Abstract
Easy shattering reduces yield due to grain loss during harvest in cereals. Shattering is also a hindrance in breeding programs that use wild accessions because the shattering habit is often linked to desirable traits. We characterized a shattering mutant line of rice, Hsh, which was derived from a nonshattering japonica variety, Hwacheong, by N-methyl-N-nitrosourea (MNU) treatment. The breaking tensile strength (BTS) of the grain pedicel was measured using a digital force gauge to evaluate the degree of shattering of rice varieties at 5, 10, 15, 20, 25, 30, 35, and 40 days after heading (DAH). The BTS of Hwacheong did not decrease with increasing DAH, maintaining a level of 180–240 gf, while that of Hsh decreased greatly during 10–20 DAH and finally stabilized at 50 gf. Optical microscopy revealed that Hsh had a well-developed abscission layer similar to the wild rice Oryza nivara (accession IRGC105706), while Hwacheong did not produce an abscission layer, indicating that the shattering of Hsh was caused by differentiation of the abscission layer. On the basis of the BTS value and morphology of the abscission layer of F1 plants and segregation data in F2 populations, it was concluded that the easy shattering of Hsh was controlled by the single recessive gene sh-h. The gene sh-h was determined to be located on rice chromosome 7 by bulked segregant analysis. Using 14 SSR markers on rice chromosome 7, the gene sh-h was mapped between the flanking markers RM8262 and RM7161 at distances of 1.6 and 2.0 cM, respectively. An SSR marker Rc17 cosegregated with the gene sh-h. The locus sh-h for shattering was tightly linked to the Rc locus conferring red pericarp, as well as a QTL qSDs-7-1 for seed dormancy, implying that this region might represent a domestication block in the evolutionary pathway of rice.
GRAIN shattering is an adaptive trait for seed dispersal in wild species of cereals. However, easy shattering causes considerable yield loss in domesticated grain crops. Through the domestication process, humans have selected crop plants with low shattering thresholds. Today, the degree of shattering that is considered favorable in a cultivar depends on the way the rice crop is harvested. Varieties with moderate shattering are favored where rice is harvested by large combined harvester–threshers, while harvesting using a small head-feeding combine is most efficient when hard-shattering to nonshattering varieties are used (Kobayashi 1990). Farmers who harvest and thresh rice by hand prefer moderate-shattering varieties, similar to those used in the most heavily mechanized systems. Easy-shattering varieties are not acceptable to any of the farmers because they cause severe yield loss no matter what type of harvesting is practiced. Shattering also causes unexpected mixing of varieties due to germination of shattered seeds in rice fields, resulting in deterioration of rice quality. In addition, breeding programs utilizing wild rice accessions are often hindered by linkage drag due to the association between easy shattering and a variety of desirable traits.
Rice grain shattering is caused by seed abscission. During the abscission process, an abscission layer differentiates first at the border of the abscising organ. Ethylene generally promotes the abscission process, while auxin inhibits it. Responding to certain signals and environmental cues, hydrolytic enzymes such as polygalacturonase and β-endo-glucanase are activated in the abscission layer cells, causing degradation of the middle lamellae and the cell walls of the cells in the abscission layer (Osborne 1989; Patterson 2001; Roberts et al. 2002).
The abscission layer forms in rice pedicel tissue 16–20 days before heading when gametophytic cells begin to differentiate in the young panicle (Jin 1986). The abscission layer in the rice pedicel is composed of one or two layers of small, round parenchyma-type cells with thin cell walls, while the surrounding pedicel and glume cells are large sclerenchyma-type cells that have thick cell walls (Jin 1986). As the seeds mature, the abscission layer cells degrade, making it easy for rice grain to disarticulate from the mother plant (Jin and Inouye 1982b). The morphology of the abscission layer differs in different rice varieties (Jin et al. 1982, 1990, 1995; Jin and Inouye 1982a,b, 1985). Some varieties do not have an abscission layer and are nonshattering. Some varieties have a nondegrading abscission layer. In easy-shattering varieties, the abscission layer is well developed from the epidermis to near the vascular bundle. Some varieties have an abscission layer that slants upward from the epidermis to the vascular bundle. In this case, the supporting zone, which is composed of a nondegrading section in between two abscission layers (Figure 1), is wider than in easy-shattering varieties, resulting in a moderate- to hard-shattering trait. Some varieties have an irregular abscission layer in which parenchyma-type abscission cells and sclerenchyma-type cells coexist. Some varieties have a partially developed abscission layer on the palea side of the pedicel while other varieties have a complete abscission layer on the palea side and an irregular abscission layer on the lemma side.
Figure 1.
Schematic of rice pedicel showing abscission layer. P, palea; L, lemma; RA, rachila; SL, sterile lemma; AL, abscission layer; SZ, supporting zone; RG, rudimentary glume; VB, vascular bundle; PE, pedicel.
Four loci governing the shattering habit have been mapped in the rice genome. They are sh1 on chromosome 11 (Nagao and Takahashi 1963), sh2 on chromosome 1 (Oba et al. 1990), Sh3 on chromosome 4 (Eiguchi and Sano 1990; Nagai et al. 2002), and Sh4 on chromosome 3 (Fukuta et al. 1994; Fukuta and Yagi 1998). The sh2 and Sh4 loci are reported to be related to the formation of the abscission layer (Fukuta et al. 1994; Oba et al. 1995; Fukuta and Yagi 1998). In addition, QTL for shattering have been reported on chromosomes 1, 3, 4, 7, 8, and 11 (Xiong et al. 1999; Cai and Morishima 2000; Bres-Patry et al. 2001; Thomson et al. 2003).
Studies to determine the genetic basis of crop domestication in other grass species have identified genes and QTL associated with the shattering habit in maize (Doebley and Stec 1991, 1993), sorghum (Paterson et al. 1995b), pearl millet (Poncet et al. 2002), wheat (Watanabe and Ikebata 2000; Jantasuriyarat et al. 2004), barley (Komatsuda et al. 2004), and American wild rice (Zizania palustris var. interior L.) (Kennard et al. 2002). Low-resolution comparative mapping revealed positional correspondences among shattering loci on linkage group C in sorghum, chromosomes 1 and 5 in maize, and chromosome 9 in rice (Paterson et al. 1995a).
Several genes related to the abscission or dehiscence phenomena have been isolated in plants. The SHATTERPROOF1, SHATTERPROOF2, and FRUITFULL genes that underlie the formation of the dehiscence zone in pods of Arabidopsis were revealed to be members of the MADS box gene family (Ferrandiz et al. 2000; Liljegren et al. 2000). In tomato, the JOINTLESS1 gene that controls the formation of an abscission layer in the pedicel was also revealed to be a MADS box gene (Mao et al. 2000). A Leu-rich repeat receptor-like kinase, HAESA, was reported to be associated with floral abscission in Arabidopsis (Jinn et al. 2000). The Arabidopsis myc/bHLH gene, ALCATRAZ (ALC), was shown to enable cell separation in Arabidopsis fruit dehiscence by promoting the differentiation of a strip of labile nonlignified cells sandwiched between layers of lignified cells (Rajani and Sundaresan 2001). Polygalacturonase and β-1,4-glucanase genes associated with abscission or dehiscence have been isolated in rapeseed, Arabidopsis, tomato, soybean, etc. (Patterson 2001; Christiansen et al. 2002; Ferrandiz 2002; Roberts et al. 2002).
In this study, an extremely easy-shattering mutant rice line was induced from the nonshattering japonica variety Hwacheong by N-methyl-N-nitrosourea treatment and genetically fixed for >10 generations. We named this mutant line Hsh. Here we report the characterization and mapping of the gene governing the shattering trait of this mutant rice line.
MATERIALS AND METHODS
Plant materials:
The shattering mutant line, Hsh, was induced and maintained at the Department of Plant Science, Seoul National University, Seoul, Korea. The seeds of Hsh were taken from an M12 line, 74336-1, the pedigree name of which was HwacheongM-8-201-1-3-2-1-2-2-2-1-B-1. To compare the degree of shattering of Hsh with other rice varieties, we selected eight accessions of Oryza sativa, O. rufopogon, and O. nivara (Table 1). The seeds were sown on January 13, 2002, and the seedlings were transplanted into 4-inch-wide pots on January 31, 2002. A total of 12–18 plants per each accession were grown in the Guterman greenhouse at Cornell University, Ithaca, New York. Hsh was crossed to five rice varieties, including Hwacheong, Ilpum, Blue&Gundil, Milyang23, and Dasan in the summer of 2001. The F1 plants from these crosses were grown in 4-inch-wide pots at the same time with the above-named rice accessions. Two F2 populations were derived by selfing F1 plants from the Hsh/Blue&Gundil and Hsh/Milyang23 crosses. A total of 240 Hsh/Blue&Gundil F2 plants and 241 Hsh/Milyang23 F2 plants were grown in 4-inch-wide pots. The seeds were sown on September 1, 2002, in the Guterman greenhouse.
TABLE 1.
Rice varieties subjected to measurement of the grain pedicel BTS
| Species | Variety name | Shattering degree |
|---|---|---|
| O. sativa (japonica) | Hwacheong | Nonshattering |
| Hsh | Very easy shattering | |
| Ilpum | Nonshattering | |
| Blue&Gundil | Nonshattering | |
| O. sativa (indica) | Milyang23 | Easy shattering |
| Dasan | Moderate shattering | |
| IR64 | Moderate shattering | |
| O. rufipogon | IRGC105491 | Easy shattering |
| O. nivara | IRGC105706 | Very easy shattering |
Measuring breaking tensile strength of rice grain pedicels:
Rice grain pedicel breaking tensile strength (BTS) was measured using a digital force gauge (FGC-1B, Shimpo, Japan). This force gauge was attached upside down to a stand (FGC-50L). Rice panicles from the main stem or primary tiller were harvested and fixed upside down to the force gauge using a flat chuck tensile grip (FG-M6FTG20U) that was attached at the top of the force gauge. Each grain was pulled down by a forcep, and the maximum tensile strength, measured at the moment of pedicel breakage, was recorded manually in gravitational unit of force (gf). We evaluated the change in BTS of grain pedicels over time measured as days after heading (DAH) in nine rice accessions (Table 1). The heading date of three or four panicles from primary tillers on each plant was measured on the basis of the emergence of the panicle from the flag leaf sheath. At 5, 10, 15, 20, 25, 30, 35, and 40 DAH, two panicles from each of two plants were harvested from each variety. On the same day that panicles were sampled, grain pedicel BTS was measured on the basis of 10 grains from the uppermost part of the panicle. So 20 observations of grain pedicel BTS were made each time that the trait was measured for a variety. The grain pedicel BTS of F1 plants was also measured. A panicle from the primary tiller of each F1 plant was harvested at 40 days after heading, when rice grains were fully ripened, and the BTS was measured on the basis of 10 grains from the uppermost part of each panicle. With 240 Hsh/Blue&Gundil F2 plants and 241 Hsh/Milyang23 F2 plants, two panicles per plant were harvested at ∼50 days after heading, and grain pedicel BTS was measured using 10 grains from the uppermost part of each panicle.
Observation of the rice abscission layer:
Panicles were harvested at heading time to obtain soft rice grain pedicel tissue. Pedicels were cut and fixed in FAA solution [ethanol (EtOH) 50%, acetic acid 5%, formaldehyde 3.7%] and stored at 4°. The pedicels were dehydrated by soaking them for 2 hr each in a graduated EtOH solution of increasing concentration, i.e., 50, 70, 85, 95, and 100%. The final soaking at 100% EtOH was done three times. The pedicel tissue was cleared by soaking it in 50% EtOH/50% Histoclear solution for 2 hr, followed by soaking three times in 100% Histoclear solution for 2 hr each time. For paraffin infiltration, the pedicel tissue was soaked at 50% Histoclear/50% paraffin solution at 58° overnight. The tissue was maintained in a 60° oven for 2–3 more days and the soaking solution was changed with 100% melted paraffin two times a day. The tissue was transferred into a plastic mold containing melted paraffin heated to 60° on a hot plate, properly positioned, and cooled down. The tissue was embedded in a paraffin block and then cut by a microtome into 10-μm thick sections and mounted on a superfrost-plus glass slide (Fisher Scientific) and dried at 40° for 2 days.
These sections were stained by hematoxylin, safranin, and fast green. Samples were deparaffinized by soaking them two times in 100% Histoclear solution for 15 min each. The pedicels were hydrated by soaking for 10 min each in graduated solutions of EtOH, i.e., 100, 95, 70, 50, and 30%, and in sterile water. Samples were soaked in iron alum mordant solution (acetic acid 1%, sulfuric acid 0.12%, ferric ammonium sulfate 3%) for 1 hr, followed by washing by sterile water for 5 min. Samples were stained in 0.5% hematoxylin solution overnight, followed by washing with sterile water for 5 min. Samples were destained in 2% ferric ammonium sulfate solution for ∼14 min, followed by washing for 30 min–1 hr with running tap water. After soaking in 30% EtOH for 10 min, samples were stained with 1% safranin (in 30% EtOH) for 30 min–1 hr, followed by washing with sterile water five times. Samples were destained by soaking in 30% acid alcohol (1 ml of 1:5 HCl in 500 cc of 30% EtOH) for 4 min, washed in 30% EtOH, and fixed with 1% NaHCO3 (in 30% EtOH) for 5 min. Samples were soaked in graduated solutions of EtOH (50, 70, and 95%) for 5 min each. Samples were stained with 0.5% fast green (in 95% EtOH) for 5–30 sec and soaked two times in 100% EtOH for 2 min each. Samples were then cleared by soaking two times in 100% Xylene for 10 min each, followed by mounting with entelan. The abscission layer region was observed by optical microscopy at ×100 and ×600 magnifications.
DNA extraction, bulked segregant analysis, and mapping:
DNA was extracted from all F2 plants and parental plants by a modified chloroform microprep method (Coburn et al. 2002). Approximately 5 cm of young leaf tissue was harvested into 2.0-ml centrifuge tubes including two metal beads and frozen on liquid nitrogen. The tissue was ground thoroughly by two passes of vigorous vortexing for 1 min each, and thawing was prevented by dipping tubes in liquid nitrogen in between vortexing. Metal beads were removed by simply cap opening and inverting tubes on paper towels, and then 800 μl of DNA extraction buffer (100 mm Tris-HCl pH 8, 50 mm EDTA pH 8, 500 mm NaCl, 1.25% SDS (w/v), 8.3 mm NaOH) was added. Samples were placed in a 65° water bath for 20–60 min. Tubes were removed from the water bath and filled with ∼750 μl of a 24:1 mixture of chloroform and isoamyl alcohol. Samples were completely mixed by shaking for 5 min and centrifuged at 13,000 rpm for 10 min using a table-top microcentrifuge. The supernatant was transferred to a new 1.5-ml Eppendorf tube, where it was mixed with 2/3 volume of cold isopropanol. DNA was precipitated by inverting tubes several times, and samples were placed at −20° for 30 min to overnight. To pellet the DNA, samples were centrifuged at 13,000 rpm for 12 min, followed by washing with 800 μl of ice-cold 70% EtOH. DNA was pelleted again, dried, and resuspended in 100 μl of TE buffer. The concentration of each DNA sample was measured on a 2% agarose gel with λDNA concentration standard solutions.
We selected 18 plants of the lowest BTS value and 18 plants of the highest BTS value in the Hsh/Blue&Gundil F2 population and extracted DNA samples from individual plants for use in bulked segregant analysis (BSA) (Michelmore et al. 1991). Three bulked samples of six plants each were prepared for the low- and high-BTS groups, respectively, by taking equal amounts of DNA from individual samples and combining them into a single bulked sample for genotyping. Seventy-seven SSR markers distributed over the 12 rice chromosomes (Temnykh et al. 2001; Coburn et al. 2002) were tested with these low- and high-BTS DNA bulks together with the parents, Hsh and Blue&Gundil. The low- and high-BTS DNA bulks were made by the same method in the Hsh/Milyang23 F2 population. Subsequently, 17 SSR markers on chromosome 7 were tested with this population. After BSA, the 240 F2 plants in the Hsh/Blue&Gundil F2 population were genotyped using 14 polymorphic SSR markers on rice chromosome 7 (Temnykh et al. 2001; McCouch et al. 2002). Combining the marker (genotypic) data and the shattering (phenotypic) data scored on the basis of grain pedicel BTS values, a linkage map of chromosome 7 was constructed using MapMaker software (Lander et al. 1987).
RESULTS
Shattering phenotype of Hsh:
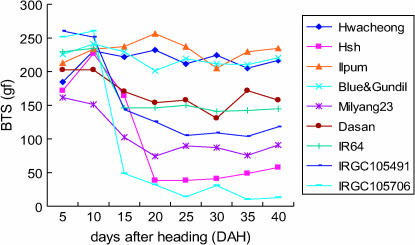
The BTS of rice grain pedicels was measured for nine varieties as summarized in Table 1. The BTS is the maximum strength of resistance to separation of the rice grain from the pedicel, and thus the BTS is inversely proportional to the degree of shattering. The changes in BTS over time are summarized in Figure 2. The BTS of the nonshattering varieties, Hwacheong, Ilpum, and Blue&Gundil, did not decrease with DAH, but retained a BTS level of 180–260 gf. In contrast, the BTS value of Hsh decreased dramatically during 10–20 DAH and finally stabilized at 50 gf at 40 DAH, a value similar to that of O. nivara (accession IRGC105706). The seeds of Hsh shattered easily with a touch of the hand, as with IRGC105706. The other rice accessions had various degrees of shattering and showed similar time course changes in BTS although their final average BTS values ranged from 90 to 160 gf. The most prominent decrease in BTS occurred during the first 10–15 DAH.
Figure 2.
Time-course change of grain pedicel BTS of rice varieties. Two panicles from primary tillers of two plants were harvested at each measuring time for each variety. Ten grains from the uppermost part of the panicle were measured for each panicle. Average value is shown in this graph.
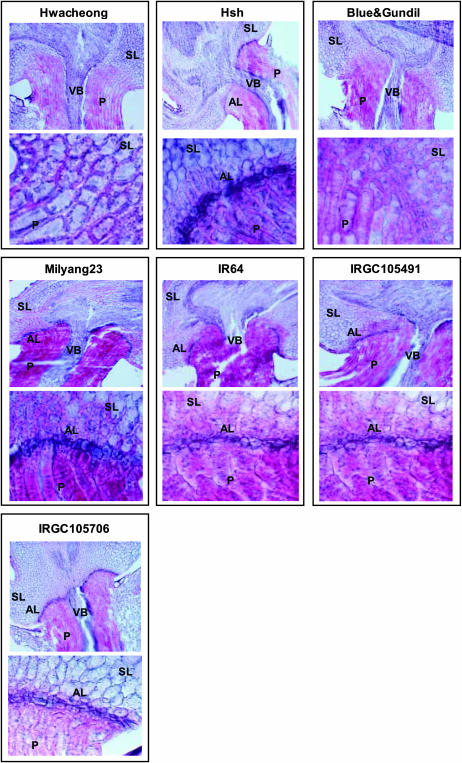
The anatomical structure of the abscission layers in rice pedicels was observed using optical microscopy (Figure 3). While Hwacheong had no abscission layer, Hsh exhibited a well-developed abscission layer. The shape of Hsh's abscission layer was very similar to that of O. nivara (accession IRGC105706). The abscission layer formed in the shape of an upward flat parabola that reached a point very close to the vascular bundle, resulting in a very narrow supporting zone. Therefore, we determined that the functional mutation in Hsh affected a gene that conditioned the formation of the abscission layer. Abscission layer cells were smaller than the surrounding pedicel and glume cells and retained their nuclei. They had thinner cell walls and were stained darker red by safranin. The parental variety, Blue&Gundil, which was nonshattering, had small cells in the region of the abscission layer. But thick cell walls developed around these cells, indicating that they were not functional abscission layer cells. IR64 had an abscission layer that slanted upward from the epidermis to the vascular bundle, but it had a wider supporting zone than the other shattering varieties tested in this experiment. Milyang23 and O. rufipogon accession IRGC105491 had similar, intermediate-type abscission layers falling between IR64 (moderate shattering) and IRGC105706 (very easy shattering).
Figure 3.
Anatomical structure of grain pedicel tissues showing variation of abscission layer morphology in seven rice varieties. Top photo for each variety: ×100 magnification. Bottom photo for each variety: ×600 magnification. Two samples from each of two plants were observed for each variety. AL, abscission layer; P, pedicel; SL, sterile lemma; VB, vascular bundle.
Shattering of Hsh is governed by a single recessive gene:
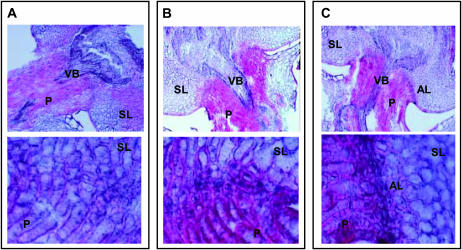
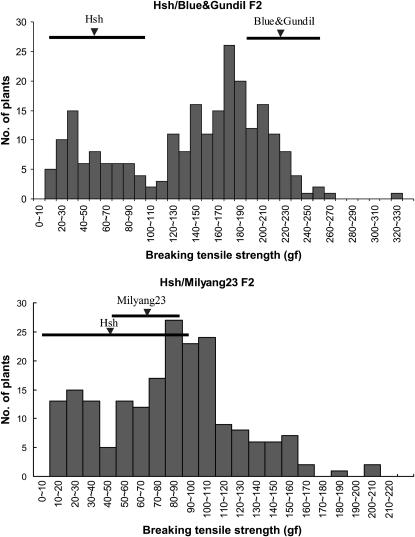
Hsh was used as the female parent in crosses with five rice varieties: Hwacheong, Ilpum, Blue&Gundil, Milyang23, and Dasan. The grain pedicel BTS of F1 plants were similar to those of the male parents in all cases, demonstrating that a recessive mutation confers shattering in Hsh (Table 2). In the crosses between Hsh and nonshattering varieties such as Hwacheong, Ilpum, and Blue&Gundil, F1 plants were all nonshattering. In crosses between Hsh and Milyang23 and Dasan, F1 plants showed a degree of shattering similar to that of Milyang23 and Dasan, respectively. The F1 plants from Hsh/Hwacheong and Hsh/Blue&Gundil crosses did not form an abscission layer in the grain pedicel, while the F1 plants from the Hsh/Milyang23 cross formed an abscission layer similar to that of Milyang23 (Figure 4). Two F2 populations from Hsh/Blue&Gundil and Hsh/Milyang23 crosses were tested for grain pedicel BTS as shown in Figure 5. In each BTS histogram, two peaks were observed and the lowest frequency interval between these two peaks was considered as the boundary between the lower and higher BTS groups. These boundary intervals were 100–110 gf in the Hsh/Blue&Gundil F2 population and 40–50 gf in the Hsh/Milyang23 F2 population. In the Hsh/Blue&Gundil cross, F2 plants were classified into two groups, Hsh type (BTS < 100–110 gf) and Blue&Gundil type (BTS > 100–110 gf). The segregation ratio of the Hsh type to the Blue&Gundil type was 1:3, indicating that the trait was governed by a single recessive gene (Figure 5 and Table 3). In the Hsh/Milyang23 cross, F2 plants were also classified into two groups, but in this case, Hsh types were classified as BTS < 40–50 gf and Milyang 23 types had BTS > 40–50 gf. The segregation ratio again corresponded to a single recessive gene (Figure 5 and Table 3). We tentatively designated the gene responsible for shattering in the Hsh mutant as sh-h.
TABLE 2.
Grain pedicel BTS of F1 plants and their parental varieties
| Grain pedicel BTS (gf)a
|
||||
|---|---|---|---|---|
| Cross combination | No. of panicles (plants) measured | F1 plant | Hsh | Male parentb |
| Hsh/Hwacheong | 10 (5) | 201 ± 21.2 | 57 ± 49.4 | 216 ± 36.5 |
| Hsh/Ilpum | 10 (5) | 217 ± 24.2 | 57 ± 49.4 | 234 ± 30.1 |
| Hsh/Blue&Gundil | 24 (14) | 209 ± 31.0 | 57 ± 49.4 | 220 ± 32.8 |
| Hsh/Milyang23 | 18 (11) | 109 ± 34.7 | 57 ± 49.4 | 91 ± 26.6 |
| Hsh/Dasan | 7 (6) | 154 ± 48.1 | 57 ± 49.4 | 158 ± 21.5 |
BTS value is the average value ± standard deviation.
Parental value was measured with two panicles.
Figure 4.
Anatomical structure of grain pedicel tissues of F1 plants. (A) Hsh/Hwacheong. (B) Hsh/Blue&Gundil. (C) Hsh/Milyang23. Top photo in A–C: ×100 magnification. Bottom photo in A–C: ×600 magnification. Two samples from each of two plants were observed for each variety. AL, abscission layer; P, pedicel; SL, sterile lemma; VB, vascular bundle.
Figure 5.
Histogram showing grain pedicel BTS value distribution in two F2 populations from the Hsh/Blue&Gundil and Hsh/Milyang23 crosses. Average BTS value and the range including average BTS ± standard deviation of parental varieties is shown by solid inverse triangles and thick lines, respectively.
TABLE 3.
Segregation of grain pedicel BTS value in two F2 populations from the Hsh/Blue&Gundil and Hsh/Milyang23 crosses
| No. of F2 plants
|
|||||
|---|---|---|---|---|---|
| Cross combination | Item | Higher BTS (110 < BTS < 340) | Lower BTS (0 < BTS < 100) | Total | |
| Hsh/Blue&Gundil | Observeda | 166 | : | 66 | 232 |
| χ2 (3:1) | 1.471 | ||||
| P | 0.23 | ||||
| No. of F2 plants
| |||||
| Cross combination | Item | Higher BTS (50 < BTS < 220) | Lower BTS (0 < BTS < 40) | Total | |
| Hsh/Milyang23 | Observeda | 157 | : | 41 | 198 |
| χ2 (3:1) | 1.946 | ||||
| P | 0.16 | ||||
Observed segregation. Plants in the boundary interval between higher and lower BTS groups are not included in this table. The boundary intevals were 100 < BTS < 110 in the Hsh/Blue&Gundil F2 population and 40 < BTS < 50 in the Hsh/Milyang23 F2 population.
Mapping of the shattering gene sh-h:
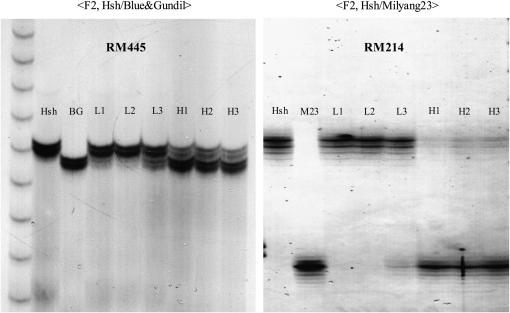
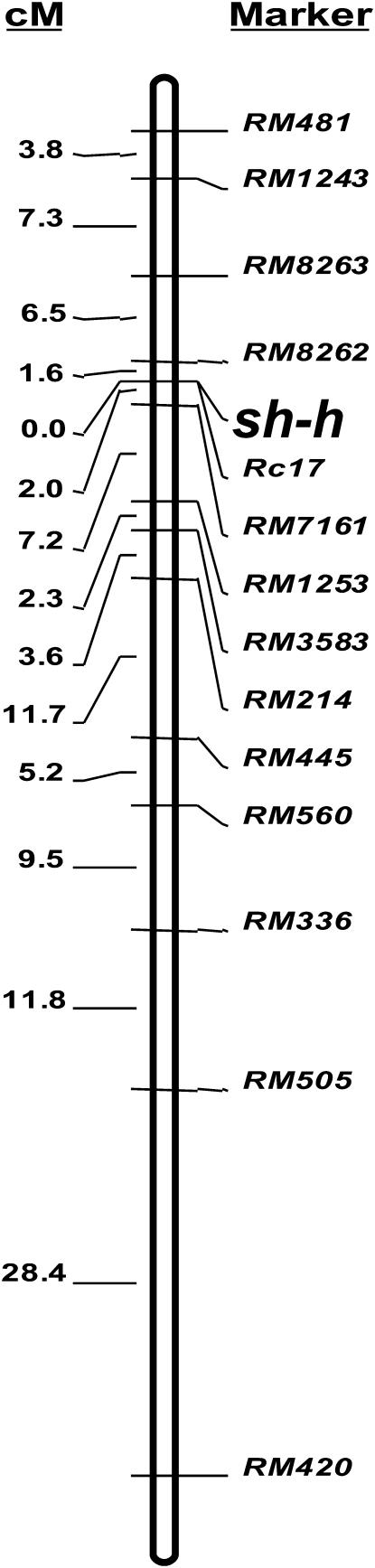
Using an F2 population consisting of 240 individuals derived from the Hsh/Blue&Gundil cross, we mapped the shattering gene sh-h, which is responsible for shattering of the mutant line Hsh. To identify markers linked to the shattering gene sh-h, BSA was used and 77 SSR markers distributed over the 12 rice chromosomes (Temnykh et al. 2001; Coburn et al. 2002) were surveyed for polymorphism on the bulked DNA samples. Significant differences in the allelic profiles of the PCR products were observed between the high-BTS and low-BTS bulks with markers RM481 and RM418 on chromosome 7. Subsequently, additional SSR markers from chromosome 7 were screened to increase the mapping resolution. RM214, RM445, RM542, and RM560 were found to be linked with sh-h. In addition, 17 SSR markers on chromosome 7 were also surveyed for polymorphism in the Hsh/Milyang23 F2 population using BSA. Most of these markers showed significant differences in molecular weight between the DNA bulks, and RM214 showed the clearest difference (Figure 6). Consequently, it was confirmed that the shattering gene sh-h was located on chromosome 7 closely linked to RM214. Next, we genotyped the 240 F2 plants of the Hsh/Blue&Gundil cross individually with 14 SSR markers from chromosome 7 and analyzed the relationship between marker genotype and the BTS value. There were 44 plants with low-BTS values (0–60 gf), 152 plants with high-BTS values (130–330 gf), 38 plants with intermediate-BTS values (60–130 gf), and 6 sterile plants. Combining the genotype data from 14 SSR markers and the phenotypic data, the shattering gene sh-h was mapped between the flanking markers RM8262 and RM7161 at a distance of 1.6 and 2.0 cM, respectively (Figure 7). Interestingly, an SSR marker closely linked to the Rc gene for red pericarp (Sweeney et al. 2006), Rc17, cosegregated with sh-h. The physical distance between RM8262 and RM7161 was 598.2 kbp. There were 58 genes encoding annotated proteins, including “expressed proteins,” and another 34 genes encoding “hypothetical proteins” in this interval in the The Institute for Genome Research Rice Genome Annotation database (http://www.tigr.org/tdb/e2k1/osa1/).
Figure 6.
Examples of BSA using markers RM445 and RM214, which belong to chromosome 7. PCR products of SSR marker primers were run on a 4% denaturing polyacrylamide gel. BG: Blue&Gundil; M23: Milyang23; L1–L3: low-BTS DNA bulks including six plant DNA samples, each selected among 18 F2 plants having the lowest grain pedicel BTS value; H1–H3: high-BTS DNA bulks including six plant DNA samples, each selected among 18 F2 plants having the highest BTS value.
Figure 7.
Molecular genetic map of rice chromosome 7 showing the location of the shattering gene sh-h (primer for Rc17: forward 5′-AGTTTGGAGGAAGCAGCAAA-3′; backward 5′-CACCAACTTGCTCTACTTGTGG-3′).
DISCUSSION
Phenotype of shattering trait:
Several methods have been used to measure grain shattering in rice. One method is to clasp the rice panicles in a clenched fist and count detached and undetached grains separately. The percentage of detached grains indicates the degree of shattering. This method is prone to error due to variation in clasping power of the person measuring the trait. The hitting method is to hit rice panicles against a drum and count detached and undetached grains separately. This method also can be affected by variation in the hitting power of the person doing the measurements. In this study, we measured grain pedicel BTS using a digital force gauge. This method has the advantage of minimizing the variation associated with the person causing the grains to shatter. However, the standard deviation was still large, in the range of 10–60 gf. This variation may be the result of differences in the pedicel diameter of individual spikelets or due to variation in the level of degradation of abscission layer cells. Sometimes when measuring BTS with shattering varieties, a few grains did not shatter and showed much higher BTS values than other grains. This resulted in high standard deviations of BTS values. Large variation in BTS values may also occur due to an unfavorable environment for grain filling. In this study, plants were grown in the greenhouse, and the grain-filling season for the parental varieties and F1 plants was spring to summer (April–July) while F2 plants ripened in winter (November–February). Grain filling during the winter season is likely to have increased the variation of BTS values more than grain filling in the summer season. The variation in BTS values may be reduced by measuring grains that have a uniform pedicel diameter, are fully ripened, and were grown to maturity in a favorable environment.
Generally, rice (O. sativa) varieties reach physiological maturity at 30–40 days after heading, and indica varieties mature slightly faster than japonica varieties (Nagato and Chaundhry 1970; Kwon and Shin 1980). The rice accessions tested in this study ripened in the greenhouse during summer without stress from low temperatures. The BTS of the grain pedicel reached the lowest level at 20–25 days after heading and did not change significantly thereafter. Therefore, the BTS value measured ∼40 days after heading is believed to represent the degree of shattering at maturity in the rice accessions tested.
BTS values decreased during the first 10–20 days after heading, as grains turned yellow and their moisture content decreased. These three aspects of seed maturation appear to be synchronized during this period. Generally, ethylene activates abscission and senescence (Osborne 1989; Patterson 2001; Ferrandiz 2002; Roberts et al. 2002). But, in cereals, it has been reported that ABA, rather than ethylene, induced seed abscission in three Gramineae species—Avena fatua, Bromus sterilis, and Aegilops squarrosa (Sargent et al. 1984). ABA causes seed desiccation, and ABA levels increase in seeds during the mid-embryo-development stage (Rock and Quatrano 1995). It is inferred that either ABA or ethylene levels increase in rice during 10–20 days after heading and then activate seed abscission, desiccation, and seed hull senescence.
The variation in morphology of the abscission layer among rice varieties observed in this experiment has also been reported by other researchers (Jin et al. 1982, 1990, 1995; Jin and Inouye 1982a,b, 1985). This variation is closely associated with the degree of shattering in rice and may be utilized as a predictor of the trait in developing varieties with a desirable degree of shattering. In rice production areas where small head-feeding combines are used, nonshattering or hard-shattering varieties will be desirable. Meanwhile, in rice areas where large combined harvester–threshers, or alternatively, where hand threshing is used, moderate-shattering varieties are preferred. Cultivars such as IR64 that have an abscission layer that slants upward along with a wider supporting zone than is seen in the easy-shattering varieties will be desirable. In this regard, efforts to isolate and characterize the genes underlying the variation in abscission layer morphology are of agronomic importance.
It was reported that the process of abscission layer development was similar in two japonica varieties, two japonica–indica hybrids bred in Korea, and six African rice (O. glaberrima) varieties and that the abscission layer became distinguishable ∼15–16 days before heading (Jin 1986; Jin et al. 1995). These results indicate that the process of abscission layer development is similar among different ecotypes and subspecies of rice, even though only a small number of accessions have been tested to date. On the other hand, the effect of the environment on the process of abscission layer development has not been well studied and further research is needed.
Identification and mapping of shattering genes:
Previously mapped shattering genes in rice, including sh2 on chromosome 1 (Oba et al. 1990, 1995) and Sh4 on chromosome 3 (Fukuta et al. 1994; Fukuta and Yagi 1998), were reported to be involved in the formation of the abscission layer. The shattering gene sh-h on chromosome 7 is newly identified and mapped in this study, but it appears to be involved in the same process. Therefore, we conclude that at least three genes are involved in abscission layer formation in rice.
The shattering gene discovered in the Hsh mutant line was mapped to the short arm of rice chromosome 7, tightly linked to the marker Rc17. Originally, this SSR marker was developed as part of an effort to fine map the red pericarp color gene Rc (Sweeney et al. 2006) and we therefore assume that the shattering gene sh-h is very close to the Rc gene. A shattering QTL was reported in a similar location on chromosome 7 in the BC2F2 population derived from a cross between the tropical japonica cultivar, Jefferson, and O. rufipogon (accession IRGC105491) (Thomson et al. 2003). Interestingly, the QTL allele for shattering that originated from O. rufipogon behaved in a dominant fashion. While we do not yet know the identity of the gene underlying the QTL, we infer that it may correspond to a different allele at the sh-h locus. We also investigated literature on genetic loci associated with the shattering habit in other grass crop species such as maize (Doebley and Stec 1991, 1993), sorghum (Paterson et al. 1995b), pearl millet (Poncet et al. 2002), wheat (Watanabe and Ikebata 2000; Jantasuriyarat et al. 2004), barley (Komatsuda et al. 2004), and American wild rice (Z. palustris var. interior L.) (Kennard et al. 2002) along with comparative genetic maps (Devos 2005) to see whether any shattering loci have been reported in a homeologous region in other grasses. To date, no shattering genes have been isolated from other cereals and comparative map analysis reveals no obvious correspondences between the map position of sh-h in rice and the locations of shattering loci in other grass species.
Interestingly, a seed dormancy QTL, qSDs-7-1, was also reported in the same region, with its peak located at RM180 on chromosome 7 (Thomson et al. 2003; Gu et al. 2004, 2005). Rc17 and RM180 share the same physical location on chromosome 7; the former was between 75409 and 75567 bp and the latter was between 75,453 and 75,562 bp on the same BAC clone (AP003943). Upon close examination, we discovered that Rc17 and RM180 represent different primer pairs flanking the same SSR motif (ATT)10. Therefore, we conclude that several genes underlying crop domestication, including seed shattering, red pericarp, and seed dormancy, are very closely linked in this region of chromosome 7. We refer to this array of colocated genes as a domestication block. Further efforts to isolate sh-h via a map-based cloning strategy are in progress. Cloning of the cluster of domestication-related genes in this region of chromosome 7 will offer new insights into the identity, function, and evolution of genes contributing to the domestication syndrome in rice and other cereals.
Acknowledgments
We thank Sandra Harrington, Fumio Onishi, Hui Jiang, and Bin Cong for their technical assistance. Also we thank Il-Doo Jin for his advice on phenotyping of the rice shattering trait. This research was supported in part by a grant (code no. 3111) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, Republic of Korea, and from the National Science Foundation (NSF00-151/011-0004). The study of Hyeon-So Ji at Cornell University was funded by the Korean Government Fellowship Program for Overseas Study.
References
- Bres-Patry, C. M. Lorieux, G. Clement, M. Bangratz and A. Ghesquiere, 2001. Heredity and genetic mapping of domestication-related traits in a temperate japonica weedy rice. Theor. Appl. Genet. 102: 118–126. [Google Scholar]
- Cai, H. W., and H. Morishima, 2000. Genomic regions affecting seed shattering and seed dormancy in rice. Theor. Appl. Genet. 100: 840–846. [Google Scholar]
- Christiansen, L. C., F. Dal Degan, P. Ulvskov and B. Borkhardt, 2002. Examination of the dehiscence zone in soybean pods and isolation of a dehiscence-related endopolygalacturonase gene. Plant Cell Environ. 25: 479–490. [Google Scholar]
- Coburn, J. R., S. V. Temnykh, E. M. Paul and S. R. McCouch, 2002. Design and application of microsatellite marker panels for semiautomated genotyping of rice (Oryza sativa L.). Crop Sci. 42: 2092–2099. [Google Scholar]
- Devos, K. M., 2005. Updating the ‘crop circle’. Curr. Opin. Plant Biol. 8: 155–162. [DOI] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1993. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiguchi, M., and Y. Sano, 1990. A gene complex responsible for seed shattering and panicle spreading found in common wild rices. Rice Genet. Newsl. 7: 105–107. [Google Scholar]
- Ferrandiz, C., 2002. Regulation of fruit dehiscence in Arabidopsis. J. Exp. Bot. 53: 2031–2038. [DOI] [PubMed] [Google Scholar]
- Ferrandiz, C., S. J. Liljegren and M. F. Yanofsky, 2000. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289: 436–438. [DOI] [PubMed] [Google Scholar]
- Fukuta, Y., and T. Yagi, 1998. Mapping of a shattering resistance gene in a mutant line SR-5 induced from an indica rice variety, Nan-jing11. Breed. Sci. 48: 345–348. [Google Scholar]
- Fukuta, Y., H. Yoshida, K. Fukui and A. Kobayashi, 1994. Analysis of shattering degrees and abscission layer development in shattering-resistant mutant lines induced from an indica rice (Oryza sativa L.) variety, nan-jing 11. Jpn. J. Breed. 44: 195–200. [Google Scholar]
- Gu, X. Y., S. F. Kianian and M. E. Foley, 2004. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166: 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X. Y., S. F. Kianian, G. A. Hareland, B. L. Hoffer and M. E. Foley, 2005. Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa). Theor. Appl. Genet. 110: 1108–1118. [DOI] [PubMed] [Google Scholar]
- Jantasuriyarat, C., M. I. Vales, C. J. Watson and O. Riera-Lizarazu, 2004. Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor. Appl. Genet. 108: 261–273. [DOI] [PubMed] [Google Scholar]
- Jin, I. D., 1986. On the formation and development of abscission layer in rice plants, Oryza sativa L. Jpn. J. Crop Sci. 55: 451–457. [Google Scholar]
- Jin, I. D., and J. Inouye, 1982. a Relation between grain shedding and pedicel morphology near the abscission layer of japonica-indica hybrid rices bred in Korea. Jpn. J. Crop Sci. 51: 271–275. [Google Scholar]
- Jin, I. D., and J. Inouye, 1982. b Relationship between grain shedding and abscission layer in pedicel of japonica-indica hybrid rices in Korea. Jpn. J. Breed. 51: 43–50. [Google Scholar]
- Jin, I. D., and J. Inouye, 1985. On the degree of grain shedding, histological peculiarity of abscission region and esterase isozyme genotype of Bulu and Tjereh rice varieties originated in Indonesia. Jpn. J. Crop Sci. 54: 373–378. [Google Scholar]
- Jin, I. D., H. Terao and J. Inouye, 1982. On the cracking of abscission layer in Asian rice cultivar (Oryza sativa L.). Jpn. J. Crop Sci. 51: 542–545. [Google Scholar]
- Jin, I. D., J. Inouye and N. N. Quat, 1990. Histological peculiarities of the abscission layers of African rice, Oryza glaberrima Steud. and its relation with degree of grain shedding. Jpn. J. Crop Sci. 59: 475–480. [Google Scholar]
- Jin, I. D., Y. H. Bae and J. Inouye, 1995. Formation and development of abscission layer between pedicel and rachilla, and changes in grain shedding during ripening in African rice, Oryza glaberrima steud. Korean J. Crop Sci. 40: 103–112. [Google Scholar]
- Jinn, T. L., J. M. Stone and J. C. Walker, 2000. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14: 108–117. [PMC free article] [PubMed] [Google Scholar]
- Kennard, C., L. Phillips and A. Porter, 2002. Genetic dissection of seed shattering, agronomic, and color traits in American wildrice (Zizania palustris var. interior L.) with a comparative map. Theor. Appl. Genet. 105: 1075–1086. [DOI] [PubMed] [Google Scholar]
- Kobayashi, A., 1990. Varietal adaptability for mechanized rice cultivation: direct seeding adaptability, shattering habit, smoothness etc. J. Agric. Sci. (Jpn.) 45: 186–189. [Google Scholar]
- Komatsuda, T., P. Maxim, N. Senthil and Y. Mano, 2004. High-density AFLP map of nonbrittle rachis 1 (btr1) and 2 (btr2) genes in barley (Hordeum vulgare L.). Theor. Appl. Genet. 109: 986–995. [DOI] [PubMed] [Google Scholar]
- Kwon, Y. W., and J. C. Shin, 1980. A study on the changes in grain weight, moisture content, shattering force, milling ratio and apparent physical quality of rice with harvesting time. J. Korean Soc. Crop Sci. 25: 1–9. [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Liljegren, S. J., G. S. Ditta, Y. Eshed, B. Savidge, J. L. Bowman et al., 2000. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770. [DOI] [PubMed] [Google Scholar]
- Mao, L., D. Begum, H. W. Chuang, M. A. Budiman, E. J. Szymkowiak et al., 2000. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406: 910–913. [DOI] [PubMed] [Google Scholar]
- McCouch, S. R., L. Teytelman, Y. Xu, K. B. Lobos, K. Clare et al., 2002. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) (supplement). DNA Res. 9: 257–279. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, Y. S., P. L. Sobrizal, T. Sanchez, K. D. Kurakazu, K. Doi et al., 2002. Sh3, a gene for seed shattering, commonly found in wild rices. Rice Genet. Newsl. 19: 74–75. [Google Scholar]
- Nagao, S., and M. Takahashi, 1963. Trial construction of twelve linkage groups in Japanese rice. J. Fac. Agric. Hokkaido Univ. 53: 72–130. [Google Scholar]
- Nagato, K., and F. M. Chaundhry, 1970. A comparative study of ripening process and kernel development in japonica and indica rice. Proc. Crop Sci. Soc. Jpn. 38: 425–433. [Google Scholar]
- Oba, S., F. Kikuchi and K. Maruyama, 1990. Genetic analysis of semidwarfness and grain shattering of Chinese rice [Oryza sativa] variety “Ai-Jio-Nan-Te.” Jpn. J. Breed. 40: 13–20. [Google Scholar]
- Oba, S., N. Sumi, F. Fujimoto and T. Yasue, 1995. Association between grain shattering habit and formation of abscission layer controlled by grain shattering gene sh-2 in rice (Oryza sativa L.). Jpn. J. Crop Sci. 64: 607–615. [Google Scholar]
- Osborne, D. J., 1989. Abscission. Crit. Rev. Plant Sci. 8: 103–129. [Google Scholar]
- Paterson, A. H., Y.-R. Lin, Z. Li, K. F. Schertz, J. F. Doebley et al., 1995. a Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714–1718. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., K. F. Schertz, Y. R. Lin, S. C. Liu and Y. L. Chang, 1995. b The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proc. Natl. Acad. Sci. USA 92: 6127–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, S. E., 2001. Cutting loose: abscission and dehiscence in Arabidopsis. Plant Physiol. 126: 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet, V., E. Martel, S. Allouis, K. M. Devos, F. Lamy et al., 2002. Comparative analysis of QTLs affecting domestication traits between two domesticated × wild pearl millet (Pennisetum glaucum L., Poaceae) crosses. Theor. Appl. Genet. 104: 965–975. [DOI] [PubMed] [Google Scholar]
- Rajani, S., and V. Sundaresan, 2001. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11: 1914–1922. [DOI] [PubMed] [Google Scholar]
- Roberts, J. A., K. A. Elliott and Z. H. Gonzalez-Carranza, 2002. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53: 131–158. [DOI] [PubMed] [Google Scholar]
- Rock, C. D., and R. S. Quatrano, 1995. The role of hormones during seed development, pp. 671–697 in Plant Hormones, edited by P. J. Davies. Kluwer Academic, Dordrecht, The Netherlands.
- Sargent, J., D. Osborne and S. Dunford, 1984. Cell separation and its hormonal control during fruit abscission in the Gramineae. J. Exp. Bot. 35: 1663–1674. [Google Scholar]
- Sweeney, M., B. Pfeil and S. McCouch, 2006. Rc, a bHLH protein conditioning red pericarp in rice, is orthologous to the maize intensifier1. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh, S., G. DeClerck, A. Lukashova, L. Lipovich, S. Cartinhour et al., 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, M. J., T. H. Tai, A. M. McClung, X. H. Lai, M. E. Hinga et al., 2003. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., and N. Ikebata, 2000. The effects of homoeologous group 3 chromosomes on grain colour dependent seed dormancy and brittle rachis in tetraploid wheat. Euphytica 115: 215–220. [Google Scholar]
- Xiong, L. Z., K. D. Liu, X. K. Dai, C. G. Xu and Q. Zhang, 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]