005. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol. Cell 18: 395–398. [DOI] [PubMed] [Google Scholar]
- Pryde, F. E., and E. J. Louis, 1997. Saccharomyces cerevisiae telomeres. A review. Biochemistry 62: 1232–1241. [PubMed] [Google Scholar]
- Riles, L., J. E. Dutchik, A. Baktha and M. V. Olsen, 1993. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics 134: 81–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. J., 1983. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Habor Laboratory Press, Cold Spring Habor, NY.
- Shnyreva, M. G., E. V. Petrova, S. N. Egorov and A. Hinnen, 1996. Biochemical properties and excretion behavior of repressible acid phosphatases with altered subunit composition. Microbiol. Res. 151: 291–300. [DOI] [PubMed] [Google Scholar]
- Southern, E. M., 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98: 503–517. [DOI] [PubMed] [Google Scholar]
- Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks and R. Sternglanz, 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 11: 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. S., L. M. Johnson and M. Grunstein, 1994. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol. Cell. Biol. 14: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Palas, M. A., E. Martin-Figueroa and F. J. Florencio, 2000. Telomeric silencing of a natural subtelomeric gene. Mol. Gen. Genet. 263: 287–291. [DOI] [PubMed] [Google Scholar]
- Wyrick, J., F. Holstege, E. Jennings, H. Causton, D. Shore et al., 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]
- Yang, C., J. F. Theis and C. S. Newlon, 1999. Conservation of ARS elements and chromosomal DNA replication origins on chromosomes III of Saccharomyces cerevisiae and S. carlsbergensis. Genetics 152: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]